Molecular Characterization of Salmonella Detected along the Broiler Production Chain in Trinidad and Tobago
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Selection
2.2. DNA Extraction and Sequencing
2.3. Genomic Data Analysis and In Silico Determination of Genetic Elements
2.4. Phenotypic Methods Used for Comparison with WGS
2.5. Statistical Analyzes
2.6. Data Deposition
3. Results
3.1. Serotyping Results
3.2. Antimicrobial Resistance Profiles
3.3. Virulence Profile
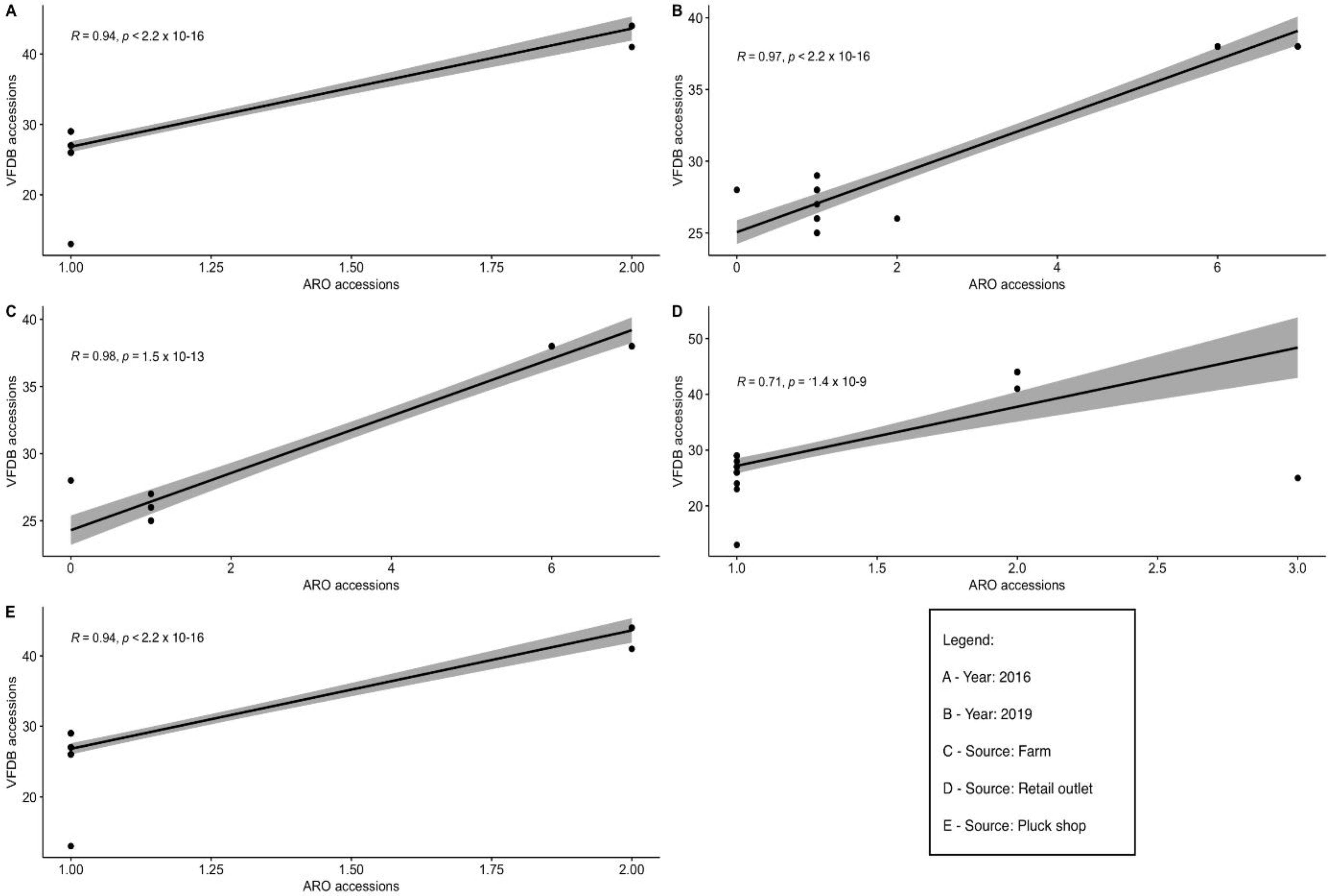
3.4. Comparison of Frequency of Detection of Resistance and Virulence Factors in Salmonella Strains
3.5. Detection of ESβL Resistance Genes and Virulence Genes in Isolates of S. Infantis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ulaya, D.W. Determination of virulence factors in Salmonella isolates of human, poultry, and dog origin in Lusaka district, Zambia. Master’s Thesis, University of Zambia, Lusaka, Zambia, 2014. [Google Scholar]
- Centers for Disease Control and Prevention. Salmonella. Available online: https://www.cdc.gov/salmonella/index.html (accessed on 3 October 2020).
- European Food Safety Authority; European Centre for Disease Prevention Control. The European Union One Health 2018 zoonoses report. EFSA J. 2019, 17, e05926. [Google Scholar] [CrossRef]
- Shilangale, R.P.; Di Giannatale, E.; Chimwamurombe, P.M.; Kaaya, G.P. Prevalence and antimicrobial resistance pattern of Salmonella in animal feed produced in Namibia. Vet. Ital. 2012, 48, 125–132. [Google Scholar] [PubMed]
- Foley, S.L.; Lynne, A.M.; Nayak, R. Salmonella challenges: Prevalence in swine and poultry and potential pathogenicity of such isolates. J. Anim. Sci. 2008, 86, E149–E162. [Google Scholar] [CrossRef] [PubMed]
- Kimera, Z.I.; Mshana, S.E.; Rweyemamu, M.M.; Mboera, L.E.G.; Matee, M.I.N. Antimicrobial use and resistance in food-producing animals and the environment: An African perspective. Antimicrob. Resist. Infect. Control. 2020, 9, 37. [Google Scholar] [CrossRef] [PubMed]
- Ma, F.; Xu, S.; Tang, Z.; Li, Z.; Zhang, L. Use of antimicrobials in food animals and impact of transmission of antimicrobial resistance on humans. Biosaf. Health 2021, 3, 32–38. [Google Scholar] [CrossRef]
- World Health Organization (WHO). Critically Important Antimicrobials for Human Medicine; World Health Organisation: Geneva, Switzerland, 2019. [Google Scholar]
- Collignon, P.J.; Conly, J.M.; Andremont, A.; McEwen, S.A.; Aidara-Kane, A.; Agerso, Y.; Andremont, A.; Collignon, P.; Conly, J.; Dang Ninh, T.; et al. World Health Organization ranking of antimicrobials according to their importance in human medicine: A critical step for developing risk management strategies to control antimicrobial resistance from food animal production. Clin. Infect. Dis. 2016, 63, 1087–1093. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Sahm, D.F.; Jacoby, G.A.; Hooper, D.C. Emerging plasmid-mediated quinolone resistance associated with the qnr gene in Klebsiella pneumoniae clinical isolates in the United States. Antimicrob. Agents Chemother. 2004, 48, 1295–1299. [Google Scholar] [CrossRef]
- Hooper, D.C.; Jacoby, G.A. Mechanisms of drug resistance: Quinolone resistance. Ann. NY Acad. Sci. 2015, 1354, 12–31. [Google Scholar] [CrossRef]
- Fàbrega, A.; Madurga, S.; Giralt, E.; Vila, J. Mechanism of action of and resistance to quinolones. Microb. Biotechnol. 2009, 2, 40–61. [Google Scholar] [CrossRef]
- Chen, S.; Zhao, S.; White, D.G.; Schroeder, C.M.; Lu, R.; Yang, H.; McDermott, P.F.; Ayers, S.; Meng, J. Characterization of multiple-antimicrobial-resistant Salmonella serovars isolated from retail meats. Appl. Environ. Microbiol. 2004, 70, 1–7. [Google Scholar] [CrossRef]
- Miko, A.; Pries, K.; Schroeter, A.; Helmuth, R. Molecular mechanisms of resistance in multidrug-resistant serovars of Salmonella enterica isolated from foods in Germany. J. Antimicrob. Chemother. 2005, 56, 1025–1033. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.Y.; Wang, Y.; Walsh, T.R.; Yi, L.X.; Zhang, R.; Spencer, J.; Doi, Y.; Tian, G.; Dong, B.; Huang, X.; et al. Emergence of plasmid-mediated colistin resistance mechanism mcr-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infect. Dis. 2016, 16, 161–168. [Google Scholar] [CrossRef]
- Carroll, L.M.; Gaballa, A.; Guldimann, C.; Sullivan, G.; Henderson, L.O.; Wiedmann, M. Identification of novel mobilized colistin resistance gene mcr-9 in a multidrug-resistant, colistin-susceptible Salmonella enterica serotype Typhimurium isolate. mBio 2019, 10, e00853-19. [Google Scholar] [CrossRef] [PubMed]
- Borowiak, M.; Baumann, B.; Fischer, J.; Thomas, K.; Deneke, C.; Hammerl, J.A.; Szabo, I.; Malorny, B. Development of a novel mcr-6 to mcr-9 multiplex PCR and assessment of mcr-1 to mcr-9 occurrence in colistin-resistant Salmonella enterica isolates from environment, feed, animals and food (2011–2018) in Germany. Front. Microbiol. 2020, 11, 80. [Google Scholar] [CrossRef]
- Rebelo, A.R.; Bortolaia, V.; Kjeldgaard, J.S.; Pedersen, S.K.; Leekitcharoenphon, P.; Hansen, I.M.; Guerra, B.; Malorny, B.; Borowiak, M.; Hammerl, J.A.; et al. Multiplex PCR for detection of plasmid-mediated colistin resistance determinants, mcr-1, mcr-2, mcr-3, mcr-4, and mcr-5 for surveillance purposes. Eurosurveillance 2018, 23, 17–00672. [Google Scholar] [CrossRef]
- Eguale, T.; Birungi, J.; Asrat, D.; Njahira, M.N.; Njuguna, J.; Gebreyes, W.A.; Gunn, J.S.; Djikeng, A.; Engidawork, E. Genetic markers associated with resistance to beta-lactam and quinolone antimicrobials in non-typhoidal Salmonella isolates from humans and animals in central Ethiopia. Antimicrob. Resist. Infect. Control. 2017, 6, 13. [Google Scholar] [CrossRef]
- Deng, W.; Quan, Y.; Yang, S.; Guo, L.; Zhang, X.; Liu, S.; Chen, S.; Zhou, K.; He, L.; Li, B.; et al. Antibiotic resistance in Salmonella from retail foods of animal origin and its association with disinfectant and heavy metal resistance. Microb. Drug Resist. 2018, 24, 782–791. [Google Scholar] [CrossRef]
- Vilela, F.P.; Gomes, C.N.; Passaglia, J.; Rodrigues, D.P.; Costa, R.G.; Tiba Casas, M.R.; Fernandes, S.A.; Falcão, J.P.; Campioni, F. Genotypic resistance to quinolone and tetracycline in Salmonella Dublin strains isolated from humans and animals in Brazil. Microb. Drug Resist. 2019, 25, 143–151. [Google Scholar] [CrossRef]
- Khan, A.S.; Georges, K.; Rahaman, S.; Abebe, W.; Adesiyun, A.A. Occurrence, risk factors, serotypes and antimicrobial resistance of Salmonella strains isolated from imported fertile hatching eggs, hatcheries, and broiler farms in Trinidad and Tobago. J. Food Prot. 2021, 85, 266–277. [Google Scholar] [CrossRef]
- Khan, A.S.; Georges, K.; Rahaman, S.; Abebe, W.; Adesiyun, A.A. Characterization of Salmonella isolates recovered from stages of the processing lines at four broiler processing plants in Trinidad and Tobago. Microorganisms 2021, 9, 1048. [Google Scholar] [CrossRef]
- Khan, A.S.; Georges, K.; Rahaman, S.; Abdela, W.; Adesiyun, A.A. Antimicrobial resistance of Salmonella isolates recovered from chickens sold at retail outlets in Trinidad. J. Food Prot. 2018, 81, 1880–1889. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.S.; Georges, K.; Rahaman, S.; Abdela, W.; Adesiyun, A.A. Prevalence and serotypes of Salmonella spp. on chickens sold at retail outlets in Trinidad. PLoS ONE 2018, 13, e0202108. [Google Scholar] [CrossRef] [PubMed]
- Rodrigo, S.; Adesiyun, A.A.; Asgarali, Z.; Swanston, W. Occurrence of selected foodborne pathogens on poultry and poultry giblets from small retail processing operations in Trinidad. J. Food Prot. 2006, 69, 1096–1105. [Google Scholar] [CrossRef] [PubMed]
- Andrews, W. Manual of food quality control. 4. Rev. 1. Microbiological analysis. Food and Drug Administration. FAO Food Nutr. Pap. 1992, 14, 1–338. [Google Scholar]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Yoshida, C.E.; Kruczkiewicz, P.; Laing, C.R.; Lingohr, E.J.; Gannon, V.P.; Nash, J.H.; Taboada, E.N. The Salmonella in silico typing resource (SISTR): An open web-accessible tool for rapidly typing and subtyping draft Salmonella genome assemblies. PLoS ONE 2016, 11, e0147101. [Google Scholar] [CrossRef]
- Hyatt, D.; Chen, G.L.; Locascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010, 11, 119. [Google Scholar] [CrossRef]
- Liu, B.; Zheng, D.; Jin, Q.; Chen, L.; Yang, J. VFDB 2019: A comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res. 2019, 47, D687–D692. [Google Scholar] [CrossRef]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.Y.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.V.; Cheng, A.A.; Liu, S.; et al. CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic. Acids Res. 2020, 48, D517–D525. [Google Scholar] [CrossRef]
- Lee, D.H.; Hyeon, J.Y.; Kim, J.; Kim, J.S.; Kim, S.J.; Jeon, S.E.; Choi, S.W.; Hong, W.T.; Song, C.S.; Lee, S.W. Close genetic relationship between Salmonella enterica serovar Enteritidis isolated from patients with diarrhoea and poultry in the Republic of Korea. Clin. Microbiol. Infect. 2015, 21, e68–e70. [Google Scholar] [CrossRef][Green Version]
- Almeida, F.; Medeiros, M.I.; Kich, J.D.; Falcao, J.P. Virulence-associated genes, antimicrobial resistance and molecular typing of Salmonella Typhimurium strains isolated from swine from 2000 to 2012 in Brazil. J. Appl. Microbiol. 2016, 120, 1677–1690. [Google Scholar] [CrossRef] [PubMed]
- Pearce, M.E.; Alikhan, N.F.; Dallman, T.J.; Zhou, Z.; Grant, K.; Maiden, M.C.J. Comparative analysis of core genome MLST and SNP typing within a European Salmonella serovar Enteritidis outbreak. Int. J. Food. Microbiol. 2018, 274, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Pightling, A.W.; Pettengill, J.B.; Luo, Y.; Baugher, J.D.; Rand, H.; Strain, E. Interpreting whole-genome sequence analyses of foodborne bacteria for regulatory applications and outbreak investigations. Front. Microbiol. 2018, 9, 1482. [Google Scholar] [CrossRef] [PubMed]
- Guibourdenche, M.; Roggentin, P.; Mikoleit, M.; Fields, P.I.; Bockemuhl, J.; Grimont, P.A.; Weill, F.X. Supplement 2003–2007 (No. 47) to the White-Kauffmann-Le Minor scheme. Res. Microbiol. 2010, 161, 26–29. [Google Scholar] [CrossRef]
- Li, P.; Liu, Q.; Luo, H.; Liang, K.; Yi, J.; Luo, Y.; Hu, Y.; Han, Y.; Kong, Q. O-serotype conversion in Salmonella Typhimurium induces protective immune responses against invasive non-typhoidal Salmonella infections. Front. Immunol. 2017, 8, 1647. [Google Scholar] [CrossRef]
- Chui, L.; Ferrato, C.; Li, V.; Christianson, S. Comparison of molecular and in silico Salmonella serotyping for Salmonella surveillance. Microorganisms 2021, 9, 955. [Google Scholar] [CrossRef]
- Yachison, C.A.; Yoshida, C.; Robertson, J.; Nash, J.H.E.; Kruczkiewicz, P.; Taboada, E.N.; Walker, M.; Reimer, A.; Christianson, S.; Nichani, A.; et al. The validation and implications of using whole-genome sequencing as a replacement for traditional serotyping for a National Salmonella reference laboratory. Front. Microbiol. 2017, 8, 1044. [Google Scholar] [CrossRef]
- Elkenany, R.; Elsayed, M.M.; Zakaria, A.I.; El-sayed, S.A.-E.-S.; Rizk, M.A. Antimicrobial resistance profiles and virulence genotyping of Salmonella enterica serovars recovered from broiler chickens and chicken carcasses in Egypt. BMC Vet. Res. 2019, 15, 124. [Google Scholar] [CrossRef]
- Duc, V.M.; Nakamoto, Y.; Fujiwara, A.; Toyofuku, H.; Obi, T.; Chuma, T. Prevalence of Salmonella in broiler chickens in Kagoshima, Japan in 2009 to 2012 and the relationship between serovars changing and antimicrobial resistance. BMC Vet. Res. 2019, 15, 108. [Google Scholar] [CrossRef]
- Wang, X.; Wang, H.; Li, T.; Liu, F.; Cheng, Y.; Guo, X.; Wen, G.; Luo, Q.; Shao, H.; Pan, Z.; et al. Characterization of Salmonella spp. isolated from chickens in Central China. BMC Vet. Res. 2020, 16, 299. [Google Scholar] [CrossRef]
- Ramirez-Hernandez, A.; Carrascal-Camacho, A.K.; Varon-Garcia, A.; Brashears, M.M.; Sanchez-Plata, M.X. Genotypic characterization of antimicrobial resistant Salmonella spp. strains from three poultry processing plants in Colombia. Foods 2021, 10, 491. [Google Scholar] [CrossRef] [PubMed]
- Tamang, M.D.; Nam, H.M.; Kim, T.S.; Jang, G.C.; Jung, S.C.; Lim, S.K. Emergence of extended-spectrum beta-lactamase (CTX-M-15 and CTX-M-14)-producing nontyphoid Salmonella with reduced susceptibility to ciprofloxacin among food animals and humans in Korea. J. Clin. Microbiol. 2011, 49, 2671–2675. [Google Scholar] [CrossRef] [PubMed]
- Aguilar-Montes de Oca, S.; Talavera-Rojas, M.; Soriano-Vargas, E.; Barba-Leon, J.; Vazquez-Navarrete, J.; Acosta-Dibarrat, J.; Salgado-Miranda, C. Phenotypic and genotypic profile of clinical and animal multidrug-resistant Salmonella enterica isolates from Mexico. J. Appl. Microbiol. 2018, 124, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Mendonça, E.P.; Melo, R.T.; Oliveira, M.R.M.; Monteiro, G.P.; Peres, P.A.B.M.; Fonseca, B.B.; Giombelli, A.; Rossi, D.A. Characteristics of virulence, resistance and genetic diversity of strains of Salmonella Infantis isolated from broiler chicken in Brazil. Braz. Vet. Res. 2020, 40, 29–38. [Google Scholar] [CrossRef]
- Brinas, L.; Moreno, M.A.; Zarazaga, M.; Porrero, C.; Saenz, Y.; Garcia, M.; Dominguez, L.; Torres, C. Detection of CMY-2, CTX-M-14, and SHV-12 beta-lactamases in Escherichia coli fecal-sample isolates from healthy chickens. Antimicrob. Agents Chemother. 2003, 47, 2056–2058. [Google Scholar] [CrossRef]
- Winokur, P.L.; Brueggemann, A.; DeSalvo, D.L.; Hoffmann, L.; Apley, M.D.; Uhlenhopp, E.K.; Pfaller, M.A.; Doern, G.V. Animal and human multidrug-resistant, cephalosporin-resistant Salmonella isolates expressing a plasmid-mediated CMY-2 AmpC beta-lactamase. Antimicrob. Agents Chemother. 2000, 44, 2777–2783. [Google Scholar] [CrossRef]
- Aviv, G.; Tsyba, K.; Steck, N.; Salmon-Divon, M.; Cornelius, A.; Rahav, G.; Grassl, G.A.; Gal-Mor, O. A unique megaplasmid contributes to stress tolerance and pathogenicity of an emergent Salmonella enterica serovar Infantis strain. Environ. Microbiol. 2014, 16, 977–994. [Google Scholar] [CrossRef]
- Bogomazova, A.N.; Gordeeva, V.D.; Krylova, E.V.; Soltynskaya, I.V.; Davydova, E.E.; Ivanova, O.E.; Komarov, A.A. Mega-plasmid found worldwide confers multiple antimicrobial resistance in Salmonella Infantis of broiler origin in Russia. Int. J. Food Microbiol. 2020, 319, 108497. [Google Scholar] [CrossRef]
- Franco, A.; Leekitcharoenphon, P.; Feltrin, F.; Alba, P.; Cordaro, G.; Iurescia, M.; Tolli, R.; D’Incau, M.; Staffolani, M.; Di Giannatale, E.; et al. Emergence of a clonal lineage of multidrug-resistant ESβL-producing Salmonella Infantis transmitted from broilers and broiler meat to humans in Italy between 2011 and 2014. PLoS ONE 2015, 10, e0144802. [Google Scholar] [CrossRef]
- Yokoyama, E.; Ando, N.; Ohta, T.; Kanada, A.; Shiwa, Y.; Ishige, T.; Murakami, K.; Kikuchi, T.; Murakami, S. A novel subpopulation of Salmonella enterica serovar Infantis strains isolated from broiler chicken organs other than the gastrointestinal tract. Vet. Microbiol. 2015, 175, 312–318. [Google Scholar] [CrossRef]
- Cartelle Gestal, M.; Zurita, J.; Paz, Y.M.A.; Ortega-Paredes, D.; Alcocer, I. Characterization of a small outbreak of Salmonella enterica serovar Infantis that harbour CTX-M-65 in Ecuador. Braz. J. Infect. Dis. 2016, 20, 406–407. [Google Scholar] [CrossRef] [PubMed]
- Palma, N.; Pons, M.J.; Gomes, C.; Mateu, J.; Riveros, M.; Garcia, W.; Jacobs, J.; Garcia, C.; Ochoa, T.J.; Ruiz, J. Resistance to quinolones, cephalosporins and macrolides in Escherichia coli causing bacteraemia in Peruvian children. J. Glob. Antimicrob. Resist. 2017, 11, 28–33. [Google Scholar] [CrossRef] [PubMed]
- Tyson, G.H.; Li, C.; Harrison, L.B.; Martin, G.; Hsu, C.H.; Tate, H.; Tran, T.T.; Strain, E.; Zhao, S. A multidrug-resistant Salmonella Infantis clone is spreading and recombining in the United States. Microb. Drug Resist. 2021, 27, 792–799. [Google Scholar] [CrossRef] [PubMed]
- Brown, A.C.; Chen, J.C.; Watkins, L.K.F.; Campbell, D.; Folster, J.P.; Tate, H.; Wasilenko, J.; Van Tubbergen, C.; Friedman, C.R. CTX-M-65 extended-spectrum beta-lactamase-producing Salmonella enterica serotype Infantis, United States. Emerg. Infect. Dis. 2018, 24, 2284–2291. [Google Scholar] [CrossRef] [PubMed]
- Bonnet, R. Growing group of extended-spectrum beta-lactamases: The CTX-M enzymes. Antimicrob. Agents Chemother. 2004, 48, 1–14. [Google Scholar] [CrossRef]
- Hofacre, C.L.; Fricke, J.A.; Inglis, T. Antimicrobial drug use in poultry. In Antimicrobial Therapy in Veterinary Medicine; Giguère, S., Prescott, J.F., Dowling, P.M., Eds.; John Wiley & Sons: Hoboken, NJ, USA, 2013; pp. 569–587. [Google Scholar]
- Dutil, L.; Irwin, R.; Finley, R.; Ng, L.K.; Avery, B.; Boerlin, P.; Bourgault, A.M.; Cole, L.; Daignault, D.; Desruisseau, A.; et al. Ceftiofur resistance in Salmonella enterica serovar Heidelberg from chicken meat and humans, Canada. Emerg. Infect. Dis. 2010, 16, 48–54. [Google Scholar] [CrossRef]
- Tate, H.; Folster, J.P.; Hsu, C.H.; Chen, J.; Hoffmann, M.; Li, C.; Morales, C.; Tyson, G.H.; Mukherjee, S.; Brown, A.C.; et al. Comparative analysis of extended-spectrum-beta-lactamase CTX-M-65-producing Salmonella enterica serovar Infantis isolates from humans, food animals, and retail chickens in the United States. Antimicrob. Agents Chemother. 2017, 61, e00488-17. [Google Scholar] [CrossRef]
- Cheddie, P.; Dziva, F.; Akpaka, P.E. Detection of a CTX-M group 2 beta-lactamase gene in a Klebsiella pneumoniae isolate from a tertiary care hospital, Trinidad and Tobago. Ann. Clin. Microbiol. Antimicrob. 2017, 16, 33. [Google Scholar] [CrossRef]
- Hindermann, D.; Gopinath, G.; Chase, H.; Negrete, F.; Althaus, D.; Zurfluh, K.; Tall, B.D.; Stephan, R.; Nuesch-Inderbinen, M. Salmonella enterica serovar Infantis from food and human infections, Switzerland, 2010–2015: Poultry-related multidrug resistant clones and an emerging ESBL producing clonal lineage. Front. Microbiol. 2017, 8, 1322. [Google Scholar] [CrossRef]
- Nógrády, N.; Király, M.; Davies, R.; Nagy, B. Multidrug resistant clones of Salmonella Infantis of broiler origin in Europe. Int. J. Food Microbiol. 2012, 157, 108–112. [Google Scholar] [CrossRef]
- Sköld, O. Resistance to trimethoprim and sulfonamides. Vet. Res. 2001, 32, 261–273. [Google Scholar] [CrossRef] [PubMed]
- Kozak, G.K.; Pearl, D.L.; Parkman, J.; Reid-Smith, R.J.; Deckert, A.; Boerlin, P. Distribution of sulfonamide resistance genes in Escherichia coli and Salmonella isolates from swine and chickens at abattoirs in Ontario and Quebec, Canada. Appl. Environ. Microbiol. 2009, 75, 5999–6001. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Lai, H.; Zou, L.; Yin, S.; Wang, C.; Han, X.; Xia, X.; Hu, K.; He, L.; Zhou, K.; et al. Antimicrobial resistance and resistance genes in Salmonella strains isolated from broiler chickens along the slaughtering process in China. Int. J. Food Microbiol. 2017, 259, 43–51. [Google Scholar] [CrossRef] [PubMed]
- Arkali, A.; Cetinkaya, B. Molecular identification and antibiotic resistance profiling of Salmonella species isolated from chickens in eastern Turkey. BMC Vet. Res. 2020, 16, 205. [Google Scholar] [CrossRef] [PubMed]
- Maguire, M.; Khan, A.S.; Adesiyun, A.A.; Georges, K.; Gonzalez-Escalona, N. Closed genome sequence of a Salmonella enterica serotype Senftenberg strain carrying the mcr-9 gene isolated from broken chicken eggshells in Trinidad and Tobago. Microbiol. Resour. Announc. 2021, 10, e0146520. [Google Scholar] [CrossRef]
- Tyson, G.H.; Li, C.; Hsu, C.H.; Ayers, S.; Borenstein, S.; Mukherjee, S.; Tran, T.T.; McDermott, P.F.; Zhao, S. The mcr-9 Gene of Salmonella and Escherichia coli is not associated with colistin resistance in the United States. Antimicrob. Agents Chemother. 2020, 64, e00573-20. [Google Scholar] [CrossRef]
- Sefton, A.M. Mechanisms of antimicrobial resistance: Their clinical relevance in the new millennium. Drugs 2002, 62, 557–566. [Google Scholar] [CrossRef]
- Sosa, A.d.J.; Byarugaba, D.K.; Amábile-Cuevas, C.F.; Hsueh, P.-R.; Kariuki, S.; Okeke, I.N. Antimicrobial Resistance in Developing Countries; Springer: Berlin, Germany, 2010; pp. 1–554. [Google Scholar]
- Alam, S.B.; Mahmud, M.; Akter, R.; Hasan, M.; Sobur, A.; Nazir, K.N.H.; Noreddin, A.; Rahman, T.; El Zowalaty, M.E.; Rahman, M. Molecular detection of multidrug resistant Salmonella species isolated from broiler farm in Bangladesh. Pathogens 2020, 9, 201. [Google Scholar] [CrossRef]
- Van, T.T.; Chin, J.; Chapman, T.; Tran, L.T.; Coloe, P.J. Safety of raw meat and shellfish in Vietnam: An analysis of Escherichia coli isolations for antibiotic resistance and virulence genes. Int. J. Food Microbiol. 2008, 124, 217–223. [Google Scholar] [CrossRef]
- Paterson, D.L.; Bonomo, R.A. Extended-spectrum beta-lactamases: A clinical update. Clin. Microbiol. Rev. 2005, 18, 657–686. [Google Scholar] [CrossRef]
- Akinola, S.A.; Mwanza, M.; Ateba, C.N. Occurrence, genetic diversities and antibiotic resistance profiles of Salmonella serovars isolated from chickens. Infect. Drug Resist. 2019, 12, 3327–3342. [Google Scholar] [CrossRef] [PubMed]
- Ammar, A.M.; Abdeen, E.E.; Abo-Shama, U.H.; Fekry, E.; Kotb Elmahallawy, E. Molecular characterization of virulence and antibiotic resistance genes among Salmonella serovars isolated from broilers in Egypt. Lett. Appl. Microbiol. 2019, 68, 188–195. [Google Scholar] [CrossRef] [PubMed]
- Kaniga, K.; Trollinger, D.; Galán, J.E. Identification of two targets of the type III protein secretion system encoded by the inv and spa loci of Salmonella Typhimurium that have homology to the Shigella IpaD and IpaA proteins. J. Bacteriol. 1995, 177, 7078–7085. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.M.; Kaniga, K.; Galán, J.E. Salmonella spp. are cytotoxic for cultured macrophages. Mol. Microbiol. 1996, 21, 1101–1115. [Google Scholar] [CrossRef] [PubMed]
- Hensel, M.; Shea, J.E.; Waterman, S.R.; Mundy, R.; Nikolaus, T.; Banks, G.; Vazquez-Torres, A.; Gleeson, C.; Fang, F.C.; Holden, D.W. Genes encoding putative effector proteins of the type III secretion system of Salmonella pathogenicity island 2 are required for bacterial virulence and proliferation in macrophages. Mol. Microbiol. 1998, 30, 163–174. [Google Scholar] [CrossRef] [PubMed]
- Cirillo, D.M.; Valdivia, R.H.; Monack, D.M.; Falkow, S. Macrophage-dependent induction of the Salmonella pathogenicity island 2 type III secretion system and its role in intracellular survival. Mol. Microbiol. 1998, 30, 175–188. [Google Scholar] [CrossRef] [PubMed]
- Galyov, E.E.; Wood, M.W.; Rosqvist, R.; Mullan, P.B.; Watson, P.R.; Hedges, S.; Wallis, T.S. A secreted effector protein of Salmonella Dublin is translocated into eukaryotic cells and mediates inflammation and fluid secretion in infected ileal mucosa. Mol. Microbiol. 1997, 25, 903–912. [Google Scholar] [CrossRef]
- Gibson, D.L.; White, A.P.; Rajotte, C.M.; Kay, W.W. AgfC and AgfE facilitate extracellular thin aggregative fimbriae synthesis in Salmonella Enteritidis. Microbiology 2007, 153, 1131–1140. [Google Scholar] [CrossRef]
- Yoo, A.Y.; Yu, J.E.; Yoo, H.; Lee, T.H.; Lee, W.H.; Oh, J.I.; Kang, H.Y. Role of sigma factor E in regulation of Salmonella Agf expression. Biochem. Biophys. Res. Commun. 2013, 430, 131–136. [Google Scholar] [CrossRef]
- Collinson, S.K.; Doig, P.C.; Doran, J.L.; Clouthier, S.; Trust, T.J.; Kay, W.W. Thin, aggregative fimbriae mediate binding of Salmonella Enteritidis to fibronectin. J. Bacteriol. 1993, 175, 12–18. [Google Scholar] [CrossRef]
- Borsoi, A.; Santin, E.; Santos, L.R.; Salle, C.T.; Moraes, H.L.; Nascimento, V.P. Inoculation of newly hatched broiler chicks with two Brazilian isolates of Salmonella Heidelberg strains with different virulence gene profiles, antimicrobial resistance, and pulsed field gel electrophoresis patterns to intestinal changes evaluation. Poult. Sci. 2009, 88, 750–758. [Google Scholar] [CrossRef] [PubMed]
- Den Bakker, H.C.; Moreno Switt, A.I.; Govoni, G.; Cummings, C.A.; Ranieri, M.L.; Degoricija, L.; Hoelzer, K.; Rodriguez-Rivera, L.D.; Brown, S.; Bolchacova, E.; et al. Genome sequencing reveals diversification of virulence factor content and possible host adaptation in distinct subpopulations of Salmonella enterica. BMC Genomics 2011, 12, 425. [Google Scholar] [CrossRef] [PubMed]
- Siddiky, N.A.; Sarker, M.S.; Khan, M.S.R.; Begum, R.; Kabir, M.E.; Karim, M.R.; Rahman, M.T.; Mahmud, A.; Samad, M.A. Virulence and antimicrobial resistance profiles of Salmonella enterica serovars isolated from chicken at wet markets in Dhaka, Bangladesh. Microorganisms 2021, 9, 952. [Google Scholar] [CrossRef] [PubMed]
- Campioni, F.; Moratto Bergamini, A.M.; Falcão, J.P. Genetic diversity, virulence genes and antimicrobial resistance of Salmonella Enteritidis isolated from food and humans over a 24-year period in Brazil. Food Microbiol. 2012, 32, 254–264. [Google Scholar] [CrossRef] [PubMed]
- Amini, K.; Salehi, T.; Nikbakht Brujeni, G.; Ranjbar, R.; Amini, J.; Ashrafganjooei, S. Molecular detection of invA and spv virulence genes in Salmonella Enteritidis isolated from human and animals in Iran. Afr. J. Microbiol. Res. 2010, 4, 2202–2210. [Google Scholar]
- Huehn, S.; La Ragione, R.M.; Anjum, M.; Saunders, M.; Woodward, M.J.; Bunge, C.; Helmuth, R.; Hauser, E.; Guerra, B.; Beutlich, J.; et al. Virulotyping and antimicrobial resistance typing of Salmonella enterica serovars relevant to human health in Europe. Foodborne Pathog. Dis. 2010, 7, 523–535. [Google Scholar] [CrossRef] [PubMed]
- Aslam, M.; Checkley, S.; Avery, B.; Chalmers, G.; Bohaychuk, V.; Gensler, G.; Reid-Smith, R.; Boerlin, P. Phenotypic and genetic characterization of antimicrobial resistance in Salmonella serovars isolated from retail meats in Alberta, Canada. Food Microbiol. 2012, 32, 110–117. [Google Scholar] [CrossRef] [PubMed]
- O’Regan, E.; Quinn, T.; Frye, J.G.; Pages, J.M.; Porwollik, S.; Fedorka-Cray, P.J.; McClelland, M.; Fanning, S. Fitness costs and stability of a high-level ciprofloxacin resistance phenotype in Salmonella enterica serotype Enteritidis: Reduced infectivity associated with decreased expression of Salmonella pathogenicity island 1 genes. Antimicrob. Agents Chemother. 2010, 54, 367–374. [Google Scholar] [CrossRef][Green Version]
- Gunn, J.S.; Ryan, S.S.; Van Velkinburgh, J.C.; Ernst, R.K.; Miller, S.I. Genetic and functional analysis of a PmrA-PmrB-regulated locus necessary for lipopolysaccharide modification, antimicrobial peptide resistance, and oral virulence of Salmonella enterica serovar Typhimurium. Infect. Immun. 2000, 68, 6139–6146. [Google Scholar] [CrossRef]
- Tamayo, R.; Ryan, S.S.; McCoy, A.J.; Gunn, J.S. Identification and genetic characterization of PmrA-regulated genes and genes involved in polymyxin B resistance in Salmonella enterica serovar Typhimurium. Infect. Immun. 2002, 70, 6770–6778. [Google Scholar] [CrossRef]
- Eswarappa, S.M.; Panguluri, K.K.; Hensel, M.; Chakravortty, D. The yejABEF operon of Salmonella confers resistance to antimicrobial peptides and contributes to its virulence. Microbiology 2008, 154, 666–678. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, A.I.; Zorzet, A.; Kanth, A.; Dahlström, S.; Berg, O.G.; Andersson, D.I. Reducing the fitness cost of antibiotic resistance by amplification of initiator tRNA genes. Proc. Natl. Acad. Sci. USA 2006, 103, 6976–6981. [Google Scholar] [CrossRef] [PubMed]
- Andersson, D.I.; Hughes, D. Antibiotic resistance and its cost: Is it possible to reverse resistance? Nat. Rev. Microbiol. 2010, 8, 260–271. [Google Scholar] [CrossRef] [PubMed]
| No. of Strains of Salmonella Detected from the Following: | |||||
|---|---|---|---|---|---|
| Serovars | Hatchery | Farm | Processing Plant | Pluck Shop a | Supermarket a |
| Aberdeen | 0 | 0 | 1 | 1 | 0 |
| Alachua | 0 | 0 | 1 | 0 | 0 |
| Albany | 0 | 4 | 8 | 1 | 0 |
| Anatomy | 0 | 0 | 5 | 0 | 0 |
| Caracas | 0 | 0 | 0 | 3 | 0 |
| Chester | 0 | 0 | 0 | 0 | 2 |
| Enteritidis | 0 | 0 | 9 | 0 | 0 |
| Fresno | 1 | 0 | 0 | 0 | 0 |
| Gaminara | 0 | 3 | 0 | 0 | 0 |
| Infantis | 0 | 11 | 9 | 0 | 0 |
| Javiana | 0 | 0 | 10 | 17 | 1 |
| Kentucky | 8 | 0 | 7 | 12 | 3 |
| Liverpool | 0 | 0 | 1 | 0 | 0 |
| Manhattan | 0 | 0 | 0 | 7 | 0 |
| Mbandaka | 0 | 0 | 1 | 0 | 0 |
| Molade | 0 | 0 | 0 | 0 | 1 |
| Montevideo | 0 | 0 | 0 | 2 | 1 |
| Oranienburg | 0 | 1 | 0 | 0 | 0 |
| Schwarzengrund | 0 | 0 | 7 | 1 | 0 |
| Senftenberg | 1 | 0 | 0 | 2 | 1 |
| Soerenga | 0 | 1 | 0 | 0 | 0 |
| Virchow | 0 | 0 | 1 | 0 | 0 |
| Weltevreden | 0 | 0 | 1 | 0 | 0 |
| Sub-total | 10 | 20 | 61 | 46 | 9 |
| Antimicrobial Class and Genes Detected a | |||||||
|---|---|---|---|---|---|---|---|
| Pattern | Aminoglycoside | Disinfectant | Cephalosporin | Peptide | Sulphonamide | Number of Isolates (%) | Serovar (n, %) |
| Pattern 1 | aph(4)-Ia | qacEDelta1 | blaCTX-M-65 | - | sul1 | 6 (4.2) | Infantis (6, 100.0) |
| aac(3)-IV | |||||||
| Pattern 2 | aph(3′)-Ia | qacEDelta1 | - | - | sul1 | 1 (0.7) | Infantis (1, 100.0) |
| aph(4)-Ia | |||||||
| aac(3)-IV | |||||||
| Pattern 3 | aph(3′)Ia | qacEDelta1 | blaCTX-M-65 | - | sul1 | 4 (2.8) | Infantis (4, 100.0) |
| aph(4)-Ia | |||||||
| aac(3)-IV | |||||||
| Pattern 4 | - | qacEDelta1 | - | - | sul1 | 1 (0.7) | Senftenberg (1, 100.0) |
| Pattern 5 | aac(6′)-Iaa | - | - | - | - | 7 (4.9) | Manhattan (7, 100.0) |
| Pattern 6 | aac(6′)-Iy | - | - | - | - | 2 (1.4) | Aberdeen (2, 100.0) |
| Pattern 7 | - | - | - | mcr-9.1 | - | 1 (0.7) | Senftenberg (1, 100.0) |
| Total | 20 (14.9) | 12 (9.0) | 10 (7.5) | 1 (0.7) | 12 (9.0) | 22 (16.1) | |
| Distribution of AROs among the Various Sampling Levels | ||||||
|---|---|---|---|---|---|---|
| ARO Name a | No. of AROs | Overall Frequency (%) b | Hatchery | Farm | Processing Plant | Retail Outlet |
| aac(3)-IV | 11 | 7.5 | 0 | 11 | 0 | 0 |
| aac(6‘)-Iaa | 7 | 4.8 | 0 | 0 | 0 | 7 |
| aac(6‘)-Iy | 2 | 1.4 | 0 | 0 | 1 | 1 |
| aph(3‘)-Ia | 5 | 3.4 | 0 | 5 | 0 | 0 |
| aph(4)-Ia | 11 | 7.5 | 0 | 11 | 0 | 0 |
| blaCTX-M-65 | 10 | 6.8 | 0 | 10 | 0 | 0 |
| mcr-9.1 | 1 | 0.7 | 1 | 0 | 0 | 0 |
| qacEDelta1 | 12 | 8.2 | 0 | 11 | 0 | 1 |
| sul1 | 12 | 8.2 | 0 | 11 | 0 | 1 |
| Total | 71 | 1 | 59 | 1 | 10 | |
| BioSample | Isolate No. a | Phenotypic AMR Using the Disk Diffusion Method b,c,d | Genotypic Characteristics e | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| P | TE | CE | AM | PH | S | F | Other Resistance Genes Detected f | Virulence Factors | ||
| SAMN25867756 | F 17 | S | R | R | R | S | R | S | qacEDelta1 | agf/csg |
| aph(4)-Ia | bcf | |||||||||
| aac(3)-IV | lpf | |||||||||
| sul1 | TTSS (SPI-1 encode) | |||||||||
| TTSS (SPI-2 encode) | ||||||||||
| TTSS-1 translocated effectors | ||||||||||
| SAMN25867757 | F 22 | S | R | R | R | S | R | S | qacEDelta1 | agf/csg |
| aph(4)-Ia | bcf | |||||||||
| aac(3)-IV | lpf | |||||||||
| sul1 | TTSS (SPI-1 encode) | |||||||||
| TTSS (SPI-2 encode) | ||||||||||
| TTSS-1 translocated effectors | ||||||||||
| SAMN14677229 | F 11 | S | R | R | R | S | R | S | qacEDelta1 | agf/csg |
| aph(4)-Ia | bcf | |||||||||
| aac(3)-IV | lpf | |||||||||
| sul1 | TTSS (SPI-1 encode) | |||||||||
| TTSS (SPI-2 encode) | ||||||||||
| TTSS-1 translocated effectors | ||||||||||
| SAMN14677211 | F 32 | S | R | R | R | S | R | S | aph(3′)-Ia | agf/csg |
| qacEDelta1 | bcf | |||||||||
| aph(4)-Ia | lpf | |||||||||
| aac(3)-IV | TTSS (SPI-1 encode) | |||||||||
| sul1 | TTSS (SPI-2 encode) | |||||||||
| TTSS-1 translocated effectors | ||||||||||
| SAMN14677232 | F 36 | S | R | R | R | S | R | S | aph(3′)-Ia | agf/csg |
| qacEDelta1 | bcf | |||||||||
| aph(4)-Ia | lpf | |||||||||
| aac(3)-IV | TTSS (SPI-1 encode) | |||||||||
| sul1 | TTSS (SPI-2 encode) | |||||||||
| TTSS-1 translocated effectors | ||||||||||
| SAMN14677210 | F 2 | S | S | S | S | S | S | S | qacEDelta1 | agf/csg |
| aph(4)-Ia | bcf | |||||||||
| aac(3)-IV | lpf | |||||||||
| sul1 | TTSS (SPI-1 encode) | |||||||||
| TTSS (SPI-2 encode) | ||||||||||
| TTSS-1 translocated effectors | ||||||||||
| SAMN14677203 | F 4 | S | R | R | R | S | S | R | qacEDelta1 | agf/csg |
| aph(4)-Ia | bcf | |||||||||
| aac(3)-IV | lpf | |||||||||
| sul1 | TTSS (SPI-1 encode) | |||||||||
| TTSS (SPI-2 encode) | ||||||||||
| TTSS-1 translocated effectors | ||||||||||
| SAMN14677209 | UWI-F30 | S | S | S | S | S | S | S | aph(3′)-Ia | agf/csg |
| qacEDelta1 | bcf | |||||||||
| aph(4)-Ia | lpf | |||||||||
| aac(3)-IV | TTSS (SPI-1 encode) | |||||||||
| sul1 | TTSS (SPI-2 encode) | |||||||||
| TTSS-1 translocated effectors | ||||||||||
| SAMN14677207 | UWI-F9 | S | R | S | R | S | S | S | qacEDelta1 | agf/csg |
| aph(4)-Ia | bcf | |||||||||
| aac(3)-IV | lpf | |||||||||
| sul1 | TTSS (SPI-1 encode) | |||||||||
| TTSS (SPI-2 encode) | ||||||||||
| TTSS-1 translocated effectors | ||||||||||
| SAMN14677208 | UWI-F31 | S | R | S | R | S | S | S | aph(3′)-Ia | agf/csg |
| qacEDelta1 | bcf | |||||||||
| aph(4)-Ia | lpf | |||||||||
| aac(3)-IV | TTSS (SPI-1 encode) | |||||||||
| sul1 | TTSS (SPI-2 encode) | |||||||||
| TTSS-1 translocated effectors | ||||||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khan, A.S.; Pierneef, R.E.; Gonzalez-Escalona, N.; Maguire, M.; Li, C.; Tyson, G.H.; Ayers, S.; Georges, K.; Abebe, W.; Adesiyun, A.A. Molecular Characterization of Salmonella Detected along the Broiler Production Chain in Trinidad and Tobago. Microorganisms 2022, 10, 570. https://doi.org/10.3390/microorganisms10030570
Khan AS, Pierneef RE, Gonzalez-Escalona N, Maguire M, Li C, Tyson GH, Ayers S, Georges K, Abebe W, Adesiyun AA. Molecular Characterization of Salmonella Detected along the Broiler Production Chain in Trinidad and Tobago. Microorganisms. 2022; 10(3):570. https://doi.org/10.3390/microorganisms10030570
Chicago/Turabian StyleKhan, Anisa Sarah, Rian Ewald Pierneef, Narjol Gonzalez-Escalona, Meghan Maguire, Cong Li, Gregory H. Tyson, Sherry Ayers, Karla Georges, Woubit Abebe, and Abiodun Adewale Adesiyun. 2022. "Molecular Characterization of Salmonella Detected along the Broiler Production Chain in Trinidad and Tobago" Microorganisms 10, no. 3: 570. https://doi.org/10.3390/microorganisms10030570
APA StyleKhan, A. S., Pierneef, R. E., Gonzalez-Escalona, N., Maguire, M., Li, C., Tyson, G. H., Ayers, S., Georges, K., Abebe, W., & Adesiyun, A. A. (2022). Molecular Characterization of Salmonella Detected along the Broiler Production Chain in Trinidad and Tobago. Microorganisms, 10(3), 570. https://doi.org/10.3390/microorganisms10030570







