Reclassification of Enterobacter sp. FY-07 as Kosakonia oryzendophytica FY-07 and Its Potential to Promote Plant Growth
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Sequences Acquisition and Phylogenetic Analysis
2.3. Biochemical Characteristics of FY-07
2.4. Comparative Genomics Analysis
2.5. Identification or Measurement of Plant Growth Promoting Traits
2.5.1. IAA Production
2.5.2. Siderophore Production
2.5.3. Phosphate Solubilization
2.5.4. Nitrogen Fixation
2.5.5. Ammonia Production
2.5.6. ACC Deaminase Production
2.6. Colonization of Rice Roots
3. Results and Discussion
3.1. Phylogenetic Analysis
3.2. Comparative Genomics Analysis
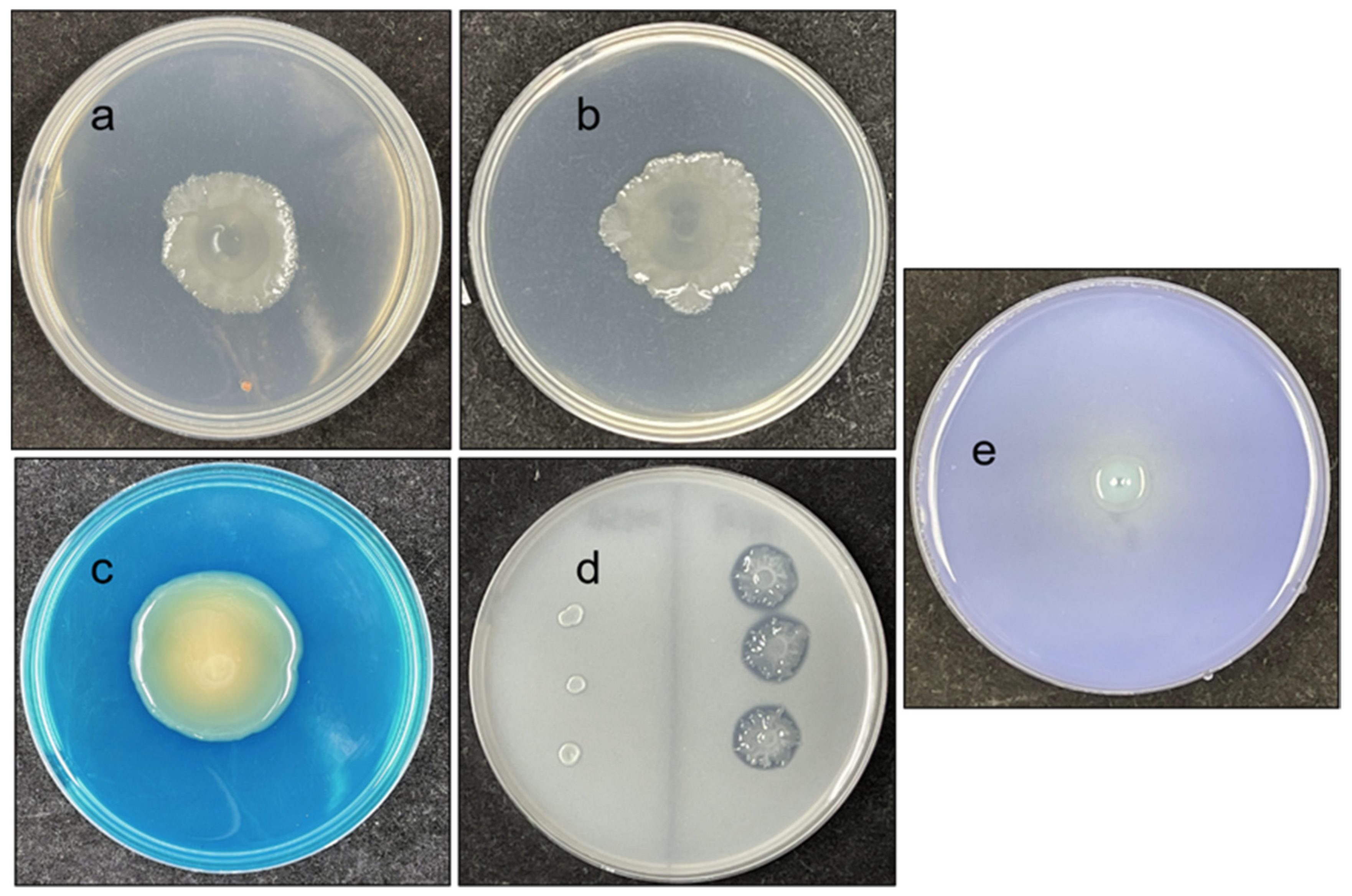
3.3. Biochemical Characteristics of FY-07
3.4. Description of Kosakonia oryzendophytica FY-07
3.5. Plant Growth-Promoting Ability of FY-07
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Stephan, R.; Grim, C.J.; Gopinath, G.R.; Mammel, M.K.; Sathyamoorthy, V.; Trach, L.H.; Chase, H.R.; Fanning, S.; Tall, B.D. Re-examination of the taxonomic status of Enterobacter helveticus, Enterobacter pulveris and Enterobacter turicensis as members of the genus Cronobacter and their reclassification in the genera Franconibacter gen. nov. and Siccibacter gen. nov. as Franconibacter helveticus comb. nov., Franconibacter pulveris comb. nov. and Siccibacter turicensis comb. nov., respectively. Int. J. Syst. Evol. Microbiol. 2014, 64, 3402–3410. [Google Scholar] [PubMed]
- Naum, M.; Brown, E.W.; Mason-Gamer, R.J. Is 16S rDNA a Reliable Phylogenetic Marker to Characterize Relationships Below the Family Level in the Enterobacteriaceae? J. Mol. Evol. 2008, 66, 630–642. [Google Scholar] [CrossRef] [PubMed]
- Brady, C.; Cleenwerck, I.; Venter, S.; Coutinho, T.; De Vos, P. Taxonomic evaluation of the genus Enterobacter based on multilocus sequence analysis (MLSA): Proposal to reclassify E. nimipressuralis and E. amnigenus into Lelliottia gen. nov. as Lelliottia nimipressuralis comb. nov. and Lelliottia amnigena comb. nov., respectively, E. gergoviae and E. pyrinus into Pluralibacter gen. nov. as Pluralibacter gergoviae comb. nov. and Pluralibacter pyrinus comb. nov., respectively, E. cowanii, E. radicincitans, E. oryzae and E. arachidis into Kosakonia gen. nov. as Kosakonia cowanii comb. nov., Kosakonia radicincitans comb. nov., Kosakonia oryzae comb. nov. and Kosakonia arachidis comb. nov., respectively, and E. turicensis, E. helveticus and E. pulveris into Cronobacter as Cronobacter zurichensis nom. nov., Cronobacter helveticus comb. nov. and Cronobacter pulveris comb. nov., respectively, and emended description of the genera Enterobacter and Cronobacter. Syst. Appl. Microbiol. 2013, 36, 309–319. [Google Scholar] [PubMed]
- Li, C.Y.; Zhou, Y.L.; Ji, J.; Gu, C.T. Reclassification of Enterobacter oryziphilus and Enterobacter oryzendophyticus as Kosakonia oryziphila comb. nov. and Kosakonia oryzendophytica comb. nov. Int. J. Syst. Evol. Microbiol. 2016, 66, 2780–2783. [Google Scholar] [CrossRef][Green Version]
- Wang, C.C.; Wu, W.J.; Wei, L.; Feng, Y.; Kang, M.; Xie, Y.; Zong, Z.Y. Kosakonia quasisacchari sp. nov. recovered from human wound secretion in China. Int. J. Syst. Evol. Microbiol. 2019, 69, 3155–3160. [Google Scholar] [CrossRef]
- Gu, C.T.; Li, C.Y.; Yang, L.J.; Huo, G.C. Enterobacter xiangfangensis sp. nov., isolated from Chinese traditional sourdough, and reclassification of Enterobacter sacchari Zhu et al. 2013 as Kosakonia sacchari comb. nov. Int. J. Syst. Evol. Microbiol. 2014, 64, 2650–2656. [Google Scholar] [CrossRef]
- Kämpfer, P.; Mclnroy, J.A.; Doijaad, S.; Chakraborty, T.; Glaeser, S.P. Kosakonia pseudosacchari sp. nov., an endophyte of Zea mays. Syst. Appl. Microbiol. 2016, 39, 1–7. [Google Scholar] [CrossRef]
- Mosquito, S.; Meng, X.; Devescovi, G.; Bertani, I.; Geller, A.; Levy, A.; Myers, M.P.; Bez, C.; Covaceuszach, S.; Venturi, V. LuxR Solos in the Plant Endophyte Kosakonia sp. Strain KO348. Appl. Environ. Microb. 2020, 86, e00622-20. [Google Scholar] [CrossRef]
- Bloch, S.E.; Clark, R.; Gottlieb, S.S.; Wood, L.K.; Shah, N.; San-Ming, M.; Lorigan, J.G.; Johnson, J.; Davis-Richardson, A.G.; Williams, L.; et al. Biological nitrogen fixation in maize: Optimizing nitrogenase expression in a root-associated diazotroph. J. Exp. Bot. 2020, 71, 4591–4603. [Google Scholar] [CrossRef]
- Beatrice, B.; Susanne, B.; Silke, R. The plant growth-promoting bacterium Kosakonia radicincitans improves fruit yield and quality of Solanum lycopersicum. J. Sci. Food Agric. 2017, 97, 4865–4871. [Google Scholar]
- Ma, T.; Ji, K.; Wang, W.; Wang, J.; Li, Z.; Ran, H.; Liu, B.; Li, G. Cellulose synthesized by Enterobacter sp. FY-07 under aerobic and anaerobic conditions. Bioresour. Technol. 2012, 126, 18–23. [Google Scholar] [CrossRef]
- Gao, G.; Cao, Y.; Zhang, Y.; Wu, M.; Ma, T.; Li, G. In situ production of bacterial cellulose/xanthan gum nanocomposites with enhanced productivity and properties using Enterobacter sp. FY-07. Carbohydr. Polym. 2020, 248, 116788. [Google Scholar] [CrossRef]
- Gao, G.; Ji, K.; Zhang, Y.; Liu, X.; Dai, X.; Zhi, B.; Cao, Y.; Liu, D.; Wu, M.; Li, G.; et al. Microbial enhanced oil recovery through deep profile control using a conditional bacterial cellulose-producing strain derived from Enterobacter sp. FY-07. Microb. Cell Fact. 2020, 19, 59. [Google Scholar] [CrossRef]
- Gao, G.; Fan, H.; Zhang, Y.; Cao, Y.; Li, T.; Qiao, W.; Wu, M.; Ma, T.; Li, G. Production of nisin-containing bacterial cellulose nanomaterials with antimicrobial properties through co-culturing Enterobacter sp. FY-07 and Lactococcus lactis N8. Carbohydr. Polym. 2021, 251, 117131. [Google Scholar] [CrossRef]
- Gao, G.; Liao, Z.T.; Cao, Y.Y.; Zhang, Y.B.; Zhang, Y.; Wu, M.M.; Li, G.Q.; Ma, T. Highly efficient production of bacterial cellulose from corn stover total hydrolysate by Enterobacter sp. FY-07. Bioresour. Technol. 2021, 341, 1225781. [Google Scholar] [CrossRef]
- Becker, M.; Patz, S.; Becker, Y.; Berger, B.; Drungowski, M.; Bunk, B.; Overmann, J.; Spröer, C.; Reetz, J.; Tchakounte, G.; et al. Comparative Genomics Reveal a Flagellar System, a Type VI Secretion System and Plant Growth-Promoting Gene Clusters Unique to the Endophytic Bacterium Kosakonia radicincitans. Front. Microbiol. 2018, 9, 1997. [Google Scholar] [CrossRef]
- Anwar, N.; Ye, Y.; Chen, C.; Tursunay, M.; Ruijun, W.; Erkin, R.; Wu, M. Nesterenkonia ebinurensis sp. nov., a Novel Actinobacterium Isolated from Populus euphratica. Curr. Microbiol. 2021, 78, 3804–3810. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Göker, M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 2019, 10, 2182. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.P.; Göker, M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef]
- Grant, J.R.; Arantes, A.S.; Stothard, P. Comparing thousands of circular genomes using the CGView Comparison Tool. BMC Genom. 2012, 13, 202. [Google Scholar] [CrossRef]
- Minkin, I.; Patel, A.; Kolmogorov, M.; Vyahhi, N.; Pham, S. Sibelia: A scalable and comprehensive synteny block generation tool for closely related microbial genomes. In International Workshop on Algorithms in Bioinformatics; Springer: Berlin, Germany, 2013; pp. 215–229. [Google Scholar]
- Rahman, A.; Sitepu, I.R.; Tang, S.-Y.; Hashidoko, Y. Salkowski’s reagent test as a primary screening index for functionalities of rhizobacteria isolated from wild dipterocarp saplings growing naturally on medium-strongly acidic tropical peat soil. Biosci. Biotech. Biochem. 2010, 74, 2202–2208. [Google Scholar] [CrossRef]
- Asghar, H.N.; Zahir, Z.A.; Abdul, K.; Muhammad, A. Isolation of rhizobacteria from different rapeseed varieties and their potential for auxin production. Pak. J. Biol. Sci. 2000, 3, 1556–1559. [Google Scholar] [CrossRef][Green Version]
- Schwyn, B.; Neilands, J.B. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 1987, 160, 47–56. [Google Scholar] [CrossRef]
- Panneerselvam, P.; Senapati, A.; Kumar, U.; Sharma, L.; Sivakumar, U. Antagonistic and plant-growth promoting novel bacillus species from long-term organic farming soils from sikkim, india. 3 Biotech 2019, 9, 416. [Google Scholar] [CrossRef]
- Ames, B.N. Assay of inorganic phosphate, total phosphate and phosphatases. Methods Enzymol. 1966, 8, 115–118. [Google Scholar]
- Jimtha, J.C.; Smitha, P.V.; Anisha, C.; Deepthi, T.; Meekha, G.; Radhakrishnan, E.K.; Gayatri, G.P.; Remakanthan, A. Isolation of endophytic bacteria from embryogenic suspension culture of banana and assessment of their plant growth promoting properties. Plant Cell Tiss. Organ Cult. 2014, 118, 57–66. [Google Scholar] [CrossRef]
- Jensen, H.L. Nitrogen fixation in leguminous plants. II. Is symbiotic nitrogen fixation influenced by Azotobacter? Proc. Linn. Soc. N. S. W. 1942, 67, 205–212. [Google Scholar]
- Cappuccino, J.C.; Sherman, N. Microbiology, Laboratory Manual; Benjamin/Cumming Pub. Co.: New York, NY, USA, 1992. [Google Scholar]
- Chrouqi, L.; Ouahmane, L.; Jadrane, I.; Koussa, T.; Feddy, M. Screening of soil rhizobacteria isolated from wheat plants grown in the marrakech region (morocco, north africa) for plant growth promoting activities. J. Environ. Sci. 2017, 8, 3382–3390. [Google Scholar]
- Dworkin, M.; Foster, J.W. Experiments with some microorganisms which utilize ethane and hydrogen. J. Bacteriol. 1958, 75, 592–603. [Google Scholar] [CrossRef]
- Huang, R.; Feng, H.; Xu, Z.; Zhang, N.; Liu, Y.; Shao, J.; Shen, Q.; Zhang, R. Identification of Adhesins in Plant Beneficial Rhizobacteria Bacillus velezensis SQR9 and Their Effect on Root Colonization. Mol. Plant Microbe Interact. 2022, 35, 64–72. [Google Scholar] [CrossRef] [PubMed]
- Goris, J.; Konstantinidis, K.T.; Klappenbach, J.A.; Coenye, T.; Vandamme, P.; Tiedje, J.M. DNA–DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 2007, 57, 81–91. [Google Scholar] [CrossRef] [PubMed]
- Ma, T.; Zhao, Q.; Ji, K.; Zeng, B.; Li, G. Homogeneous and porous modified bacterial cellulose achieved by in situ modification with low amounts of carboxymethyl cellulose. Cellulose 2014, 21, 2637–2646. [Google Scholar] [CrossRef]
- Ji, K.; Wang, W.; Zeng, B.; Chen, S.; Zhao, Q.; Chen, Y.; Li, G.; Ma, T. Bacterial cellulose synthesis mechanism of facultative anaerobe Enterobacter sp. FY-07. Sci. Rep. 2016, 6, 21863. [Google Scholar] [CrossRef]
- Liu, D.; Cao, Y.; Qu, R.; Gao, G.; Chen, S.; Zhang, Y.; Wu, M.; Ma, T.; Li, G. Production of bacterial cellulose hydrogels with tailored crystallinity from Enterobacter sp. FY-07 by the controlled expression of colanic acid synthetic genes. Carbohydr. Polym. 2019, 207, 563–570. [Google Scholar] [CrossRef]
- Hardoim, P.R.; Nazir, R.; Sessitsch, A.; Elhottová, D.; Korenblum, E.; van Overbeek, L.S.; Elsas, J.D. The new species Enterobacter oryziphilus sp. nov. and Enterobacter oryzendophyticus sp. nov. are key inhabitants of the endosphere of rice. BMC Microbiol. 2013, 13, 164. [Google Scholar] [CrossRef]
- Sherpa, M.T.; Sharma, L.; Bag, N.; Das, S. Isolation, Characterization, and Evaluation of Native Rhizobacterial Consortia Developed from the Rhizosphere of Rice Grown in Organic State Sikkim, India, and Their Effect on Plant Growth. Front. Microbiol. 2021, 12, 713660. [Google Scholar] [CrossRef]
- Matsumoto, H.; Fan, X.; Wang, Y.; Kusstatscher, P.; Cernava, T.; Wang, M. Bacterial seed endophyte shapes disease resistance in rice. Nat. Plants 2021, 7, 60–72. [Google Scholar] [CrossRef]
- Brock, A.K.; Berger, B.; Schreiner, M.; Ruppel, S.; Mewis, I. Plant growth-promoting bacteria Kosakonia radicincitans mediate anti-herbivore defense in Arabidopsis thaliana. Planta 2018, 248, 1383–1392. [Google Scholar] [CrossRef]
- Nag, P.; Shriti, S.; Das, S. Microbiological strategies for enhancing biological nitrogen fixation in nonlegumes. J. Appl. Microbiol. 2019, 129, 186–198. [Google Scholar] [CrossRef]
- Li, Y.; Li, Y.; Zhang, H.; Wang, M.; Chen, S. Diazotrophic Paenibacillus beijingensis BJ-18 Provides Nitrogen for Plant and Promotes Plant Growth, Nitrogen Uptake and Metabolism. Front. Microbiol. 2019, 10, 1119. [Google Scholar] [CrossRef]
- Carvalho, T.L.G.; Balsemão-Pires, E.; Saraiva, R.M.; Ferreira, P.C.G.; Hemerly, A.S. Nitrogen signalling in plant interactions with associative and endophytic diazotrophic bacteria. J. Exp. Bot. 2014, 65, 5631–5642. [Google Scholar] [CrossRef]

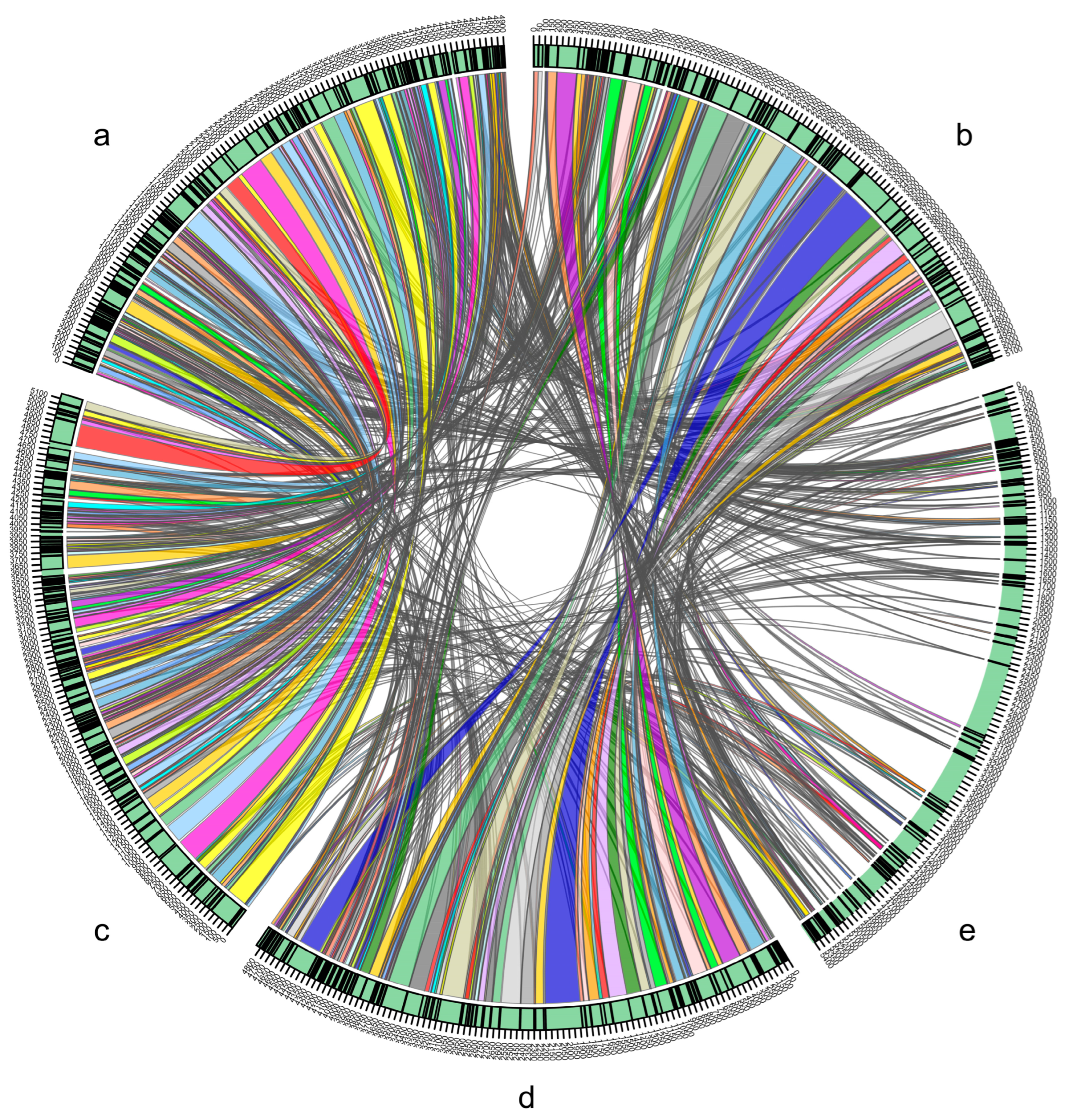

| = ANI ≥ 95% or isDDH ≥ 70% | Enterobacter sp. FY-07 | Kosakonia oryzendophytica LMG 26432T | Kosakonia sacchari LMG 26783T | Kosakonia pseudosacchari JM-387T | Kosakonia oryzae LMG 24251T | Kosakonia quasisacchari WCHEs 120001T | Kosakonia radicincitans LMG 23767T | Kosakonia oryziphila LMG 26429T | Kosakonia arachidis KCTC 22375T |
|---|---|---|---|---|---|---|---|---|---|
| Enterobacter sp. FY-07 | 96.10 | 26.50 | 26.40 | 26.40 | 26.10 | 26.50 | 25.60 | 25.30 | |
| Kosakonia oryzendophytica LMG 26432T | 99.45 | 26.20 | 26.30 | 26.20 | 26.10 | 26.20 | 25.60 | 25.30 | |
| Kosakonia sacchari LMG 26783T | 84.13 | 84.21 | 59.10 | 28.00 | 50.90 | 27.80 | 26.70 | 26.60 | |
| Kosakonia pseudosacchari JM-387T | 84.07 | 84.08 | 94.94 | 27.80 | 48.60 | 27.70 | 27.00 | 26.50 | |
| Kosakonia oryzae LMG 24251T | 83.89 | 83.89 | 84.95 | 84.74 | 27.40 | 66.40 | 53.10 | 51.30 | |
| Kosakonia quasisacchari WCHEs 120001T | 83.83 | 83.90 | 93.29 | 92.71 | 84.45 | 27.40 | 26.60 | 26.40 | |
| Kosakonia radicincitans LMG 23767T | 83.78 | 83.78 | 84.66 | 84.52 | 95.85 | 84.36 | 54.30 | 52.40 | |
| Kosakonia oryziphila LMG 26429T | 83.08 | 83.15 | 83.99 | 84.13 | 92.92 | 83.80 | 93.04 | 54.80 | |
| Kosakonia arachidis KCTC 22375T | 82.78 | 82.99 | 83.86 | 83.80 | 92.62 | 83.69 | 92.80 | 93.36 | |
| Kosakonia cowanii LMG 23569T | 82.37 | 82.48 | 83.66 | 83.71 | 83.17 | 83.27 | 82.98 | 82.39 | 82.10 |
| Characteristic | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
|---|---|---|---|---|---|---|---|---|---|---|
| β-Galactosidase | + | + | + | + | + | + | + | + | + | + |
| Arginine dihydrolase | + | + | − | + | + | + | + | + | + | + |
| Lysine decarboxylase | − | − | − | − | − | − | − | − | − | − |
| Ornithine decarboxylase | + | + | + | − | + | − | − | − | + | − |
| Citrate utilization | + | + | + | + | + | + | + | + | + | + |
| H2S production | − | − | − | − | − | − | − | − | − | − |
| Urea hydrolysis | − | − | − | − | − | − | − | − | − | − |
| Indole production | − | − | − | − | − | − | − | − | − | − |
| Voges-Proskauer reaction | + | + | + | + | + | + | + | + | + | + |
| Gelatinase | − | − | − | − | − | − | − | − | − | − |
| d-Glucose | + | + | + | + | + | + | + | + | + | + |
| d-Mannitol | + | + | + | + | + | + | + | + | + | + |
| Inositol | − | + | − | − | − | − | − | + | + | − |
| d-Sorbitol | + | + | + | + | + | + | + | + | + | + |
| Sucrose | + | + | + | + | + | + | + | + | + | + |
| Melibiose | + | + | + | − | + | − | − | − | − | + |
| Traits | Genes |
|---|---|
| Nitrogen fixation and metabolism | amtB, narB, narG, narH, narI, narJ, nark, narL, narX, nasD, nifA, nifB, nifD, nifE, nifF, nifH, nifI, nifJ, nifK, nifL, nifK, nifL, nifM, nifN, nifQ, nifS, nifT, nifU, nifV, nifW, nifX, nifZ, nirB, nirD, ntrA, ntrB, ntrC |
| Phosphate solubilization | phoA, phoB, phoC, phoE, phoH, phoR, phoU, pqqE-like, pstA, pstB, pstC, pstS |
| Plant hormone synthesis | ipdC, gabD, puuE |
| Siderophore production | acrB, efeB, efeO, efeU, entA, entB, entC, entE, entF, entH, entS, exbB, exbD, feoA, feoB, feoC, fepA, fepB, fepC, fepD, fepE, fepG, fes, fhuB, fhuC, fhuE, fhuF, fptA, mbtH-like, mdtA, mdtB, mdtC, mexA, mexB, tonB, yusV |
| ACC deaminase production | dcyD |
| Betaine production | opuA, opuB, opuC, osmY, proV, proW, proX |
| Acetoin and butanodiol sythesis | als, budA, budB, budC, poxB |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, G.; Zhang, Y.; Niu, S.; Chen, Y.; Wang, S.; Anwar, N.; Chen, S.; Li, G.; Ma, T. Reclassification of Enterobacter sp. FY-07 as Kosakonia oryzendophytica FY-07 and Its Potential to Promote Plant Growth. Microorganisms 2022, 10, 575. https://doi.org/10.3390/microorganisms10030575
Gao G, Zhang Y, Niu S, Chen Y, Wang S, Anwar N, Chen S, Li G, Ma T. Reclassification of Enterobacter sp. FY-07 as Kosakonia oryzendophytica FY-07 and Its Potential to Promote Plant Growth. Microorganisms. 2022; 10(3):575. https://doi.org/10.3390/microorganisms10030575
Chicago/Turabian StyleGao, Ge, Yan Zhang, Shaofang Niu, Yu Chen, Shaojing Wang, Nusratgul Anwar, Shuai Chen, Guoqiang Li, and Ting Ma. 2022. "Reclassification of Enterobacter sp. FY-07 as Kosakonia oryzendophytica FY-07 and Its Potential to Promote Plant Growth" Microorganisms 10, no. 3: 575. https://doi.org/10.3390/microorganisms10030575
APA StyleGao, G., Zhang, Y., Niu, S., Chen, Y., Wang, S., Anwar, N., Chen, S., Li, G., & Ma, T. (2022). Reclassification of Enterobacter sp. FY-07 as Kosakonia oryzendophytica FY-07 and Its Potential to Promote Plant Growth. Microorganisms, 10(3), 575. https://doi.org/10.3390/microorganisms10030575






