Paromomycin Reduces Vairimorpha (Nosema) ceranae Infection in Honey Bees but Perturbs Microbiome Levels and Midgut Cell Function
Abstract
:1. Introduction
2. Materials and Methods
2.1. Honey Bee Colonies and Caging Experiments
2.2. Isolation and Quantification of V. ceranae Spores
2.3. Infections and Chemical Treatments
2.4. DNA Extraction and qPCR
2.5. RNA Isolation, Reverse Transcription, and Quantitative PCR for Gene Expression Analysis
2.6. Statistical Analysis
3. Results
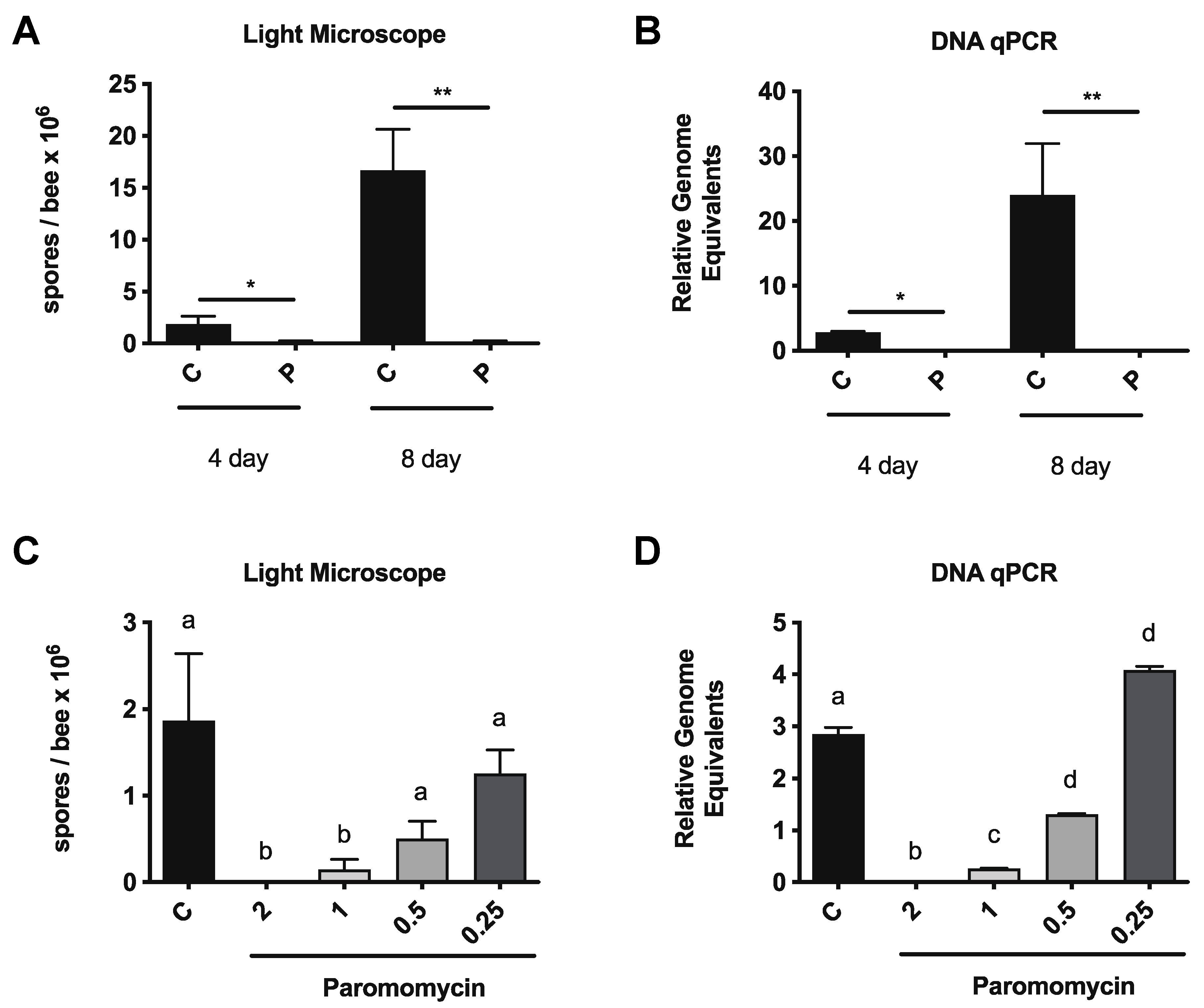
3.1. Paromomycin Reduces vs. Ceranae Infection Intensity in Honey Bees
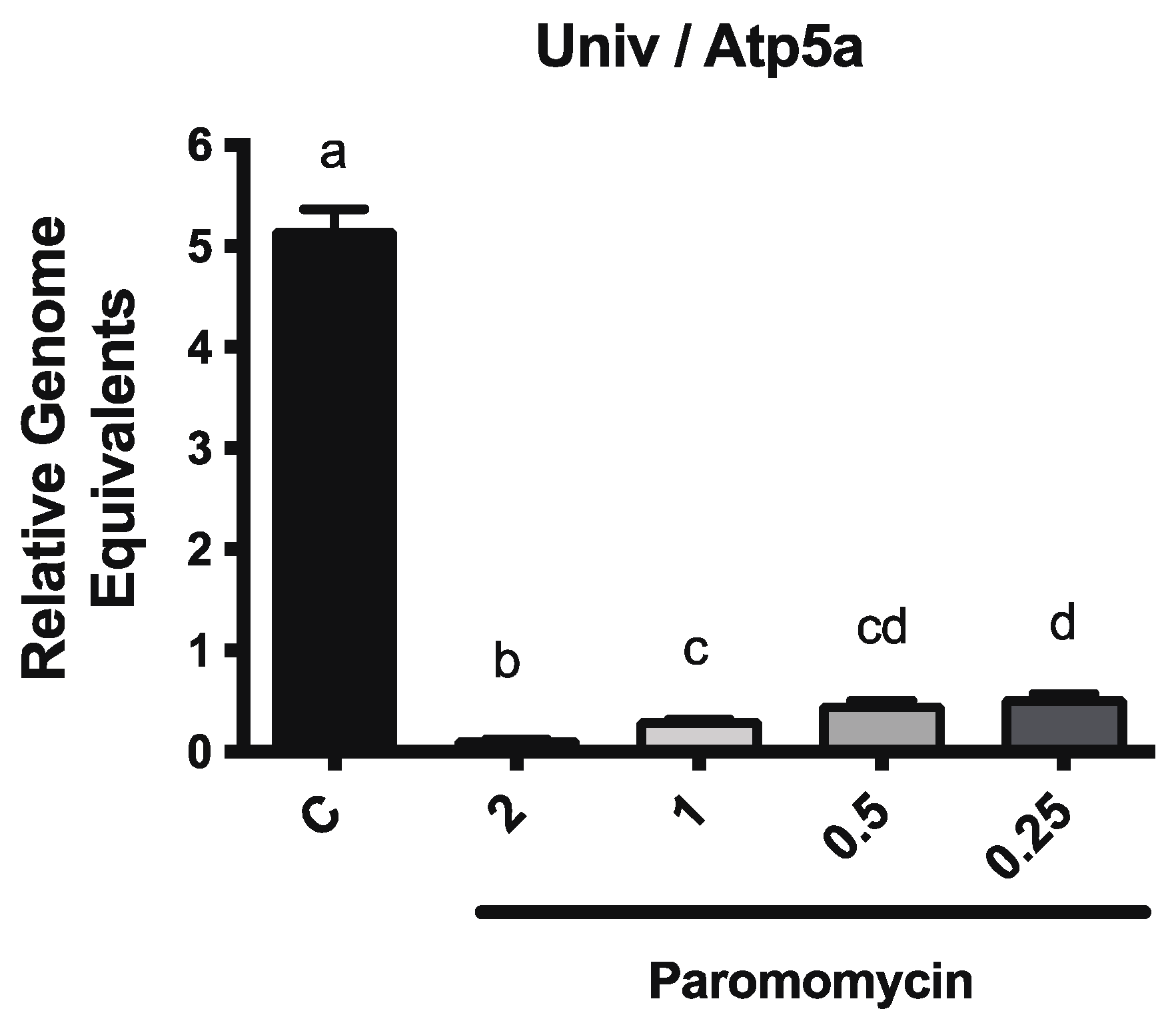
3.2. Paromomycin Impacts on Microbiome Levels
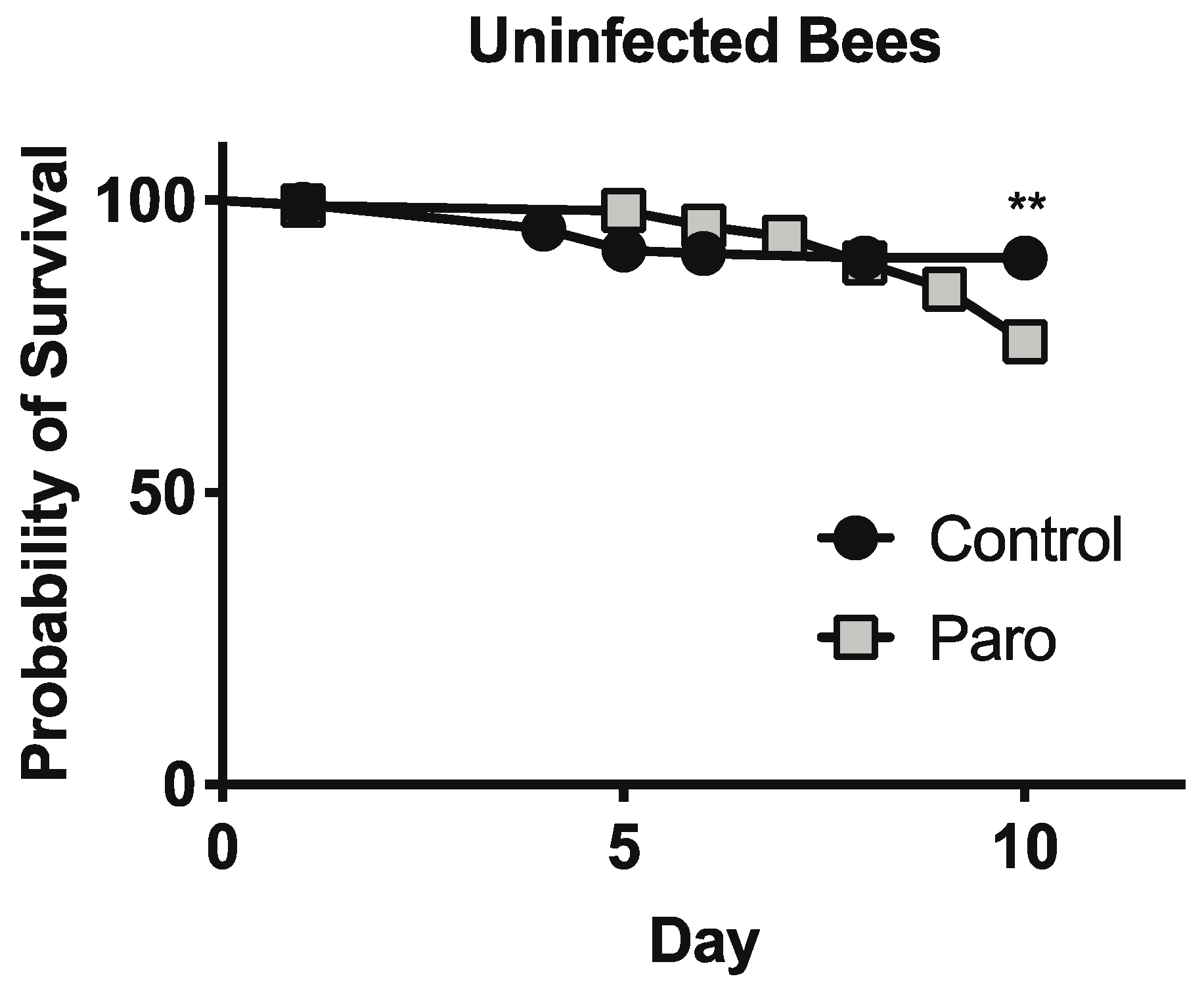
3.3. Paromomycin Impacts on Host Survival
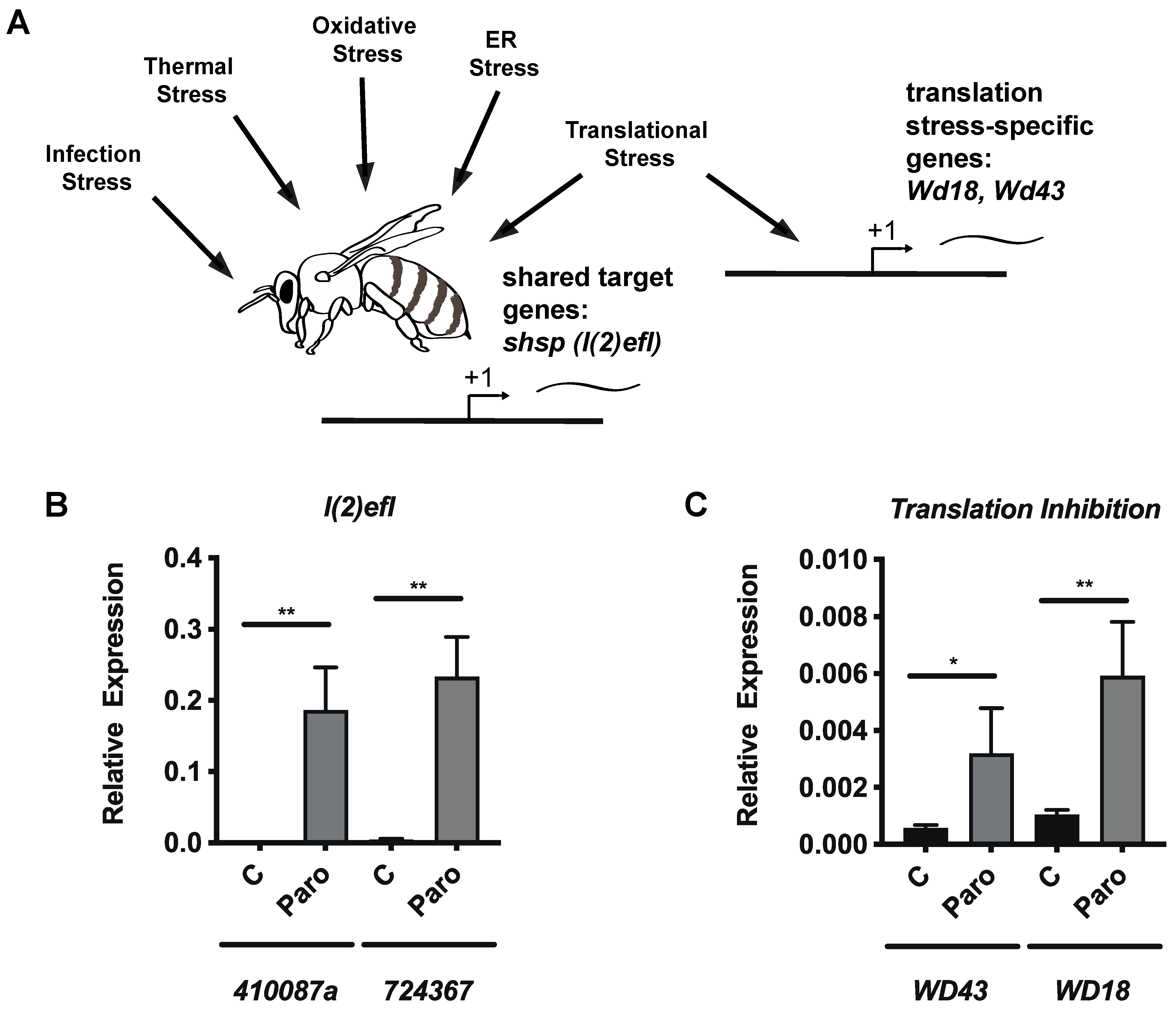
3.4. Paromomycin Impacts on the Expression of General Stress Biomarker Genes and Specific Genes for Translation Inhibition
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Weiss, L.M. Microsporidia: Pathogens of Opportunity, 1st ed.; Wiley: Hoboken, NJ, USA, 2014; pp. 371–401. [Google Scholar] [CrossRef]
- Tokarev, Y.S.; Huang, W.-F.; Solter, L.F.; Malysh, J.M.; Becnel, J.J.; Vossbrinck, C.R. A Formal Redefinition of the Genera Nosema and Vairimorpha (Microsporidia: Nosematidae) and Reassignment of Species Based on Molecular Phylogenetics. J. Invertebr. Pathol. 2020, 169, 107279. [Google Scholar] [CrossRef] [PubMed]
- Goblirsch, M. Nosema ceranae Disease of the Honey Bee (Apis mellifera). Ann. Abeille. 2017, 49, 131–150. [Google Scholar] [CrossRef] [Green Version]
- Martín-Hernández, R.; Bartolomé, C.; Chejanovsky, N.; Conte, Y.L.; Dalmon, A.; Dussaubat, C.; García-Palencia, P.; Meana, A.; Pinto, M.A.; Soroker, V.; et al. Nosema ceranae in Apis mellifera: A 12 Years Postdetection Perspective. Environ. Microbiol. 2018, 20, 1302–1329. [Google Scholar] [CrossRef] [Green Version]
- Snow, J.W. Nosema apis and N. ceranae Infection in Honey Bees: A Model for Host-Pathogen Interactions in Insects. In Microsporidia, Current Advances in Biology; Weiss, L.M., Reinke, A.W., Eds.; Experientia Supplementum; Academic Press: Cambridge, MA, USA, 2022; Volume 114, pp. 153–177. ISBN 9783030933050. [Google Scholar]
- Solter, L.F.; Becnel, J.J.; Oi, D.H. Insect Pathology, 2nd ed.; Academic Press: San Diego, CA, USA, 2012; pp. 221–263. [Google Scholar] [CrossRef]
- Van den Heever, J.P.; Thompson, T.S.; Curtis, J.M.; Ibrahim, A.; Pernal, S.F. Fumagillin: An Overview of Recent Scientific Advances and Their Significance for Apiculture. J. Agric. Food Chem. 2014, 62, 2728–2737. [Google Scholar] [CrossRef]
- Mendoza, Y.; Diaz-Cetti, S.; Ramallo, G.; Santos, E.; Porrini, M.; Invernizzi, C. Nosema ceranae Winter Control: Study of the Effectiveness of Different Fumagillin Treatments and Consequences on the Strength of Honey Bee (Hymenoptera: Apidae) Colonies. J. Econ. Entomol. 2017, 110, 1–5. [Google Scholar] [CrossRef]
- Huang, W.-F.; Solter, L.F.; Yau, P.M.; Imai, B.S. Nosema ceranae Escapes Fumagillin Control in Honey Bees. PLoS Pathog. 2013, 9, e1003185. [Google Scholar] [CrossRef] [Green Version]
- Huntsman, E.M.; Cho, R.M.; Kogan, H.V.; McNamara-Bordewick, N.K.; Tomko, R.J.; Snow, J.W. Proteasome Inhibition Is an Effective Treatment Strategy for Microsporidia Infection in Honey Bees. Biomolecules 2021, 11, 1600. [Google Scholar] [CrossRef]
- Krause, K.M.; Serio, A.W.; Kane, T.R.; Connolly, L.E. Aminoglycosides: An Overview. Cold Spring Harb. Perspect. Med. 2016, 6, a027029-19. [Google Scholar] [CrossRef] [Green Version]
- Katiyar, S.K.; Visvesvara, G.S.; Edlind, T.D. Comparisons of Ribosomal RNA Sequences from Amitochondrial Protozoa: Implications for Processing, MRNA Binding and Paromomycin Susceptibility. Gene 1995, 152, 27–33. [Google Scholar] [CrossRef]
- Moffett, J.O.; Lackett, J.J.; Hitchcock, J.D. Compounds Tested for Control of Nosema in Honey Bees. J. Econ. Entomol. 1969, 62, 886–889. [Google Scholar] [CrossRef]
- Beauvais, B.; Sarfati, C.; Challier, S.; Derouin, F. In Vitro Model to Assess Effect of Antimicrobial Agents on Encephalitozoon cuniculi. Antimicrob. Agents Chemother. 1994, 38, 2440–2448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Gool, T.; Snijders, F.; Reiss, P.; Schattenkerk, J.K.E.; van den Bergh Weerman, M.A.; Bartelsman, J.F.; Bruins, J.J.; Canning, E.U.; Dankert, J. Diagnosis of Intestinal and Disseminated Microsporidial Infections in Patients with HIV by a New Rapid Fluorescence Technique. J. Clin. Pathol. 1993, 46, 694–699. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gisder, S.; Genersch, E. Identification of Candidate Agents Active against N. ceranae Infection in Honey Bees: Establishment of a Medium Throughput Screening Assay Based on N. ceranae Infected Cultured Cells. PLoS ONE 2015, 10, e0117200. [Google Scholar] [CrossRef] [PubMed]
- Gisder, S.; Moeckel, N.; Linde, A.; Genersch, E. A Cell Culture Model for Nosema ceranae and Nosema apis Allows New Insights into the Life Cycle of These Important Honey Bee-Pathogenic Microsporidia. Environ. Microbiol. 2011, 13, 404–413. [Google Scholar] [CrossRef] [PubMed]
- Snow, J.W. Prolyl-tRNA Synthetase Inhibition Reduces Microsporidia Infection Intensity in Honey Bees. Apidologie 2020, 51, 557–569. [Google Scholar] [CrossRef]
- Holt, H.L.; Aronstein, K.A.; Grozinger, C.M. Chronic Parasitization by Nosema Microsporidia Causes Global Expression Changes in Core Nutritional, Metabolic and Behavioral Pathways in Honey Bee Workers (Apis mellifera). BMC Genom. 2013, 14, 799. [Google Scholar] [CrossRef] [Green Version]
- Fries, I.; Chauzat, M.-P.; Chen, Y.P.; Doublet, V.; Genersch, E.; Gisder, S.; Higes, M.; McMahon, D.P.; Martín-Hernández, R.; Natsopoulou, M.; et al. Standard Methods for Nosema Research. J. Apic. Res. 2013, 52, 1–28. [Google Scholar] [CrossRef] [Green Version]
- Cantwell, G.E. Standard Methods for Counting Nosema Spores. Am. Bee J. 1970, 110, 222–223. [Google Scholar]
- Snow, J.W.; Koydemir, H.C.; Karinca, D.K.; Liang, K.; Tseng, D.; Ozcan, A. Rapid Imaging, Detection, and Quantification of Nosema ceranae spores in Honey Bees Using Mobile Phone-Based Fluorescence Microscopy. Lab A Chip 2019, 19, 789–797. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing Real-Time PCR Data by the Comparative C(T) Method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Kešnerová, L.; Mars, R.A.T.; Ellegaard, K.M.; Troilo, M.; Sauer, U.; Engel, P. Disentangling Metabolic Functions of Bacteria in the Honey Bee Gut. PLoS Biol. 2017, 15, e2003467. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shih, S.R.; Bach, D.M.; Rondeau, N.C.; Sam, J.; Lovinger, N.L.; Lopatkin, A.J.; Snow, J.W. Honey Bee SHSP Are Responsive to Diverse Proteostatic Stresses and Potentially Promising Biomarkers of Honey Bee Stress. Sci. Rep 2021, 11, 22087. [Google Scholar] [CrossRef] [PubMed]
- McAfee, A.; Milone, J.; Chapman, A.; Foster, L.J.; Pettis, J.S.; Tarpy, D.R. Candidate Stress Biomarkers for Queen Failure Diagnostics. BMC Genom. 2020, 21, 571. [Google Scholar] [CrossRef] [PubMed]
- McAfee, A.; Chapman, A.; Higo, H.; Underwood, R.; Milone, J.; Foster, L.J.; Guarna, M.M.; Tarpy, D.R.; Pettis, J.S. Vulnerability of Honey Bee Queens to Heat-Induced Loss of Fertility. Nat. Sustain. 2020, 3, 367–376. [Google Scholar] [CrossRef]
- Flores, M.E.; McNamara-Bordewick, N.K.; Lovinger, N.L.; Snow, J.W. Halofuginone Triggers a Transcriptional Program Centered on Ribosome Biogenesis and Function in Honey Bees. Insect Biochem. Molec. 2021, 139, 103667. [Google Scholar] [CrossRef]
- Fan-Minogue, H.; Bedwell, D.M. Eukaryotic Ribosomal RNA Determinants of Aminoglycoside Resistance and Their Role in Translational Fidelity. RNA 2008, 14, 148–157. [Google Scholar] [CrossRef] [Green Version]
- De Loubresse, N.G.; Prokhorova, I.; Holtkamp, W.; Rodnina, M.V.; Yusupova, G.; Yusupov, M. Structural Basis for the Inhibition of the Eukaryotic Ribosome. Nature 2014, 513, 517–522. [Google Scholar] [CrossRef] [Green Version]
- Prokhorova, I.; Altman, R.B.; Djumagulov, M.; Shrestha, J.P.; Urzhumtsev, A.; Ferguson, A.; Chang, C.-W.T.; Yusupov, M.; Blanchard, S.C.; Yusupova, G. Aminoglycoside Interactions and Impacts on the Eukaryotic Ribosome. Proc. Natl. Acad. Sci. USA 2017, 114, E10899–E10908. [Google Scholar] [CrossRef] [Green Version]
- Kamita, M.; Kimura, Y.; Ino, Y.; Kamp, R.M.; Polevoda, B.; Sherman, F.; Hirano, H. Nα-Acetylation of Yeast Ribosomal Proteins and Its Effect on Protein Synthesis. J. Proteom. 2011, 74, 431–441. [Google Scholar] [CrossRef]
- Fujii, K.; Susanto, T.T.; Saurabh, S.; Barna, M. Decoding the Function of Expansion Segments in Ribosomes. Mol. Cell 2018, 72, 1013–1020.e6. [Google Scholar] [CrossRef] [Green Version]
- Rakwalska, M.; Rospert, S. The Ribosome-Bound Chaperones RAC and Ssb1/2p Are Required for Accurate Translation in Saccharomyces Cerevisiae. Mol. Cell. Biol. 2004, 24, 9186–9197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Melnikov, S.V.; Rivera, K.D.; Ostapenko, D.; Makarenko, A.; Sanscrainte, N.D.; Becnel, J.J.; Solomon, M.J.; Texier, C.; Pappin, D.J.; Söll, D. Error-Prone Protein Synthesis in Parasites with the Smallest Eukaryotic Genome. Proc. Natl. Acad. Sci. USA 2018, 115, E6245–E6253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peyretaillade, E.; Biderre, C.; Peyret, P.; Duffieux, F.; Méténier, G.; Gouy, M.; Michot, B.; Vivarès, C.P. Microsporidian Encephalitozoon cuniculi, a Unicellular Eukaryote with an Unusual Chromosomal Dispersion of Ribosomal Genes and a LSU RRNA Reduced to the Universal Core. Nucleic Acids Res. 1998, 26, 3513–3520. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vossbrinck, C.R.; Maddox, J.V.; Friedman, S.; Debrunner-Vossbrinck, B.A.; Woese, C.R. Ribosomal-Rna Sequence Suggests Microsporidia Are Extremely Ancient Eukaryotes. Nature 1987, 326, 411–414. [Google Scholar] [CrossRef] [PubMed]
- Melnikov, S.; Manakongtreecheep, K.; Rivera, K.; Makarenko, A.; Pappin, D.; Söll, D. Muller’s Ratchet and Ribosome Degeneration in the Obligate Intracellular Parasites Microsporidia. Int. J. Mol. Sci. 2018, 19, 4125. [Google Scholar] [CrossRef] [Green Version]
- Barandun, J.; Hunziker, M.; Vossbrinck, C.R.; Klinge, S. Evolutionary Compaction and Adaptation Visualized by the Structure of the Dormant Microsporidian Ribosome. Nat. Microbiol. 2019, 4, 1798–1804. [Google Scholar] [CrossRef]
- Shalev-Benami, M.; Zhang, Y.; Rozenberg, H.; Nobe, Y.; Taoka, M.; Matzov, D.; Zimmerman, E.; Bashan, A.; Isobe, T.; Jaffe, C.L.; et al. Atomic Resolution Snapshot of Leishmania Ribosome Inhibition by the Aminoglycoside Paromomycin. Nat. Commun. 2017, 8, 1589. [Google Scholar] [CrossRef] [Green Version]
- Li, J.H.; Evans, J.D.; Li, W.F.; Zhao, Y.Z.; DeGrandi-Hoffman, G.; Huang, S.K.; Li, Z.G.; Hamilton, M.; Chen, Y.P. New Evidence Showing That the Destruction of Gut Bacteria by Antibiotic Treatment Could Increase the Honey Bee’s Vulnerability to Nosema Infection. PLoS ONE 2017, 12, e0187505. [Google Scholar] [CrossRef]
- Engel, P.; Moran, N.A. The Gut Microbiota of Insects—Diversity in Structure and Function. FEMS Microbiol. Rev. 2013, 37, 699–735. [Google Scholar] [CrossRef]
- Kwong, W.K.; Medina, L.A.; Koch, H.; Sing, K.-W.; Soh, E.J.Y.; Ascher, J.S.; Jaffé, R.; Moran, N.A. Dynamic Microbiome Evolution in Social Bees. Sci. Adv. 2017, 3, e1600513. [Google Scholar] [CrossRef] [Green Version]
- Raymann, K.; Moran, N.A. The Role of the Gut Microbiome in Health and Disease of Adult Honey Bee Workers. Curr. Opin. Insect Sci. 2018, 26, 97–104. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Powell, J.E.; Steele, M.I.; Dietrich, C.; Moran, N.A. Honeybee Gut Microbiota Promotes Host Weight Gain via Bacterial Metabolism and Hormonal Signaling. Proc. Natl. Acad. Sci. USA 2017, 114, 4775–4780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kwong, W.K.; Mancenido, A.L.; Moran, N.A. Immune System Stimulation by the Native Gut Microbiota of Honey Bees. R. Soc. Open Sci. 2017, 4, 170003. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raymann, K.; Shaffer, Z.; Moran, N.A. Antibiotic Exposure Perturbs the Gut Microbiota and Elevates Mortality in Honeybees. PLoS Biol. 2017, 15, e2001861. [Google Scholar] [CrossRef] [PubMed]
- Maes, P.W.; Rodrigues, P.A.P.; Oliver, R.; Mott, B.M.; Anderson, K.E. Diet Related Gut Bacterial Dysbiosis Correlates with Impaired Development, Increased Mortality and Nosema Disease in the Honey Bee (Apis mellifera). Mol. Ecol. 2016, 25, 5439–5450. [Google Scholar] [CrossRef] [PubMed]
- Rubanov, A.; Russell, K.A.; Rothman, J.A.; Nieh, J.C.; Mcfrederick, Q.S. Intensity of Nosema ceranae Infection Is Associated with Specific Honey Bee Gut Bacteria and Weakly Associated with Gut Microbiome Structure. Sci. Rep. 2019, 9, 3820–3828. [Google Scholar] [CrossRef]
- Huang, S.K.; Ye, K.T.; Huang, W.F.; Ying, B.H.; Su, X.; Lin, L.H.; Li, J.H.; Chen, Y.P.; Li, J.L.; Bao, X.L.; et al. Influence of Feeding Type and Nosema ceranae Infection on the Gut Microbiota of Apis cerana Workers. mSystems 2018, 3, e00177-18. [Google Scholar] [CrossRef] [Green Version]
- Schwarz, R.S.; Moran, N.A.; Evans, J.D. Early Gut Colonizers Shape Parasite Susceptibility and Microbiota Composition in Honey Bee Workers. Proc. Natl. Acad. Sci. USA 2016, 113, 9345–9350. [Google Scholar] [CrossRef] [Green Version]
- Ptaszyńska, A.A.; Paleolog, J.; Borsuk, G. Nosema ceranae Infection Promotes Proliferation of Yeasts in Honey Bee Intestines. PLoS ONE 2016, 11, e0164477. [Google Scholar] [CrossRef]
- Tauber, J.P.; Nguyen, V.; Lopez, D.; Evans, J.D. Effects of a Resident Yeast from the Honeybee Gut on Immunity, Microbiota, and Nosema Disease. Insects 2019, 10, 296. [Google Scholar] [CrossRef] [Green Version]
- Heinsen, F.-A.; Knecht, H.; Neulinger, S.C.; Schmitz, R.A.; Knecht, C.; Kühbacher, T.; Rosenstiel, P.C.; Schreiber, S.; Friedrichs, A.K.; Ott, S.J. Dynamic Changes of the Luminal and Mucosa-Associated Gut Microbiota during and after Antibiotic Therapy with Paromomycin. Gut Microbes 2015, 6, 243–254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sagastume, S.; Martín-Hernández, R.; Higes, M.; Henriques-Gil, N. Genotype Diversity in the Honey Bee Parasite Nosema ceranae: Multi-Strain Isolates, Cryptic Sex or Both? BMC Evol. Biol. 2016, 16, 216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gillespie, J.J.; Johnston, J.S.; Cannone, J.J.; Gutell, R.R. Characteristics of the Nuclear (18S, 5.8S, 28S and 5S) and Mitochondrial (12S and 16S) RRNA Genes of Apis mellifera (Insecta: Hymenoptera): Structure, Organization, and Retrotransposable Elements. Insect Mol. Biol. 2006, 15, 657–686. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bach, D.M.; Holzman, M.A.; Wague, F.; Miranda, J.; Lopatkin, A.J.; Mansfield, J.H.; Snow, J.W. Thermal Stress Induces Tissue Damage and a Broad Shift in Regenerative Signaling Pathways in the Honey Bee Digestive Tract. J. Exp. Biol. 2021, 224, jeb242262. [Google Scholar] [CrossRef] [PubMed]
- Fischer, N.L.; Naseer, N.; Shin, S.; Brodsky, I.E. Effector-Triggered Immunity and Pathogen Sensing in Metazoans. Nat. Microbiol. 2020, 5, 14–26. [Google Scholar] [CrossRef]
- Melo, J.A.; Ruvkun, G. Inactivation of Conserved C. elegans Genes Engages Pathogen- and Xenobiotic-Associated Defenses. Cell 2012, 149, 452–466. [Google Scholar] [CrossRef] [Green Version]
- Dunbar, T.L.; Yan, Z.; Balla, K.M.; Smelkinson, M.G.; Troemel, E.R. C. elegans Detects Pathogen-Induced Translational Inhibition to Activate Immune Signaling. Cell Host Microbe 2012, 11, 375–386. [Google Scholar] [CrossRef] [Green Version]
- McEwan, D.L.; Kirienko, N.V.; Ausubel, F.M. Host Translational Inhibition by Pseudomonas aeruginosa Exotoxin A Triggers an Immune Response in Caenorhabditis elegans. Cell Host Microbe 2012, 11, 364–374. [Google Scholar] [CrossRef] [Green Version]
- Chakrabarti, S.; Liehl, P.; Buchon, N.; Lemaitre, B. Infection-Induced Host Translational Blockage Inhibits Immune Responses and Epithelial Renewal in the Drosophila Gut. Cell Host Microbe 2012, 12, 60–70. [Google Scholar] [CrossRef] [Green Version]
- Even, N.; Devaud, J.-M.; Barron, A. General Stress Responses in the Honey Bee. Insects 2012, 3, 1271–1298. [Google Scholar] [CrossRef] [Green Version]
- López-Uribe, M.M.; Ricigliano, V.A.; Simone-Finstrom, M. Defining Pollinator Health: Assessing Bee Ecological, Genetic, and Physiological Factors at the Individual, Colony, and Population Levels. Annu. Rev. Anim. Biosci. 2019, 8, 269–294. [Google Scholar] [CrossRef] [PubMed]
- Brutscher, L.M.; Daughenbaugh, K.F.; Flenniken, M.L. Virus and DsRNA-Triggered Transcriptional Responses Reveal Key Components of Honey Bee Antiviral Defense. Sci. Rep. 2017, 7, 6448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McMenamin, A.; Daughenbaugh, K.; Parekh, F.; Pizzorno, M.; Flenniken, M. Honey Bee and Bumble Bee Antiviral Defense. Viruses 2018, 10, 395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cho, R.M.; Kogan, H.V.; Elikan, A.B.; Snow, J.W. Paromomycin Reduces Vairimorpha (Nosema) ceranae Infection in Honey Bees but Perturbs Microbiome Levels and Midgut Cell Function. Microorganisms 2022, 10, 1107. https://doi.org/10.3390/microorganisms10061107
Cho RM, Kogan HV, Elikan AB, Snow JW. Paromomycin Reduces Vairimorpha (Nosema) ceranae Infection in Honey Bees but Perturbs Microbiome Levels and Midgut Cell Function. Microorganisms. 2022; 10(6):1107. https://doi.org/10.3390/microorganisms10061107
Chicago/Turabian StyleCho, Rachel M., Helen V. Kogan, Annabelle B. Elikan, and Jonathan W. Snow. 2022. "Paromomycin Reduces Vairimorpha (Nosema) ceranae Infection in Honey Bees but Perturbs Microbiome Levels and Midgut Cell Function" Microorganisms 10, no. 6: 1107. https://doi.org/10.3390/microorganisms10061107
APA StyleCho, R. M., Kogan, H. V., Elikan, A. B., & Snow, J. W. (2022). Paromomycin Reduces Vairimorpha (Nosema) ceranae Infection in Honey Bees but Perturbs Microbiome Levels and Midgut Cell Function. Microorganisms, 10(6), 1107. https://doi.org/10.3390/microorganisms10061107





