The Effects of Freeze-Thaw and UVC Radiation on Microbial Survivability in a Selected Mars-like Environment
Abstract
:1. Introduction
2. Materials and Methods
2.1. Test Micro-Organisms and Culture Preparation
2.2. Freeze-Thaw Treatment
2.3. UV Treatment
2.4. Freezing Treatment Combined with UV Treatment
2.5. Data Analysis
3. Results
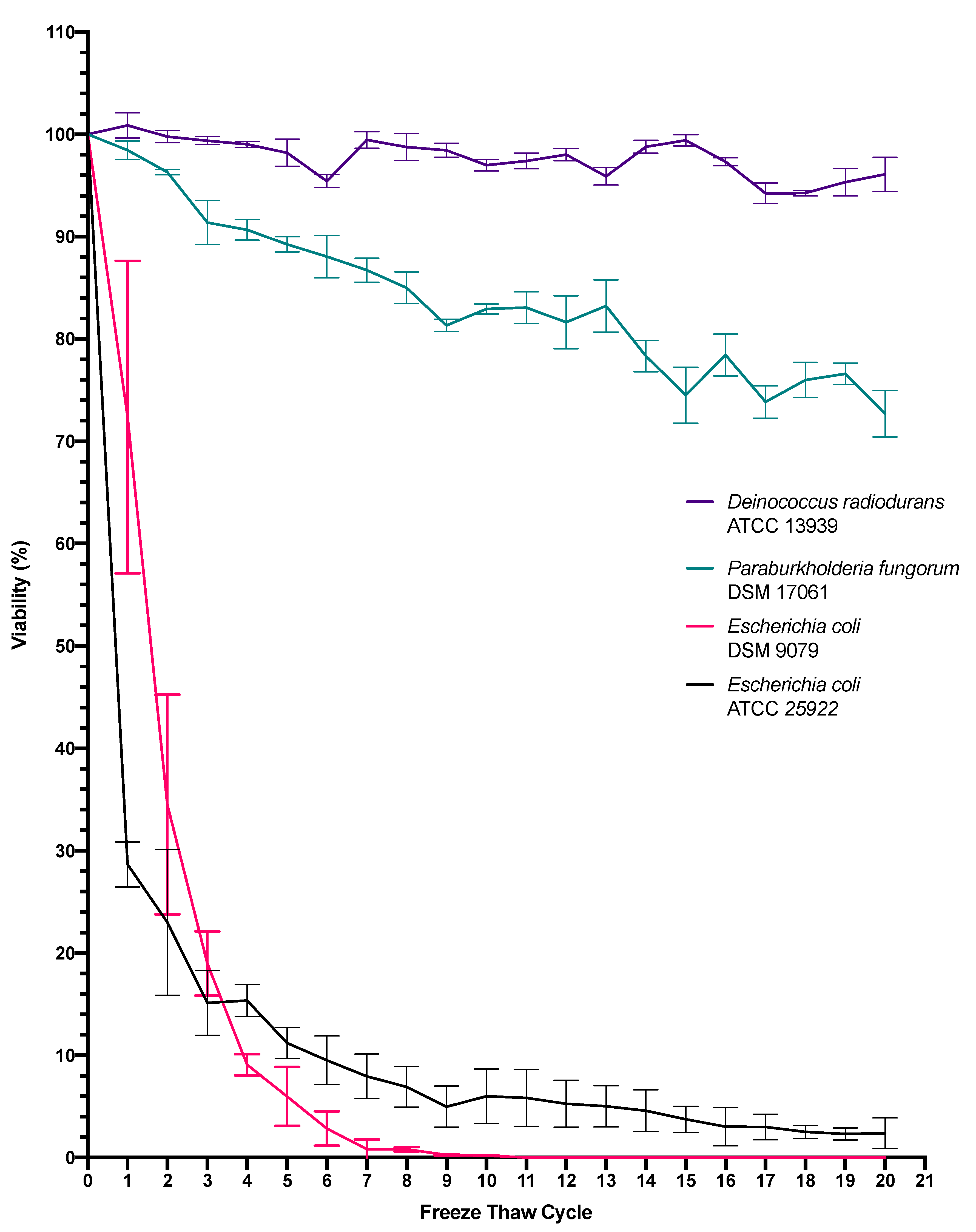
3.1. The Effect of Freeze-Thawing on Bacterial Viability
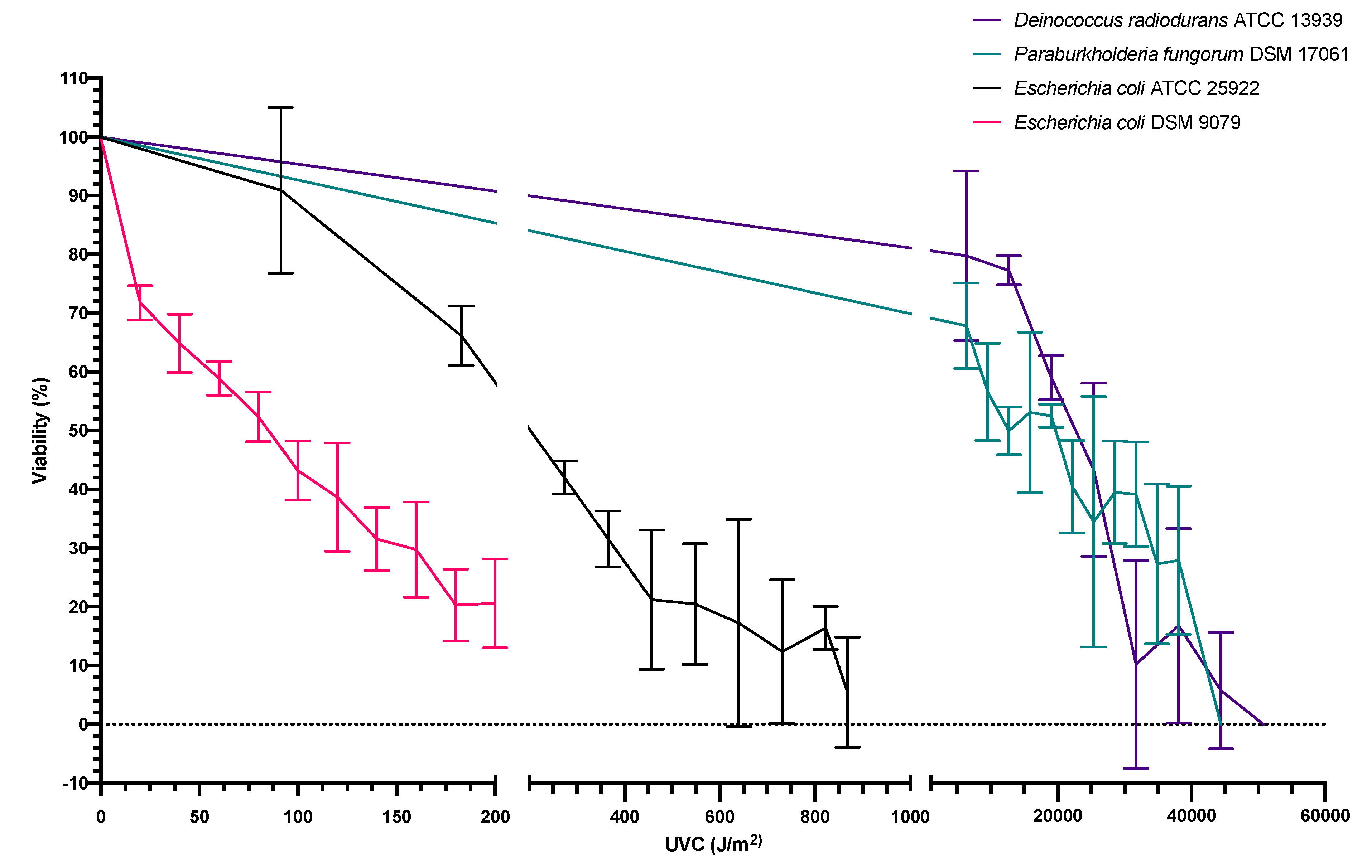
3.2. Dose Response of Bacterial Viability following UVC Exposure
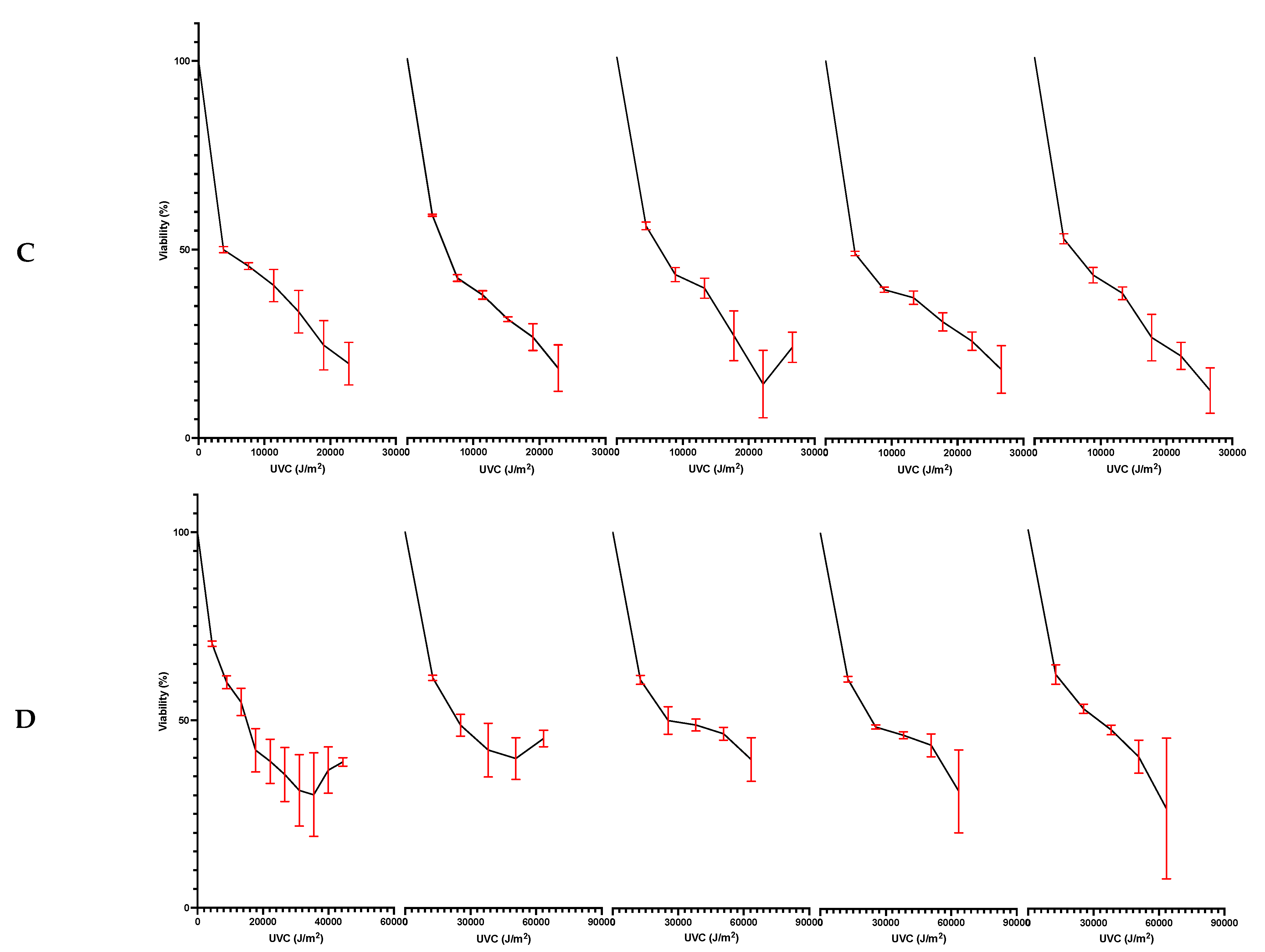
3.3. Bacterial Viability following Dual Exposure to Freeze-Thawing and UVC Radiation
4. Discussion
4.1. Freeze-Thaw Cycle Exposure
4.2. Molecular Influences Associated with Freeze-Thaw Cycle Exposure
4.3. UV Exposure
4.4. Molecular Influences Associated with UV Exposure
4.5. Combined Freeze-Thaw and UV Radiation
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sinton, W.M. Taking the temperatures of the moon and planets. Leafl. Astron. Soc. Pac. 1958, 7, 361. [Google Scholar]
- Moissl-Eichinger, C.; Cockell, C.; Rettberg, P. Venturing into new realms? Microorganisms in spacea. FEMS Microbiol. Rev. 2016, 40, 722–737. [Google Scholar] [CrossRef]
- NASA. Mars Facts|All About Mars—NASA’s Mars Exploration Program. 2020. Available online: NASA.gov (accessed on 13 April 2020).
- Catling, D.C.; Cockell, C.S.; Mckay, C.P. Ultraviolet radiation on the surface of mars. Orig. Life Evol. Biosph. 2000, 30, 467–500. [Google Scholar]
- Cockell, C.S.; Catling, D.C.; Davis, W.L.; Snook, K.; Kepner, R.L.; Lee, P.; McKay, C.P. The Ultraviolet Environment of Mars: Biological Implications Past, Present, and Future. Icarus 2000, 146, 343–359. [Google Scholar] [CrossRef] [Green Version]
- Pattison, D.I.; Davies, M.J. Actions of Ultraviolet Light on Cellular Structures. In Cancer: Cell Structures, Carcinogens and Genomic Instability; Birkhäuser-Verlag: Basel, Switzerland, 2006; pp. 131–157. [Google Scholar]
- Peplow, M. How Mars got its rust. Nature 2004. [Google Scholar] [CrossRef]
- Hecht, M.; Kounaves, S.; Quinn, R.; West, S.; Young, S.; Ming, D.; Catling, D.; Clark, B.; Boynton, W.; Hoffman, J.; et al. Detection of perchlorate and the soluble chemistry of martian soil at the Phoenix lander site. Science 2009, 325, 64–67. [Google Scholar] [CrossRef] [Green Version]
- Davila, A.F.; Willson, D.; Coates, J.D.; McKay, C.P.; Davila, A.F.; Willson, D.; Coates, J.D.; McKay, C.P. Perchlorate on Mars: A chemical hazard and a resource for humans. Int. J. Astrobiol. 2013, 12, 321–325. [Google Scholar] [CrossRef]
- Srinivasan, A.; Viraraghavan, T. Perchlorate: Health effects and technologies for its removal from water resources. Int. J. Environ. Res. Public Health 2009, 6, 1418–1442. [Google Scholar] [CrossRef]
- Haynes, R.H.; McKay, C.P. The implantation of life on Mars: Feasibility and motivation. Adv. Space Res. 1992, 12, 133–140. [Google Scholar] [CrossRef]
- Maeda, T.; Horinouchi, T.; Sakata, N.; Sakai, A.; Furusawa, C. High-throughput identification of the sensitivities of an Escherichia coli ΔrecA mutant strain to various chemical compounds. J. Antibiot. 2019, 72, 566–573. [Google Scholar] [CrossRef]
- Selbitschka, W.; Arnold, W.; Priefer, U.B.; Rottschäfer, T.; Schmidt, M.; Simon, R.; Pühler, A. Characterization of recA genes and recA mutants of Rhizobium meliloti and Rhizobium leguminosarum biovar viciae. Mol. Gen. Genet. 1991, 229, 86–95. [Google Scholar] [CrossRef]
- Oteiza, J.M.; Caturla, M.Y.R.; do Prado-Silva, L.; Câmara, A.A.; Barril, P.A.; Sant’Ana, A.S.; Giannuzzi, L.; Zaritzky, N. Adaptation of O157:H7 and non-O157 Escherichia coli strains in orange juice and subsequent resistance to UV-C radiation. LWT 2022, 157, 113107. [Google Scholar] [CrossRef]
- Gao, W.; Leung, K. Sensitivity of a Shiga Toxin-Producing and an Uropathogenic Escherichia coli to UV Irradiation after Freeze-Thaw. Water Air Soil Pollut. 2015, 226, 298. [Google Scholar] [CrossRef]
- Harsojo Kityama, S.; Matsuyama, A. Genome Multiplicity and Radiation Resistance in Micrococcus radiodurans1. J. Biochem. 1981, 90, 877–880. [Google Scholar] [CrossRef]
- Hansen, M.T. Multiplicity of genome equivalents in the radiation-resistant bacterium Micrococcus radiodurans. J. Bacteriol. 1978, 134, 71–75. [Google Scholar] [CrossRef] [Green Version]
- Daly, M.J.; Minton, K.W. Interchromosomal recombination in the extremely radioresistant bacterium Deinococcus radiodurans. J. Bacteriol. 1995, 177, 5495–5505. [Google Scholar] [CrossRef] [Green Version]
- Brim, H.; McFarlan, S.C.; Fredrickson, J.K.; Minton, K.W.; Zhai, M.; Wackett, L.P.; Daly, M.J. Engineering Deinococcus radiodurans for metal remediation in radioactive mixed waste environments. Nat. Biotechnol. 2000, 18, 85–90. [Google Scholar] [CrossRef]
- Choi, M.H.; Jeong, S.-W.; Shim, H.E.; Yun, S.-J.; Mushtaq, S.; Choi, D.S.; Jang, B.-S.; Yang, J.E.; Choi, Y.J.; Jeon, J. Efficient bioremediation of radioactive iodine using biogenic gold nanomaterial-containing radiation-resistant bacterium, Deinococcus radiodurans R1. Chem. Commun. 2017, 53, 3937–3940. [Google Scholar] [CrossRef]
- Al-Taee, A. Biosorption of some Heavy Metals by Deinococcus radiodurans Isolated from Soil in Basra Governorate-Iraq. J. Bioremediation Biodegrad. 2016, 7, 2. [Google Scholar] [CrossRef] [Green Version]
- Andreolli, M.; Lampis, S.; Zenaro, E.; Salkinoja-Salonen, M.; Vallini, G. Burkholderia fungorum DBT1: A promising bacterial strain for bioremediation of PAHs-contaminated soils. FEMS Microbiol. Lett. 2011, 319, 11–18. [Google Scholar] [CrossRef]
- Coohill, T.P.; Sagripanti, J.-L. Overview of the Inactivation by 254nm Ultraviolet Radiation of Bacteria with Particular Relevance to Biodefense. Photochem. Photobiol. 2008, 84, 1084–1090. [Google Scholar] [CrossRef]
- Gao Critser, J. Mechanisms of cryoinjury in living cells. ILAR J. 2000, 41, 187–196. [Google Scholar]
- Cox, C.; Heckly, R. Effects of oxygen upon freeze-dried and freeze-thawed bacteria: Viability and free radical studies. Can J. Microbiol. 1973, 19, 189–194. [Google Scholar] [CrossRef]
- Packer, E.L.; Ingraham, J.L.; Scher, S. Factors Affecting the Rate of Killing of Escherichia coli by Repeated Freezing and Thawing. J. Bacferiol. 1965, 89, 718–724. [Google Scholar] [CrossRef] [Green Version]
- Sawicka, J.E.; Robador, A.; Hubert, C.; Jørgensen, B.B.; Brüchert, V. Effects of freeze–thaw cycles on anaerobic microbial processes in an Arctic intertidal mud flat. ISME J. 2010, 4, 585–594. [Google Scholar] [CrossRef] [Green Version]
- Alur, M.D.; Grecz, N. Mechanism of injury of Escherichiacoli by freezing and thawing. Biochem. Biophys. Res. Commun. 1975, 62, 308–312. [Google Scholar] [CrossRef]
- Lyscov, V.N.; Moshkovsky, Y.S. DNA cryolysis. Biochim. Biophys. Acta-Nucleic Acids Protein Synth. 1969, 190, 101–110. [Google Scholar] [CrossRef]
- Shao, W.; Khin, S.; Kopp, W.C. Characterization of Effect of Repeated Freeze and Thaw Cycles on Stability of Genomic DNA Using Pulsed Field Gel Electrophoresis. Biopreserv Biobank 2012, 10, 4–11. [Google Scholar] [CrossRef]
- Sleight, S.C.; Wigginton, N.S.; Lenski, R.E. Increased susceptibility to repeated freeze-thaw cycles in Escherichia coli following long-term evolution in a benign environment. BMC Evol. Biol. 2006, 6, 104. [Google Scholar] [CrossRef] [Green Version]
- Loong, S.K.; Tan, K.-K.; Zulkifle, N.-I.; AbuBakar, S. Draft genome of Paraburkholderia fungorum sequence type 868 recovered from human synovial tissues. Data Br. 2019, 25, 104159. [Google Scholar] [CrossRef]
- Minogue, T.D.; Daligault, H.A.; Davenport, K.W.; Bishop-Lilly, K.A.; Broomall, S.M.; Bruce, D.C.; Chain, P.S.; Chertkov, O.; Coyne, S.R.; Freitas, T.; et al. Complete Genome Assembly of Escherichia coli ATCC 25922, a Serotype O6 Reference Strain. Genome Announc. 2014, 2, e00969-14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yeager, C.M.; Bottomley, P.J.; Arp, D.J. Requirement of DNA Repair Mechanisms for Survival of Burkholderia cepacia G4 upon Degradation of Trichloroethylene. Appl. Environ. Microbiol. 2001, 67, 5384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Slade, D.; Lindner, A.B.; Paul, G.; Radman, M. Recombination and Replication in DNA Repair of Heavily Irradiated Deinococcus radiodurans. Cell 2009, 136, 1044–1055. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morley, C.R.; Trofymow, J.A.; Coleman, D.C.; Cambardella, C. Effects of freeze-thaw stress on bacterial populations in soil microcosms. Microb. Ecol. 1983, 9, 329–340. [Google Scholar] [CrossRef]
- Moolenaar, G.F.; Moorman, C.; Goosen, N. Role of the Escherichia coli nucleotide excision repair proteins in DNA replication. J. Bacteriol. 2000, 182, 5706–5714. [Google Scholar] [CrossRef] [Green Version]
- Guo, C.; Song, Y.; Wang, L.; Sun, L.; Sun, Y.; Peng, C.; Li, Z. Atomic Force Microscopic Study of Low Temperature Induced Disassembly of RecA−dsDNA Filaments. J. Phys. Chem. B 2008, 112, 1022–1027. [Google Scholar] [CrossRef]
- Nguyen, H.H.; de la Tour, C.B.; Toueille, M.; Vannier, F.; Sommer, S.; Servant, P. The essential histone-like protein HU plays a major role in Deinococcus radiodurans nucleoid compaction. Mol. Microbiol. 2009, 73, 240–252. [Google Scholar] [CrossRef]
- Dennehy, R.; Dignam, S.; Mccormack, S.; Romano, M.; Hou, Y.; Ardill, L.; Whelan, M.X.; Drulis-Kawa, Z.; Cróinín, T.Ó.; Valvano, M.A.; et al. The DNA mimic protein BCAS0292 is involved in the regulation of 1 virulence of Burkholderia cenocepacia. bioRxiv 2020, 2, 3. [Google Scholar] [CrossRef]
- Wynn-Williams, D.D. Ecological Aspects of Antarctic Microbiology; Springer: Boston, MA, USA, 1990; pp. 71–146. [Google Scholar]
- Wynn-Williams, D.D. Cold adaptation of microorganisms. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1990, 326, 595–611. [Google Scholar]
- Crowe, J.H.; Hoekstra, F.A.; Crowe, L.M. Anhydrobiosis. Annu. Rev. Physiol. 1992, 54, 579–599. [Google Scholar] [CrossRef]
- Hassan, N.; Rafiq, M.; Hayat, M.; Shah, A.A.; Hasan, F. Psychrophilic and psychrotrophic fungi: A comprehensive review. Rev. Environ. Sci. Bio. Technol. 2016, 15, 147–172. [Google Scholar] [CrossRef]
- Hassan, N.; Anesio, A.M.; Rafiq, M.; Holtvoeth, J.; Bull, I.; Haleem, A.; Shah, A.A.; Hasan, F. Temperature Driven Membrane Lipid Adaptation in Glacial Psychrophilic Bacteria. Front. Microbiol. 2020, 11, 824. [Google Scholar] [CrossRef] [PubMed]
- Delong, E.F.; Yayanos, A.A. Biochemical function and ecological significance of novel bacterial lipids in deep-sea procaryotes. Appl. Environ. Microbiol. 1986, 51, 730–737. [Google Scholar] [CrossRef] [Green Version]
- Keto-Timonen, R.; Hietala, N.; Palonen, E.; Hakakorpi, A.; Lindström, M.; Korkeala, H. Cold Shock Proteins: A Minireview with Special Emphasis on Csp-family of Enteropathogenic Yersinia. Front. Microbiol. 2016, 7, 1151. [Google Scholar] [CrossRef] [PubMed]
- Seyer, K.; Lessard, M.; Piette, G.; Lacroix, M.; Saucier, L. Escherichia coli heat shock protein DnaK: Production and consequences in terms of monitoring cooking. Appl. Environ. Microbiol. 2003, 69, 3231–3237. [Google Scholar] [CrossRef] [Green Version]
- Li, R.; Shi, Y.; Ling, B.; Cheng, T.; Huang, Z.; Wang, S. Thermo-tolerance and heat shock protein of Escherichia coli ATCC 25922 under thermal stress using test cell method. Emir. J. Food Agric. 2017, 29, 91. [Google Scholar] [CrossRef] [Green Version]
- Deming, J.W. Extremophiles: Cold Environments. In Encyclopedia of Microbiology; Elsevier: Amsterdam, The Netherlands, 2009; pp. 147–158. [Google Scholar]
- Jones, P.G.; VanBogelen, R.A.; Neidhardt, F.C. Induction of proteins in response to low temperature in Escherichia coli. J. Bacteriol. 1987, 169, 2092–2095. [Google Scholar] [CrossRef] [Green Version]
- Airo, A.; Chan, S.L.; Martinez, Z.; Platt, M.O.; Trent, J.D. Heat Shock and Cold Shock in Deinococcus radiodurans. Cell Biochem. Biophys. 2004, 40, 277–288. [Google Scholar] [CrossRef]
- Woh Choo, S.; Yun Tan, K.; Dutta, A.; King Tan, T.; Hari, R.; Othman, R.Y. Comprehensive genome analysis of a pangolin-associated Paraburkholderia fungorum provides new insights into its secretion systems and virulence. PeerJ 2020, 8, e9733. [Google Scholar] [CrossRef]
- Ryan, R.P.; An, S.; Allan, J.H.; McCarthy, Y.; Dow, J.M. The DSF Family of Cell–Cell Signals: An Expanding Class of Bacterial Virulence Regulators. PLoS Pathog. 2015, 11, e1004986. [Google Scholar] [CrossRef] [PubMed]
- Janßen, H.; Steinbüchel, A. Fatty acid synthesis in Escherichia coli and its applications towards the production of fatty acid based biofuels. Biotechnol. Biofuels 2014, 7, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Slade, D.; Radman, M. Oxidative stress resistance in Deinococcus radiodurans. Microbiol. Mol. Biol. Rev. 2011, 75, 133–191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hazel, J.R.; Eugene Williams, E. The role of alterations in membrane lipid composition in enabling physiological adaptation of organisms to their physical environment. Prog. Lipid Res. 1990, 29, 167–227. [Google Scholar] [CrossRef]
- Suutari, M.; Laakso, S. Microbial Fatty Acids and Thermal Adaptation. Crit. Rev. Microbiol. 1994, 20, 285–328. [Google Scholar] [CrossRef]
- Fulco, A.J. Fatty acid metabolism in bacteria. Prog. Lipid Res. 1983, 22, 133–160. [Google Scholar] [CrossRef]
- Oguma, K.; Katayama, H.; Ohgaki, S. Photoreactivation of Escherichia coli after low- or medium-pressure UV disinfection determined by an endonuclease sensitive site assay. Appl. Environ. Microbiol. 2002, 68, 6029–6035. [Google Scholar] [CrossRef] [Green Version]
- Hutchinson, F.; Fuller, W.; Mullins, L.J. IUPAB Biophysics Series sponsored by the International Union of Pure and Applied Biophysics Editors-Biological Effects of Ultraviolet Radiation Walter Harm Frontmatter More information. 1980. Available online: www.cambridge.org (accessed on 12 January 2021).
- Montelone, B.A. DNA Repair and Mutagenesis, 2nd ed.; Errol, C.F., Graham, C.W., Wolfram, S., Richard, D.W., Roger, A.S., Tom, E., Eds.; ASM Press: Washington, DC, USA, 2006; Volume 81, p. 273. ISBN 1-55581-319-4. [Google Scholar]
- Hitomi, K.; Okamoto, K.; Daiyasu, H.; Miyashita, H.; Iwai, S.; Toh, H.; Ishiura, M.; Todo, T. Bacterial cryptochrome and photolyase: Characterization of two photolyase-like genes of Synechocystis sp. PCC6803. Nucleic. Acids Res. 2000, 28, 2353–2362. [Google Scholar] [CrossRef] [Green Version]
- Paulino-Lima, I.G.; Fujishima, K.; Navarrete, J.U.; Galante, D.; Rodrigues, F.; Azua-Bustos, A.; Rothschild, L.J. Extremely high UV-C radiation resistant microorganisms from desert environments with different manganese concentrations. J. Photochem. Photobiol. B 2016, 163, 327–336. [Google Scholar] [CrossRef]
- Krisko, A.; Radman, M. Protein damage and death by radiation in Escherichia coli and Deinococcus radiodurans. Proc. Natl. Acad. Sci. USA 2010, 107, 14373–14377. [Google Scholar] [CrossRef] [Green Version]
- Sun, H.; Xu, G.; Zhan, H.; Chen, H.; Sun, Z.; Tian, B.; Hua, Y. Identification and evaluation of the role of the manganese efflux protein in Deinococcus radiodurans. BMC Microbiol. 2010, 10, 319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Si, M.; Zhao, C.; Burkinshaw, B.; Zhang, B.; Wei, D.; Wang, Y.; Dong, T.G.; Shen, X. Manganese scavenging and oxidative stress response mediated by type VI secretion system in Burkholderia thailandensis. Proc. Natl. Acad. Sci. USA 2017, 114, E2233–E2242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin, J.E.; Waters, L.S.; Storz, G.; Imlay, J.A. The Escherichia coli small protein MntS and exporter MntP optimize the intracellular concentration of manganese. PLoS Genet. 2015, 11, e1004977. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hancock, L.C.; Hassang, H.M. Regulation of the Manganese-containing Superoxide Dismutase Is Independent of the Inducible DNA Repair System in Escherichia coZi. J. Biol. Chem. 1985, 260, 12954–12956. [Google Scholar] [CrossRef]
- Chang, J.C.; Ossoff, S.F.; Lobe, D.C.; Dorfman, M.H.; Dumais, C.M.; Qualls, R.G.; Johnson, J.D. UV inactivation of pathogenic and indicator microorganisms. Appl. Environ. Microbiol. 1985, 49, 1361–1365. [Google Scholar] [CrossRef] [Green Version]
- Dai, T.; Vrahas, M.S.; Murray, C.K.; Hamblin, M.R. Ultraviolet C irradiation: An alternative antimicrobial approach to localized infections? Expert Rev. Anti. Infect. Ther. 2012, 10, 185. [Google Scholar] [CrossRef] [Green Version]
- Janissen, R.; Arens, M.M.A.; Vtyurina, N.N.; Rivai, Z.; Sunday, N.D.; Eslami-Mossallam, B.; Gritsenko, A.A.; Laan, L.; de Ridder, D.; Artsimovitch, I.; et al. Global DNA Compaction in Stationary-Phase Bacteria Does Not Affect Transcription. Cell 2018, 174, 1188–1199.e14. [Google Scholar] [CrossRef] [Green Version]
- Dantur, K.I.; Pizarro, R.A. Effect of growth phase on the Escherichia coli response to ultraviolet-A radiation: Influence of conditioned media, hydrogen peroxide and acetate. J. Photochem. Photobiol. B Biol. 2004, 75, 33–39. [Google Scholar] [CrossRef]
- Abedi-Moghaddam, N.; Bulić, A.; Henderson, L.; Lam, E. Survival of Escherichia coli to UV Irradiation during Exponential and Stationary Phases of Growth. J. Exp. Microbiol. Immunol. 2004, 5, 44–49. [Google Scholar]
- Rolfe, M.D.; Rice, C.J.; Lucchini, S.; Pin, C.; Thompson, A.; Cameron, A.D.S.; Alston, M.; Stringer, M.F.; Betts, R.P.; Baranyi, J.; et al. Lag phase is a distinct growth phase that prepares bacteria for exponential growth and involves transient metal accumulation. J. Bacteriol. 2012, 194, 686–701. [Google Scholar] [CrossRef] [Green Version]
- Holland, A.D.; Rothfuss, H.M.; Lidstrom, M.E. Development of a defined medium supporting rapid growth for Deinococcus radiodurans and analysis of metabolic capacities. Appl. Microbiol. Biotechnol. 2006, 72, 1074–1082. [Google Scholar] [CrossRef] [PubMed]
- Mckenney, D.; Allison, D.G. Effects of Growth Rate and Nutrient Limitation on Virulence Factor Production in Burkholderia cepacian. The influence of growth rate and oxygen availability on siderophore, protease, and lipase production in Burkholderia cepacia was assessed for cells grown in a chemostat under iron limitation. Whereas siderophore and protease production increased with growth rate and oxygen yet decreased under oxygen depletion, lipase production demonstrated the opposite trend. J. Bacteriol. 1995, 177, 4140–4143. [Google Scholar] [PubMed] [Green Version]
- Sezonov, G.; Joseleau-Petit, D.; D’Ari, R. Escherichia coli physiology in Luria-Bertani broth. J. Bacteriol. 2007, 189, 8746–8749. [Google Scholar] [CrossRef] [PubMed] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Keaney, D.; Lucey, B.; Quinn, N.; Finn, K. The Effects of Freeze-Thaw and UVC Radiation on Microbial Survivability in a Selected Mars-like Environment. Microorganisms 2022, 10, 576. https://doi.org/10.3390/microorganisms10030576
Keaney D, Lucey B, Quinn N, Finn K. The Effects of Freeze-Thaw and UVC Radiation on Microbial Survivability in a Selected Mars-like Environment. Microorganisms. 2022; 10(3):576. https://doi.org/10.3390/microorganisms10030576
Chicago/Turabian StyleKeaney, Daniel, Brigid Lucey, Noreen Quinn, and Karen Finn. 2022. "The Effects of Freeze-Thaw and UVC Radiation on Microbial Survivability in a Selected Mars-like Environment" Microorganisms 10, no. 3: 576. https://doi.org/10.3390/microorganisms10030576
APA StyleKeaney, D., Lucey, B., Quinn, N., & Finn, K. (2022). The Effects of Freeze-Thaw and UVC Radiation on Microbial Survivability in a Selected Mars-like Environment. Microorganisms, 10(3), 576. https://doi.org/10.3390/microorganisms10030576





