Revising the Freshwater Thelohania to Astathelohania gen. et comb. nov., and Description of Two New Species
Abstract
:1. Introduction
2. Materials and Methods
2.1. Crayfish Locality and Collection
2.2. Histopathology
2.3. Transmission Electron Microscopy
2.4. Molecular Diagnostics
2.5. Phylogenetics and Genetic Comparisons
3. Results
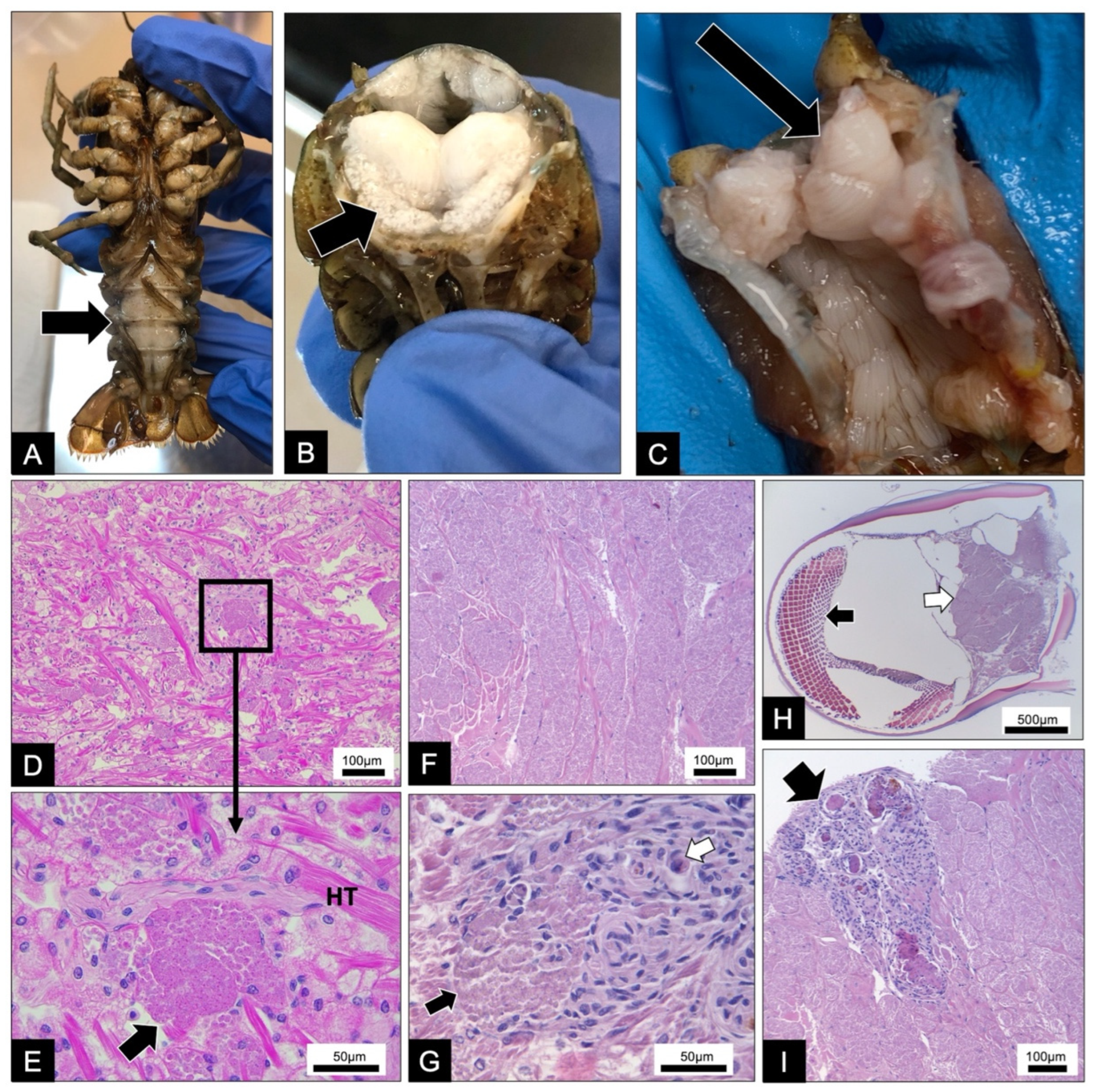
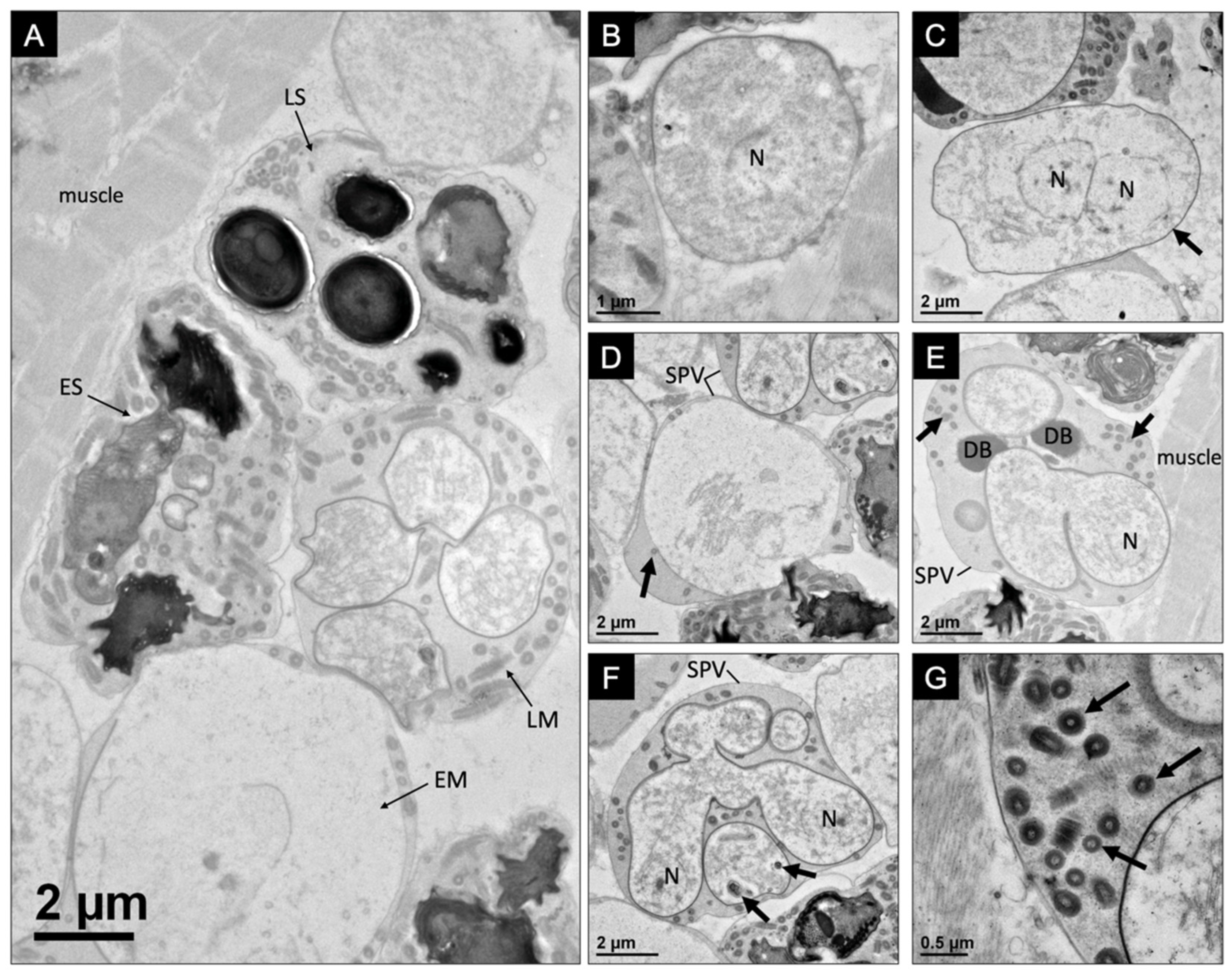
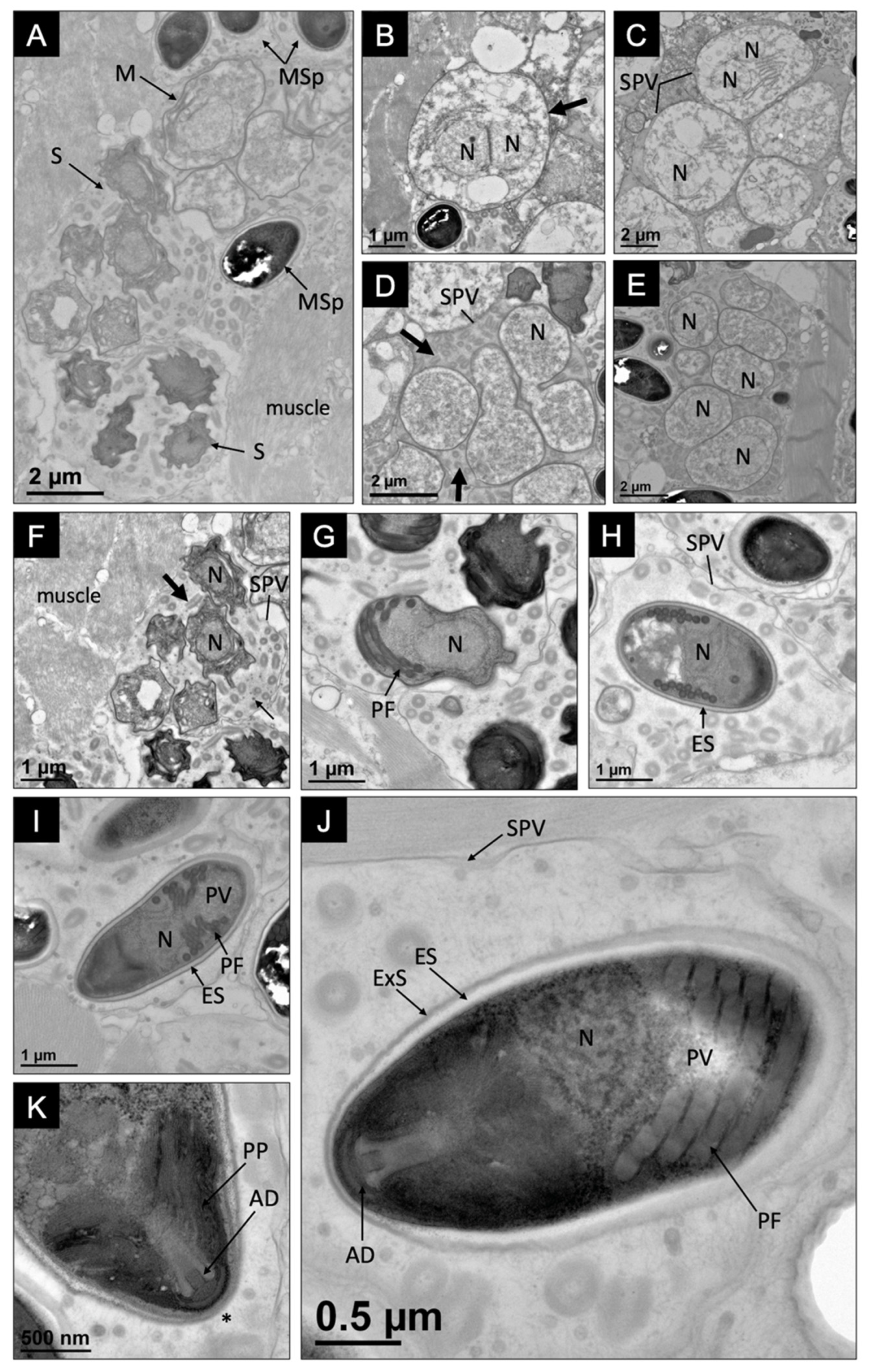
3.1. Pathology, Ultrastructure, and Development for Microsporidiosis in Faxonius virilis
3.2. Pathology, Ultrastructure, and Development for Microsporidiosis in Faxonius rusticus
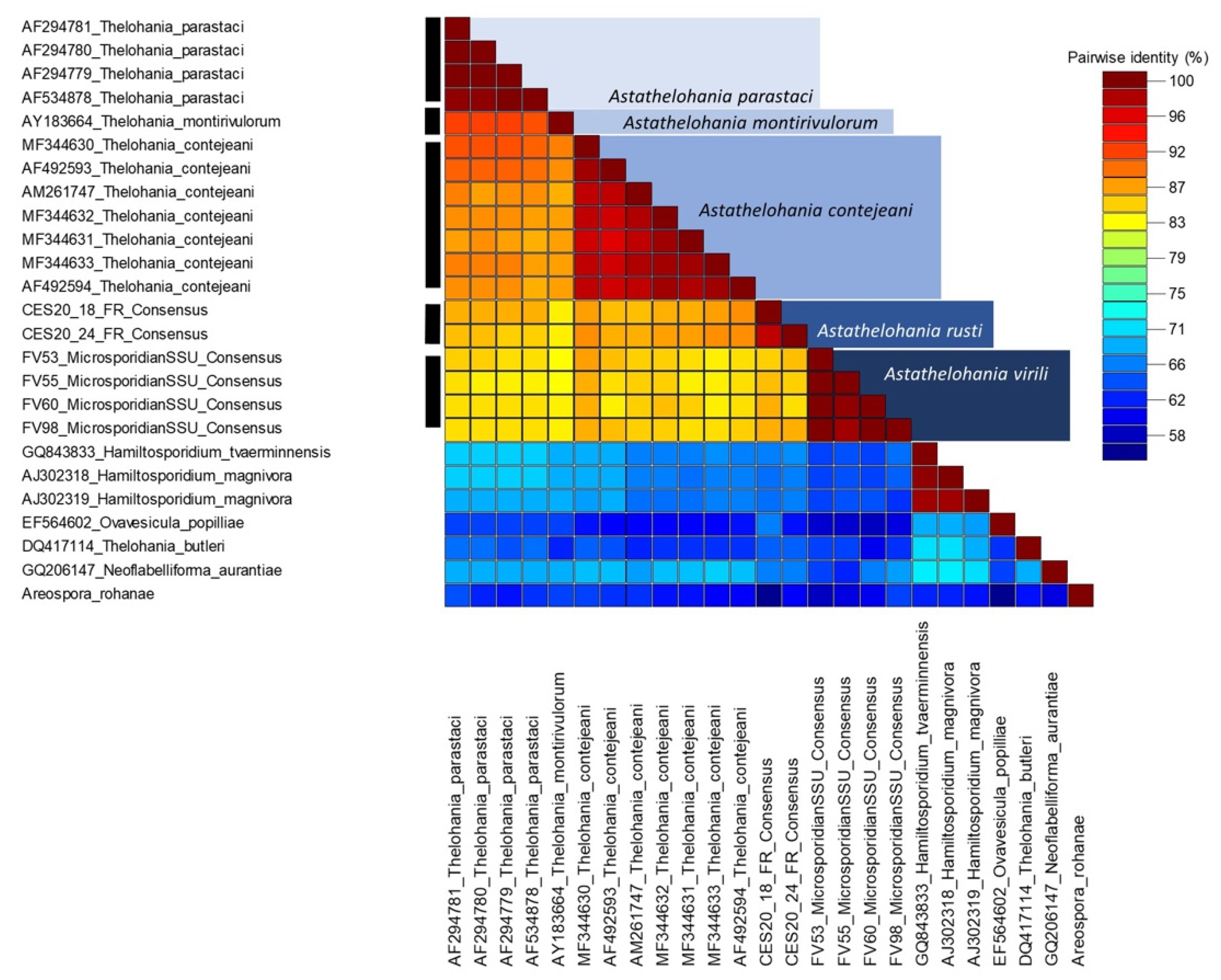
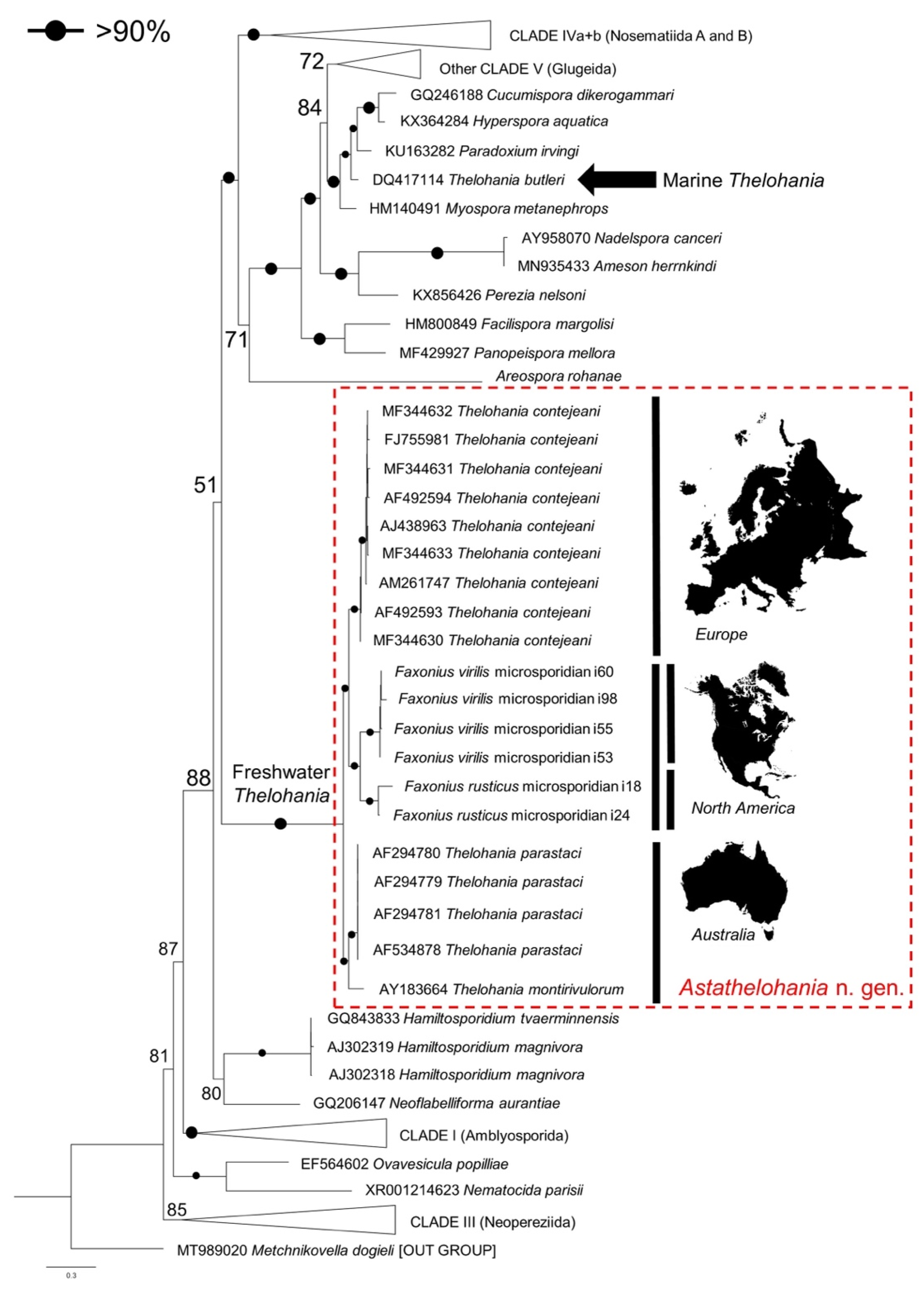
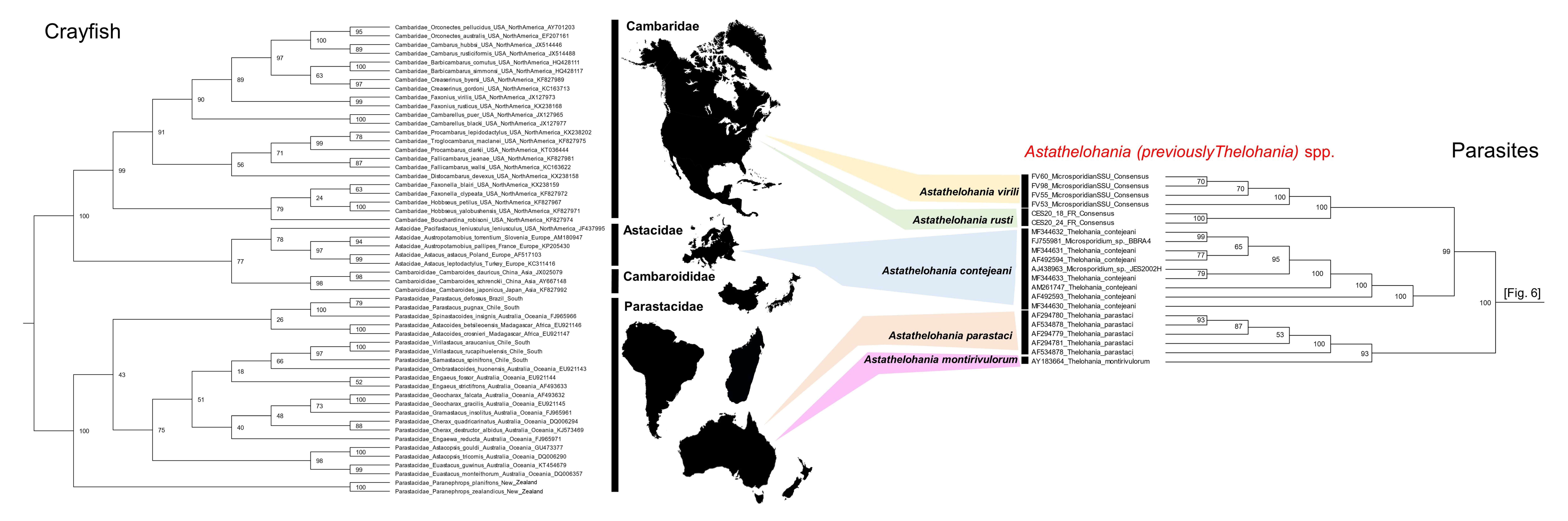
3.3. Genetic Similarity and Phylogenetic Placement of the Novel Microsporidians
4. Taxonomic Summary
4.1. Higher Taxonomy
4.2. Astathelohania virili n. sp. Stratton, Reisinger, Behringer, Bojko 2022
4.3. Astathelohania rusti n. sp. Stratton, Reisinger, Behringer, Bojko 2022
4.4. Novel and Redescribed Astathelohania Species
5. Discussion
5.1. Renaming the Freshwater Thelohania to Astathelohania n. gen.
5.2. Two Novel Crayfish Parasites in the USA
5.3. Host–Parasite Co-Evolution
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Stentiford, G.D.; Feist, S.W.; Stone, D.M.; Bateman, K.S.; Dunn, A.M. Microsporidia: Diverse, Dynamic, and Emergent Pathogens in Aquatic Systems. Trends Parasitol. 2013, 29, 567–578. [Google Scholar] [CrossRef]
- Freeman, M.A.; Turnbill, J.F.; Yeomans, W.E.; Bean, C.W. Prospects for Management Strategies of Invasive Crayfish Populations with an Emphasis on Biological Control. Aquat. Conserv. Mar. Freshw. Ecosyst. 2010, 20, 211–223. [Google Scholar] [CrossRef]
- Edgerton, B.F.; Evans, L.H.; Stephens, F.J.; Overstreet, R.M. Synopsis of Freshwater Crayfish Diseases and Commensal Organisms. Aquaculture 2002, 206, 57–135. [Google Scholar] [CrossRef] [Green Version]
- Longshaw, M. Diseases of Crayfish: A Review. J. Invertebr. Pathol. 2011, 106, 54–70. [Google Scholar] [CrossRef] [PubMed]
- Bojko, J.; Behringer, D.C.; Moler, P.; Stratton, C.E.L.; Reisinger, L. A New Lineage of Crayfish-Infecting Microsporidia: The Cambaraspora floridanus n. gen. n. sp. (Glugeida: Glugeidae) Complex from Floridian Freshwaters (USA). J. Invertebr. Pathol. 2020, 171, 107345. [Google Scholar] [CrossRef] [PubMed]
- Bojko, J.; Behringer, D.C.; Moler, P.; Reisinger, L. Ovipleistophora diplostomuri, a Parasite of Fish and Their Trematodes, also Infects the Crayfish Procambarus bivittatus. J. Invertebr. Pathol. 2020, 169, 107306. [Google Scholar] [CrossRef] [PubMed]
- Stentiford, G.D.; Ross, S.H.; Kerr, R.; Bass, D.; Bateman, K.S. Paradoxium irvingi n. gen. n. sp. (Microsporidia) Infecting the Musculature of European Pink Shrimp Pandalus montagui. J. Invertebr. Pathol. 2015, 130, 1–8. [Google Scholar] [CrossRef]
- Henneguy, G.; Thélohan, P. Myxosporides Parasites Des Muscles Chez Quelques Crustaces Decapodes. Ann Microgr. 1892, 4, 617–641. [Google Scholar]
- Moodie, E.G.; Le Jambre, L.F.; Katz, M.E. Thelohania montirivulorum sp. nov. (Microspora: Thelohaniidae), a Parasite of the Australian Freshwater Crayfish, Cherax destructor (Decapoda: Parastacidae): Fine Ultrastructure, Molecular Characteristics and Phylogenetic Relationships. Parasitol. Res. 2003, 91, 215–228. [Google Scholar] [CrossRef]
- Moodie, E.G.; Le Jambre, L.F.; Katz, M.E. Thelohania parastaci sp. nov. (Microspora: Thelohaniidae), a Parasite of the Australian Freshwater Crayfish, Cherax destructor (Decapoda: Parastacidae). Parasitol. Res. 2003, 91, 151–165. [Google Scholar] [CrossRef]
- Cossins, A.R.; Bowler, K. A Histological and Ultrastructural Study of Thelohania contejeani Henneguy, 1892 (Nosematidae), Microsporidian Parasite of Austropotamobius pallipes Lereboullet. Parasitology 1974, 68, 81–91. [Google Scholar] [CrossRef]
- Vivares, C.P. Etude Comparative Faite En Microscopies Photonique et Electronique de Trois Especes de Microsporidies Appartenant Au Genre Thelohania Henneguy, 1892, Parasites de Crustaces Decapodes Marins. Ann. Sci. Nat. 1975, 17, 141–178. [Google Scholar]
- Lom, J.; Nilsen, F.; Dyková, I. Thelohania contejeani Henneguy, 1892: Dimorphic Life Cycle and Taxonomic Affinities, as Indicated by Ultrastructural and Molecular Study. Parasitol. Res. 2001, 87, 860–872. [Google Scholar] [CrossRef] [PubMed]
- Dunn, J.C.; McClymont, H.E.; Christmas, M.; Dunn, A.M. Competition and Parasitism in the Native White Clawed Crayfish Austropotamobius pallipes and the Invasive Signal Crayfish Pacifastacus leniusculus in the UK. Biol. Invasions 2009, 11, 315–324. [Google Scholar] [CrossRef] [Green Version]
- Pretto, T.; Montesi, F.; Ghia, D.; Berton, V.; Abbadi, M.; Gastaldelli, M.; Manfrin, A.; Fea, G. Ultrastructural and Molecular Characterization of Vairimorpha austropotamobii sp. nov. (Microsporidia: Burenellidae) and Thelohania contejeani (Microsporidia: Thelohaniidae), Two Parasites of the White-Clawed Crayfish, Austropotamobius pallipes Complex (Decapoda: Astacidae). J. Invertebr. Pathol. 2018, 151, 59–75. [Google Scholar] [CrossRef] [PubMed]
- France, R.; Graham, L. Increased Microsporidian Parasitism of the Crayfish Orconectes virilis in an Experimentally Acidified Lake. Water Air Soil Pollut. 1985, 26, 129–1336. [Google Scholar] [CrossRef]
- Graham, L.; France, R. Attempts To Transmit Experimentally the Microsporidian Thelohania contejeani in Freshwater Crayfish (Orconectes virilis). Crustaceana 1986, 51, 208–211. [Google Scholar] [CrossRef]
- McGriff, D.; Modin, J. Thelohania contejeani Parasitism of the Crayfish, Pacifastacus leniusculus, in California. Calif. Fish Game 1983, 69, 178–183. [Google Scholar]
- Sprague, V. Thelohania cambarii n. sp., a Microsporidian Parasite of North American Crayfish. J. Parasitol. 1950, 36, 46. [Google Scholar]
- Stentiford, G.D.; Bateman, K.S.; Feist, S.W.; Chambers, E.; Stone, D.M. Plastic Parasites: Extreme Dimorphism Creates a Taxonomic Conundrum in the Phylum Microsporidia. Int. J. Parasitol. 2013, 43, 339–352. [Google Scholar] [CrossRef] [PubMed]
- Brown, A.M.V.; Adamson, M.L. Phylogenetic Distance of Thelohania butleri (Microsporidia; Thelohaniidae), a Parasite of the Smooth Pink Shrimp Pandalus jordani, from Its Congeners Suggests Need for Major Revision of the Genus Thelohania Henneguy, 1892. J. Eukaryot. Microbiol. 2006, 53, 445–455. [Google Scholar] [CrossRef] [PubMed]
- Sprague, V. Annotated List of Species of Microsporidia. In Comparative Pathology, 2nd ed.; Bulla, L.A., Cheng, T.C., Eds.; Plenum Press: New York, NY, USA, 1977; pp. 1–333. [Google Scholar]
- Vavra, J.; Hylis, M.; Vossbrinck, C.R.; Pilarska, D.K.; Linde, A.; Weiser, J.; Mcmanus, M.L.; Hoch, G.; Solter, L.F. Vairimorpha disparis n. comb. (Microsporidia: Burenellidae): A Redescription and Taxonomic Revision of Thelohania disparis Timofejeva 1956, a Microsporidian Parasite of the Gypsy Moth Lymantria dispar (L.) (Lepidoptera: Lymantriidae). J. Eukaryot. Microbiol. 2006, 53, 292–304. [Google Scholar] [CrossRef] [PubMed]
- Sokolova, Y.Y.; Fuxa, J.R. Biology and Life-cycle of the Microsporidium Kneallhazia solenopsae Knell Allan Hazard 1977 gen. n., comb. n., from the Fire Ant Solenopsis invicta. Parasitology 2008, 135, 903–929. [Google Scholar] [CrossRef]
- Cormier, A.; Wattier, R.; Giraud, I.; Teixeira, M.; Grandjean, F.; Rigaud, T.; Cordaux, R. Draft Genome Sequences of Thelohania contejeani and Cucumispora dikerogammari, Pathogenic Microsporidia of Freshwater Crustaceans. Microbiol. Resour. Announc. 2021, 10, e01346-20. [Google Scholar] [CrossRef] [PubMed]
- Morris, D.J.; Freeman, M.A. Hyperparasitism has Wide-Ranging Implications for Studies on the Invertebrate Phase of Myxosporean (Myxozoa) Life Cycles. Int. J. Parasitol. 2010, 40, 357–369. [Google Scholar] [CrossRef] [PubMed]
- Haag, K.L.; Larsson, J.I.R.; Refardt, D.; Ebert, D. Cytological and Molecular Description of Hamiltosporidium tvaerminnensis gen. et sp. nov., a Microsporidian Parasite of Daphnia magna, and Establishment of Hamiltosporidium magnivora comb. nov. Parasitology 2011, 138, 447–462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stentiford, G.D.; Bateman, K.S.; Feist, S.W.; Oyarzún, S.; Uribe, J.C.; Palacios, M.; Stone, D.M. Areospora rohanae n. gen. n. sp. (Microsporidia; Areosporiidae n. fam.) Elicits Multi-Nucleate Giant-Cell Formation in Southern King Crab (Lithodes santolla). J. Invertebr. Pathol. 2014, 118, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Dubuffet, A.; Chauvet, M.; Moné, A.; Debroas, D.; Lepère, C. A Phylogenetic Framework to Investigate the Microsporidian Communities through Metabarcoding and Its Application to Lake Ecosystems. Environ. Microbiol. 2021, 23, 4344–4359. [Google Scholar] [CrossRef] [PubMed]
- Bojko, J.; Reinke, A.W.; Stentiford, G.D.; Williams, B.; Rogers, M.S.J.; Bass, D. Microsporidia: A Taxonomic, Evolutionary, and Ecological Synthesis. Trends Parasitol. 2022, in press. [Google Scholar]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 Years of Image Analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Ovcharenko, M.O.; Bacela, K.; Wilkinson, T.; Ironside, J.E.; Rigaud, T.; Wattier, R.A. Cucumispora dikerogammari n. gen. (Fungi: Microsporidia) Infecting the Invasive Amphipod Dikerogammarus villosus: A Potential Emerging Disease in European Rivers. Parasitology 2010, 137, 191–204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. The CIPRES Science Gateway: A Community Resource for Phylogenetic Analyses. In Proceedings of the 2011 TeraGrid Conference on Extreme Digital Discovery—TG’11, Salt Lake City, UT, USA, 18–21 July 2011; ACM Press: Salt Lake City, UT, USA, 2011; p. 1. [Google Scholar]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Muhire, B.M.; Varsani, A.; Martin, D.P. SDT: A Virus Classification Tool Based on Pairwise Sequence Alignment and Identity Calculation. PLoS ONE 2014, 9, e108277. [Google Scholar] [CrossRef] [PubMed]
- Karpov, S.A.; Mamkaeva, M.A.; Aleoshin, V.V.; Nassonova, E.; Lilje, O.; Gleason, F.H. Morphology, Phylogeny, and Ecology of the Aphelids (Aphelidea, Opisthokonta) and Proposal for the New Superphylum Opisthosporidia. Front. Microbiol. 2014, 5, 112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tedersoo, L.; Sánchez-Ramírez, S.; Kõljalg, U.; Bahram, M.; Döring, M.; Schigel, D.; May, T.; Ryberg, M.; Abarenkov, K. High-Level Classification of the Fungi and a Tool for Evolutionary Ecological Analyses. Fungal Divers. 2018, 90, 135–159. [Google Scholar] [CrossRef] [Green Version]
- Balbiani, E. Sur Les Microsporidies Ou Psorospermies Des Articules. Compt. Rend. Acad. Sco. Paris 1882, 95, 1168–1171. [Google Scholar]
- Wijayawardene, N.N.; Hyde, K.D.; Dai, D.Q.; Sánchez-García, M.; Goto, B.T.; Saxena, R.K.; Erdoğdu, M.; Selçuk, F.; Rajeshkumar, K.C.; Aptroot, A.; et al. Outline of Fungi and fungus-like taxa—2021. Mycosphere 2022, 13, 53–453. [Google Scholar] [CrossRef]
- Vossbrinck, C.R.; Debrunner-Vossbrinck, B.A. Molecular Phylogeny of the Microsporidia: Ecological, Ultrastructural and Taxonomic Considerations. Folia Parasitol. 2005, 52, 131–142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tokarev, Y.S.; Huang, W.-F.; Solter, L.F.; Malysh, J.M.; Becnel, J.J.; Vossbrinck, C.R. A Formal Redefinition of the Genera Nosema and Vairimorpha (Microsporidia: Nosematidae) and Reassignment of Species Based on Molecular Phylogenetics. J. Invertebr. Pathol. 2020, 169, 107279. [Google Scholar] [CrossRef] [PubMed]
- Hazard, E.I.; Oldacre, S.W. Revision of Microsporidia (Protozoa) Close to Thelohania, with Descriptions of One New Family, Eight New Genera, and Thirteen New Species; US Department of Agriculture, Agricultural Research Service: Washington, DC, USA, 1976. [CrossRef]
- Weiser, J. Contribution to the Classification of Microsporidia. Vestn. Cesk. Spol. Zool. 1977, 41, 308–320. [Google Scholar]
- Baker, M.D.; Vossbrinck, C.R.; Becnel, J.J.; Andreadis, T.G. Phylogeny of Amblyospora (Microsporida: Amblyosporidae) and Related Genera Based on Small Subunit Ribosomal DNA Data: A Possible Example of Host Parasite Cospeciation. J. Invertebr. Pathol. 1998, 71, 199–206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sokolova, Y.; Pelin, A.; Hawke, J.; Corradi, N. Morphology and Phylogeny of Agmasoma penaei (Microsporidia) from the Type Host, Litopenaeus setiferus, and the Type Locality, Louisiana, USA. Int. J. Parasitol. 2015, 45, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Crandall, K.A.; De Grave, S. An Updated Classification of the Freshwater Crayfishes (Decapoda: Astacidea) of the World, with a Complete Species List. J. Crustac. Biol. 2017, 37, 615–653. [Google Scholar] [CrossRef] [Green Version]
- Moodie, E.G.; Le Jambre, L.F.; Katz, M.E. Ultrastructural Characteristics and Small Subunit Ribosomal DNA Sequence of Vairimorpha cheracis sp. nov., (Microspora: Burenellidae), a Parasite of the Australian Yabby, Cherax destructor (Decapoda: Parastacidae). J. Invertebr. Pathol. 2003, 84, 198–213. [Google Scholar] [CrossRef] [PubMed]
- Stern, D.B.; Breinholt, J.; Pedraza-Lara, C.; López-Mejía, M.; Owen, C.L.; Bracken-Grissom, H.; Fetzner, J.W.; Crandall, K.A. Phylogenetic Evidence from Freshwater Crayfishes That Cave Adaptation Is Not an Evolutionary Dead-end. Evolution 2017, 71, 2522–2532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bracken-Grissom, H.D.; Ahyong, S.T.; Wilkinson, R.D.; Feldmann, R.M.; Schweitzer, C.E.; Breinholt, J.W.; Bendall, M.; Palero, F.; Chan, T.-Y.; Felder, D.L.; et al. The Emergence of Lobsters: Phylogenetic Relationships, Morphological Evolution and Divergence Time Comparisons of an Ancient Group (Decapoda: Achelata, Astacidea, Glypheidea, Polychelida). Syst. Biol. 2014, 63, 457–479. [Google Scholar] [CrossRef] [Green Version]
- Reynolds, J.D.; Souty-Grosset, C. Management of Freshwater Biodiversity: Crayfish as Bioindicators; Cambridge University Press: Cambridge, UK; New York, NY, USA, 2012; ISBN 978-0-521-51400-2. [Google Scholar]

| Host Species | Site | Coordinates | Collection Date | Sex | Carapace Length (mm) | Microsporidian Species | SSU | Histology | Electron Microscopy | Accession Number |
|---|---|---|---|---|---|---|---|---|---|---|
| F. rusticus | Darby Creek, OH | 40.013388, −83.383180 | 30 June 2021 | MII | 27 | A. rusti n. sp. | ✓ | ✓ | — | OM630066 |
| F. rusticus | Darby Creek, OH | 40.013388, −83.383180 | 30 June 2021 | MI | 32 | A. rusti n. sp. | ✓ | ✓ | ✓ | OM630067 |
| F. virilis | South Turtle Lake, WI | 46.217698, −89.891143 | 09 July 2019 | MII | 51 | A. virili n. sp. | ✓ | ✓ | ✓ | OM630068 |
| F. virilis | South Turtle Lake, WI | 46.217698, −89.891143 | 09 July 2019 | MII | 50 | A. virili n. sp. | ✓ | ✓ | — | OM630069 |
| F. virilis | South Turtle Lake, WI | 46.217698, −89.891143 | 09 July 2019 | MII | 43 | A. virili n. sp. | ✓ | ✓ | — | OM630070 |
| F. virilis | Crab Lake, WI | 46.203368, −89.729255 | 19 July 2019 | MII | 40 | A. rusti n. sp. | ✓ | ✓ | — | OM630071 |
| Morphological feature | A. rusti n. sp. | A. virili n. sp. | A. montirivulorum | A. parastaci | A. contejeani | A. contejeani | “T. contejeani” | “T. contejeani” | “T. cambari” | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Moodie et al. [9] | Moodie et al. [10] | Lom et al. [13] | Pretto et al. [15] | Graham and France [17] | McGriff and Modin [18] | Sprague [19] | |||||||
| Shore shape | Oval, wider posterior end | Oval, wider posterior end | Lozenge, round ends | Lozenge, round ends | Oval, wider posterior end | Oval, wider posterior end | Oval | Oval | Oval, wider posterior end | ||||
| Uninucleate spore length (μm) | 3.2 ± 0.5 1 | n = 10 | 3.4 ± 0.1 1 | n = 7 | n/a | n/a | 4.2 2 | 3.6 ± 0.4 2 | n = 50 | 3.3 (2.8–3.6) | n = 50 | 3.0–3.8 | 4.6 |
| Uninucleate spore width (μm) | 1.7 ± 0.31 | n = 10 | 2.0 ± 0.3 1 | n = 10 | n/a | n/a | 2.1 2 | 2.3 ± 0.3 2 | n = 50 | 2.2 (2.0–2.6) | n = 50 | 1.8–2.4 | 2.2 |
| Binucleate spore length (μm) | n/a | n/a | 5.9 (4.9–7.2) 2 | 3.9 (3.2–4.9) 2 | 3.8 2 | 3.3 ± 0.5 2 | n = 50 | n/a | n/a | n/a | |||
| Binucleate spore width (μm) | n/a | n/a | 2.6 (2.0–3.1) 2 | 2.0 (1.5–2.7) 2 | 1.8 2 | 1.7 ± 0.2 2 | n = 50 | n/a | n/a | n/a | |||
| Uninucleate—no. coils in polar filament | 13–14 | 16–17 | 20–22 | 11–20 | 9–10 | 9–12 | n/a | n/a | n/a | ||||
| Uninucleate—polar filament diameter (nm) | 141 ± 14 | n = 10 | 118 ± 3 | n = 10 | 98 (82–111) | 59 (53–74) | 120–180 3 | 77 | n = 10 | n/a | n/a | n/a | |
| Binucleate—no. coils in polar filament | n/a | n/a | 20–22 | 6–8 | 5–7 | 5–6 | n/a | n/a | n/a | ||||
| Binucleate—polar filament diameter (nm) | n/a | n/a | 107 (90–140) | 83 (65–102) | n/a | 108 | n = 10 | n/a | n/a | n/a | |||
| SPV diameter (μm) | 5.2 ± 0.6 1 | n = 10 | 8.1 ± 0.7 1 | n = 10 | 8.4 (7.0–9.6) 2 | 8.8 (7.4–10.5) 2 | 8–9 3 | 9.4 ± 0.6 2 | n = 20 | 7.9 (6.4–8.1) | n = 10 | n/a | n/a |
| SPV tubular-like structure diameter (nm) | 244 ± 32 | n = 10 | 241 ± 26 | n = 10 | 171 (130–249) | 249 (205–307) | 220 | 155–185 | n = 20 | n/a | n/a | n/a | |
| SPV microtubular-like structure diameter (nm) | 70 ± 9 | n = 10 | 73 ± 10 | n = 10 | 85 (63–117) | 73 (50–99) | 80–100 | 75–85 | n = 20 | n/a | n/a | n/a | |
| Lateral exospore thickness of uninucleate spores (nm) | 25 ± 6 | n = 10 | 25 ± 3 | n = 10 | 31 (30–40) | 24 (20–40) | 24–30 4 | 28 | n = 15 | n/a | n/a | n/a | |
| Lateral endospore thickness of uninucleate spores (nm) | 57 ± 18 | n = 10 | 82 ± 12 | n = 10 | 108 (80–130) | 73 (56–110) | 60–90 4 | 78 | n = 15 | n/a | n/a | n/a | |
| Lateral exospore thickness of binucleate spores (nm) | n/a | n/a | 22 (17–30) | 34 (30–40) | n/a | 32 | n = 8 | n/a | n/a | n/a | |||
| Lateral endospore thickness of binucleate spores (nm) | n/a | n/a | 65 (40–80) | 58 (50–60) | n/a | 55 | n = 8 | n/a | n/a | n/a | |||
| Dimorphic sporogony? | No | No | Yes | Yes | Yes | Yes | n/a | n/a | n/a | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stratton, C.E.; Reisinger, L.S.; Behringer, D.C.; Bojko, J. Revising the Freshwater Thelohania to Astathelohania gen. et comb. nov., and Description of Two New Species. Microorganisms 2022, 10, 636. https://doi.org/10.3390/microorganisms10030636
Stratton CE, Reisinger LS, Behringer DC, Bojko J. Revising the Freshwater Thelohania to Astathelohania gen. et comb. nov., and Description of Two New Species. Microorganisms. 2022; 10(3):636. https://doi.org/10.3390/microorganisms10030636
Chicago/Turabian StyleStratton, Cheyenne E., Lindsey S. Reisinger, Donald C. Behringer, and Jamie Bojko. 2022. "Revising the Freshwater Thelohania to Astathelohania gen. et comb. nov., and Description of Two New Species" Microorganisms 10, no. 3: 636. https://doi.org/10.3390/microorganisms10030636
APA StyleStratton, C. E., Reisinger, L. S., Behringer, D. C., & Bojko, J. (2022). Revising the Freshwater Thelohania to Astathelohania gen. et comb. nov., and Description of Two New Species. Microorganisms, 10(3), 636. https://doi.org/10.3390/microorganisms10030636






