RNase III Participates in the Adaptation to Temperature Shock and Oxidative Stress in Escherichia coli
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Growth Conditions
2.2. Promoter Replacement and Strain Construction
2.3. RNA Extraction and Northern Blot Analysis
2.4. Western Blot
2.5. In Vitro Processing by RNase III
2.6. Data Analysis
3. Results
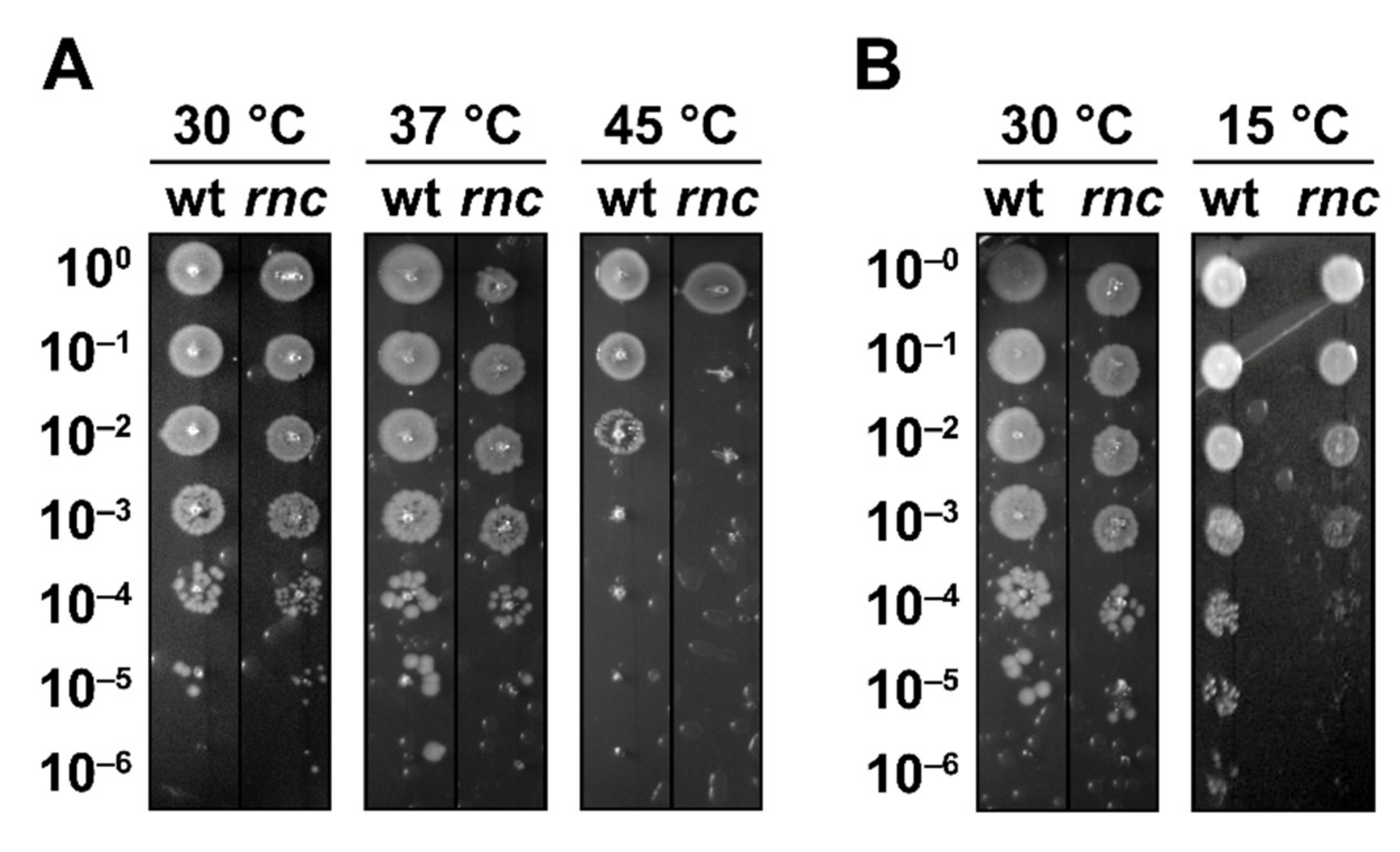
3.1. Survival under Extreme Temperatures Depends on RNase III
3.2. RNase III Inactivation Increases Sensitivity to Hydrogen Peroxide
3.3. The Induction of rpoH at High Temperature Depends on RNase III
3.4. RNase III and PNPase Stabilize rpoH mRNA during a Heat Shock
3.5. RNase III Acts Independently from Transcription Factors CRP and CytR
3.6. Induction of Three Genes of the RpoH Regulon Is Defective in the rnc Mutant
3.7. RNase III Positively Regulates sodA Expression
3.8. RNase III and PNPase Stabilize sodA mRNA
3.9. Transcriptional and Post-Transcriptional Regulation of the sodA Gene by RNase III
3.10. Overexpression of SodA in the rnc Mutant Supports Growth during Oxidative Stress
4. Discussion
4.1. Role of RNase III in Thermotolerance
4.2. Importance of RNase III in the Positive Regulation of Gene Expression
4.3. RNase III Is a General Stress Response Regulator and a Potential Target to Control Bacterial Virulence
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hör, J.; Matera, G.; Vogel, J.; Gottesman, S.; Storz, G. Trans-Acting Small RNAs and Their Effects on Gene Expression in Escherichia coli and Salmonella enterica. EcoSal Plus 2020, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kavita, K.; de Mets, F.; Gottesman, S. New aspects of RNA-based regulation by Hfq and its partner sRNAs. Curr. Opin. Microbiol. 2018, 42, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Holmqvist, E.; Vogel, J. RNA-binding proteins in bacteria. Nat. Rev. Microbiol. 2018, 16, 601–615. [Google Scholar] [CrossRef] [PubMed]
- Trinquier, A.; Durand, S.; Braun, F.; Condon, C. Regulation of RNA processing and degradation in bacteria. Biochim. Biophys. Acta Gene Regul. Mech. 2020, 1863, 194505. [Google Scholar] [CrossRef]
- Lejars, M.; Kobayashi, A.; Hajnsdorf, E. RNase III, Ribosome Biogenesis and Beyond. Microorganisms 2021, 9, 2608. [Google Scholar] [CrossRef]
- Nicholson, A.W. Ribonuclease III mechanisms of double-stranded RNA cleavage. WIREs RNA 2014, 5, 31–48. [Google Scholar] [CrossRef]
- Court, D.L.; Gan, J.; Liang, Y.H.; Shaw, G.X.; Tropea, J.E.; Costantino, N.; Waugh, D.S.; Ji, X. RNase III: Genetics and function; structure and mechanism. Annu. Rev. Genet. 2013, 47, 405–431. [Google Scholar] [CrossRef]
- Lee, D.; Shin, C. Emerging roles of DROSHA beyond primary microRNA processing. RNA Biol. 2018, 15, 186–193. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Deutscher, M.P. Exoribonucleases and Endoribonucleases. EcoSal Plus 2004, 1. [Google Scholar] [CrossRef]
- Gegenheimer, P.; Apirion, D. Precursors to 16S and 23S ribosomal RNA from a ribonuclear III-strain of Escherichia coli contain intact RNase III processing sites. Nucleic Acids Res. 1980, 8, 1873–1891. [Google Scholar] [CrossRef] [Green Version]
- King, T.C.; Schlessinger, D. S1 nuclease mapping analysis of ribosomal RNA processing in wild type and processing deficient Escherichia coli. J. Biol. Chem. 1983, 258, 12034–12042. [Google Scholar] [CrossRef]
- Srivastava, R.K.; Miczak, A.; Apirion, D. Maturation of precursor 10Sa RNA in Escherichia coli is a two-step process: The first reaction is catalyzed by RNase III in presence of Mn2+. Biochimie 1990, 72, 791–802. [Google Scholar] [CrossRef]
- Srivastava, R.A.; Srivastava, N.; Apirion, D. Characterization of the RNA processing enzyme RNase III from wild type and overexpressing Escherichia coli cells in processing natural RNA substrates. Int. J. Biochem. 1992, 24, 737–749. [Google Scholar] [CrossRef]
- Robertson, H.D.; Dunn, J.J. Ribonucleic acid processing activity of Escherichia coli Ribonuclease III. J. Biol. Chem. 1975, 250, 3050–3056. [Google Scholar] [CrossRef]
- Kameyama, L.; Fernandez, L.; Court, D.L.; Guarneros, G. RNase lll activation of bacteriophage λ N synthesis. Mol. Microbiol. 1991, 5, 2953–2963. [Google Scholar] [CrossRef]
- Bardwell, J.C.; Régnier, P.; Chen, S.M.; Nakamura, Y.; Grunberg-Manago, M.; Court, D.L. Autoregulation of RNase III operon by mRNA processing. EMBO J. 1989, 8, 3401–3407. [Google Scholar] [CrossRef]
- Takata, R.; Izuhara, M.; Planta, R.J. Differential degradation of the Escherichia coli polynucleotide phosphorylase mRNA. Nucleic Acids Res. 1989, 17, 7441–7451. [Google Scholar] [CrossRef] [Green Version]
- Portier, C.; Dondon, L.; Grunberg-Manago, M.; Régnier, P. The first step in the functional inactivation of the Escherichia coli polynucleotide phosphorylase messenger is a ribonuclease III processing at the 5′ end. EMBO J. 1987, 6, 2165–2170. [Google Scholar] [CrossRef]
- Barry, G.; Squires, C.; Squires, C.L. Attenuation and processing of RNA from the rplJL-rpoBC transcription unit of Escherichia coli. Proc. Natl. Acad. Sci. USA 1980, 77, 3331–3335. [Google Scholar] [CrossRef] [Green Version]
- Régnier, P.; Portier, C. Initiation, attenuation and RNase III processing of transcripts from the Escherichia coli operon encoding ribosomal protein S15 and polynucleotide phosphorylase. J. Mol. Biol. 1986, 187, 23–32. [Google Scholar] [CrossRef]
- Régnier, P.; Grunberg-Manago, M. RNase III cleavages in non-coding leaders of Escherichia coli transcripts control mRNA stability and genetic expression. Biochimie 1990, 72, 825–834. [Google Scholar] [CrossRef]
- Hajnsdorf, E.; Carpousis, A.J.; Regnier, P. Nucleolytic inactivation and degradation of the RNase III processed pnp message encoding polynucleotide phosphorylase of Escherichia coli. J. Mol. Biol. 1994, 239, 439–454. [Google Scholar] [CrossRef] [PubMed]
- Robert-Le Meur, M.; Portier, C.E. coli polynucleotide phosphorylase expression is autoregulated through an RNase III-dependent mechanism. EMBO J. 1992, 11, 2633–2641. [Google Scholar] [CrossRef] [PubMed]
- Aiso, T.; Kamiya, S.; Yonezawa, H.; Gamou, S. Overexpression of an antisense RNA, ArrS, increases the acid resistance of Escherichia coli. Microbiology 2014, 160, 954–961. [Google Scholar] [CrossRef] [Green Version]
- Aristarkhov, A.; Mikulskis, A.; Belasco, J.G.; Lin, E.C. Translation of the adhE transcript to produce ethanol dehydrogenase requires RNase III cleavage in Escherichia coli. J. Bacteriol. 1996, 178, 4327–4332. [Google Scholar] [CrossRef] [Green Version]
- Lee, M.; Joo, M.; Sim, M.; Sim, S.H.; Kim, H.L.; Lee, J.; Ryu, M.; Yeom, J.H.; Hahn, Y.; Ha, N.C.; et al. The coordinated action of RNase III and RNase G controls enolase expression in response to oxygen availability in Escherichia coli. Sci. Rep. 2019, 9, 17257. [Google Scholar] [CrossRef] [Green Version]
- Altuvia, S.; Kornitzer, D.; Kobi, S.; Oppenheim, A.B. Functional and structural elements of the mRNA of the cIII gene of bacteriophage lambda. J. Mol. Biol. 1991, 218, 723–733. [Google Scholar] [CrossRef]
- Dunn, J.J.; Studier, F.W. T7 Early RNAs and Escherichia coli Ribosomal RNAs are Cut from Large Precursor RNAs In Vivo by Ribonuclease III. Proc. Natl. Acad. Sci. USA 1973, 70, 3296–3300. [Google Scholar] [CrossRef] [Green Version]
- Sim, S.-H.; Yeom, J.-H.; Shin, C.; Song, W.-S.; Shin, E.; Kim, H.-M.; Cha, C.-J.; Han, S.H.; Ha, N.-C.; Kim, S.W.; et al. Escherichia coli ribonuclease III activity is downregulated by osmotic stress: Consequences for the degradation of bdm mRNA in biofilm formation. Mol. Microbiol. 2010, 75, 413–425. [Google Scholar] [CrossRef]
- Stead, M.B.; Marshburn, S.; Mohanty, B.K.; Mitra, J.; Castillo, L.P.; Ray, D.; van Bakel, H.; Hughes, T.R.; Kushner, S.R. Analysis of Escherichia coli RNase E and RNase III activity in vivo using tiling microarrays. Nucleic Acids Res. 2011, 39, 3188–3203. [Google Scholar] [CrossRef] [Green Version]
- Huang, L.; Deighan, P.; Jin, J.; Li, Y.; Cheung, H.-C.; Lee, E.; Mo, S.S.; Hoover, H.; Abubucker, S.; Finkel, N.; et al. Tombusvirus p19 Captures RNase III-Cleaved Double-Stranded RNAs Formed by Overlapping Sense and Antisense Transcripts in Escherichia coli. mBio 2020, 11, e00485-20. [Google Scholar] [CrossRef] [PubMed]
- Gordon, G.C.; Cameron, J.C.; Pfleger, B.F. RNA Sequencing Identifies New RNase III Cleavage Sites in Escherichia coli and Reveals Increased Regulation of mRNA. mBio 2017, 8, e00128-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Altuvia, Y.; Bar, A.; Reiss, N.; Karavani, E.; Argaman, L.; Margalit, H. In vivo cleavage rules and target repertoire of RNase III in Escherichia coli. Nucleic Acids Res. 2018, 46, 10380–10394. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sim, M.; Lim, B.; Sim, S.H.; Kim, D.; Jung, E.; Lee, Y.; Lee, K. Two tandem RNase III cleavage sites determine betT mRNA stability in response to osmotic stress in Escherichia coli. PLoS ONE 2014, 9, e100520. [Google Scholar] [CrossRef] [Green Version]
- Kavalchuk, K.; Madhusudan, S.; Schnetz, K. RNase III initiates rapid degradation of proU mRNA upon hypo-osmotic stress in Escherichia coli. RNA Biol. 2012, 9, 98–109. [Google Scholar] [CrossRef] [Green Version]
- Lim, B.; Lee, K. Stability of the Osmoregulated Promoter-Derived proP mRNA Is Posttranscriptionally Regulated by RNase III in Escherichia coli. J. Bacteriol. 2015, 197, 1297–1305. [Google Scholar] [CrossRef] [Green Version]
- Kim, K.S.; Manasherob, R.; Cohen, S.N. YmdB: A stress-responsive ribonuclease-binding regulator of E. coli RNase III activity. Genes Dev. 2008, 22, 3497–3508. [Google Scholar] [CrossRef] [Green Version]
- Apirion, D.; Neil, J.; Watson, N. Consequences of losing ribonuclease III on the Escherichia coli cell. Mol. Gen. Genet. 1976, 144, 185–190. [Google Scholar] [CrossRef]
- Song, W.; Kim, Y.-H.; Sim, S.-H.; Hwang, S.; Lee, J.-H.; Lee, Y.; Bae, J.; Hwang, J.; Lee, K. Antibiotic stress-induced modulation of the endoribonucleolytic activity of RNase III and RNase G confers resistance to aminoglycoside antibiotics in Escherichia coli. Nucleic Acids Res. 2014, 42, 4669–4681. [Google Scholar] [CrossRef]
- Apirion, D.; Watson, N. Ribonuclease III is involved in motility of Escherichia coli. J. Bacteriol. 1978, 133, 1543–1545. [Google Scholar] [CrossRef] [Green Version]
- Keseler, I.M.; Mackie, A.; Santos-Zavaleta, A.; Billington, R.; Bonavides-Martínez, C.; Caspi, R.; Fulcher, C.; Gama-Castro, S.; Kothari, A.; Krummenacker, M.; et al. The EcoCyc database: Reflecting new knowledge about Escherichia coli K-12. Nucleic Acids Res. 2016, 45, 543–550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baba, T.; Ara, T.; Hasegawa, M.; Takai, Y.; Okumura, Y.; Baba, M.; Datsenko, K.A.; Tomita, M.; Wanner, B.L.; Mori, H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol. Syst. Biol. 2006, 2, 2006.0008. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dasgupta, S.; Fernandez, L.; Kameyama, L.; Inada, T.; Nakamura, Y.; Pappas, A.; Court, D.L. Genetic uncoupling of the dsRNA-binding and RNA cleavage activities of the Escherichia coli endoribonuclease RNase III-the effect of dsRNA binding on gene expression. Mol. Microbiol. 1998, 28, 629–640. [Google Scholar] [CrossRef] [PubMed]
- Korea, C.-G.; Badouraly, R.; Prevost, M.-C.; Ghigo, J.-M.; Beloin, C. Escherichia coli K-12 possesses multiple cryptic but functional chaperone–usher fimbriae with distinct surface specificities. Environ. Microbiol. 2010, 12, 1957–1977. [Google Scholar] [CrossRef]
- Guillier, M.; Gottesman, S. Remodelling of the Escherichia coli outer membrane by two small regulatory RNAs. Mol. Microbiol. 2006, 59, 231–247. [Google Scholar] [CrossRef]
- Braun, F.; Hajnsdorf, E.; Regnier, P. Polynucleotide phosphorylase is required for the rapid degradation of the RNase E-processed rpsO mRNA of Escherichia coli devoid of its 3′ hairpin. Mol. Microbiol. 1996, 19, 997–1005. [Google Scholar] [CrossRef]
- Hajnsdorf, E.; Regnier, P.E. coli rpsO mRNA decay: RNase E processing at the beginning of the coding sequence stimulates poly(A)-dependent degradation of the mRNA. J. Mol. Biol. 1999, 286, 1033–1043. [Google Scholar] [CrossRef]
- Hajnsdorf, E.; Regnier, P. Host factor Hfq of Escherichia coli stimulates elongation of poly(A) tails by poly(A) polymerase I. Proc. Natl. Acad. Sci. USA 2000, 97, 1501–1505. [Google Scholar] [CrossRef] [Green Version]
- Folichon, M.; Allemand, F.; Régnier, P.; Hajnsdorf, E. Stimulation of poly(A) synthesis by E. coli poly(A)polymerase I is correlated with Hfq binding to poly(A) tails. FEBS J. 2005, 272, 454–463. [Google Scholar] [CrossRef]
- Fontaine, F.; Gasiorowski, E.; Gracia, C.; Ballouche, M.; Caillet, J.; Marchais, A.; Hajnsdorf, E. The small RNA SraG participates in PNPase homeostasis. RNA 2016, 22, 1560–1573. [Google Scholar] [CrossRef] [Green Version]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Awano, N.; Inouye, M.; Phadtare, S. RNase Activity of Polynucleotide Phosphorylase Is Critical at Low Temperature in Escherichia coli and Is Complemented by RNase II. J. Bacteriol. 2008, 190, 5924–5933. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paudyal, S.; Alfonso-Prieto, M.; Carnevale, V.; Redhu, S.K.; Klein, M.L.; Nicholson, A.W. Combined computational and experimental analysis of a complex of ribonuclease III and the regulatory macrodomain protein, YmdB. Proteins 2015, 83, 459–472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Imlay, J.A. Oxidative Stress. EcoSal Plus 2009, 3. [Google Scholar] [CrossRef] [PubMed]
- Nonaka, G.; Blankschien, M.; Herman, C.; Gross, C.A.; Rhodius, V.A. Regulon and promoter analysis of the E. coli heat-shock factor, σ32, reveals a multifaceted cellular response to heat stress. Genes Dev. 2006, 20, 1776–1789. [Google Scholar] [CrossRef] [Green Version]
- Jenkins, D.E.; Auger, E.A.; Matin, A. Role of RpoH, a heat shock regulator protein, in Escherichia coli carbon starvation protein synthesis and survival. J. Bacteriol. 1991, 173, 1992–1996. [Google Scholar] [CrossRef] [Green Version]
- Guo, M.S.; Updegrove, T.B.; Gogol, E.B.; Shabalina, S.A.; Gross, C.A.; Storz, G. MicL, a new sigmaE-dependent sRNA, combats envelope stress by repressing synthesis of Lpp, the major outer membrane lipoprotein. Genes Dev. 2014, 28, 1620–1634. [Google Scholar] [CrossRef] [Green Version]
- Morita, M.T.; Tanaka, Y.; Kodama, T.S.; Kyogoku, Y.; Yanagi, H.; Yura, T. Translational induction of heat shock transcription factor sigma32: Evidence for a built-in RNA thermosensor. Genes Dev. 1999, 13, 655–665. [Google Scholar] [CrossRef] [Green Version]
- Morita, M.; Kanemori, M.; Yanagi, H.; Yura, T. Heat-Induced Synthesis of ς32 in Escherichia coli: Structural and Functional Dissection of rpoH mRNA Secondary Structure. J. Bacteriol. 1999, 181, 401–410. [Google Scholar] [CrossRef] [Green Version]
- Arsène, F.; Tomoyasu, T.; Bukau, B. The heat shock response of Escherichia coli. Int. J. Food Microbiol. 2000, 55, 3–9. [Google Scholar] [CrossRef]
- Erickson, J.W.; Vaughn, V.; Walter, W.A.; Neidhardt, F.C.; Gross, C.A. Regulation of the promoters and transcripts of rpoH, the Escherichia coli heat shock regulatory gene. Genes Dev. 1987, 1, 419–432. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Straus, D.B.; Walter, W.A.; Gross, C.A. The heat shock response of E. coli is regulated by changes in the concentration of σ32. Nature 1987, 329, 348–351. [Google Scholar] [CrossRef] [PubMed]
- Afonyushkin, T.; Večerek, B.; Moll, I.; Bläsi, U.; Kaberdin, V.R. Both RNase E and RNase III control the stability of sodB mRNA upon translational inhibition by the small regulatory RNA RyhB. Nucleic Acids Res. 2005, 33, 1678–1689. [Google Scholar] [CrossRef]
- Bianchi, A.A.; Baneyx, F. Hyperosmotic shock induces the σ32 and σE stress regulons of Escherichia coli. Mol. Microbiol. 1999, 34, 1029–1038. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taglicht, D.; Padan, E.; Oppenheim, A.B.; Schuldiner, S. An alkaline shift induces the heat shock response in Escherichia coli. J. Bacteriol. 1987, 169, 885–887. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- VanBogelen, R.A.; Kelley, P.M.; Neidhardt, F.C. Differential induction of heat shock, SOS, and oxidation stress regulons and accumulation of nucleotides in Escherichia coli. J. Bacteriol. 1987, 169, 26–32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lüders, S.; Fallet, C.; Franco-Lara, E. Proteome analysis of the Escherichia coli heat shock response under steady-state conditions. Proteome Sci. 2009, 7, 36. [Google Scholar] [CrossRef] [Green Version]
- Riehle, M.M.; Bennett, A.F.; Long, A.D. Changes in Gene Expression Following High-Temperature Adaptation in Experimentally Evolved Populations of E. coli. Physiol. Biochem. Zool. 2005, 78, 299–315. [Google Scholar] [CrossRef] [Green Version]
- Rasouly, A.; Ron, E.Z. Interplay between the heat shock response and translation in Escherichia coli. Res. Microbiol. 2009, 160, 288–296. [Google Scholar] [CrossRef]
- Al Refaii, A.; Alix, J.-H. Ribosome biogenesis is temperature-dependent and delayed in Escherichia coli lacking the chaperones DnaK or DnaJ. Mol. Microbiol. 2009, 71, 748–762. [Google Scholar] [CrossRef]
- Mayer, J.E.; Schweiger, M. RNase III is positively regulated by T7 protein kinase. J. Biol. Chem. 1983, 258, 5340–5343. [Google Scholar] [CrossRef]
- Kim, M.; Kim, M.; Kim, K.S. YmdB-mediated down-regulation of sucA inhibits biofilm formation and induces apramycin susceptibility in Escherichia coli. Biochem. Biophys. Res. Commun. 2017, 483, 252–257. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Qiao, M.; Meng, Q.; Qiao, J.; Wu, Y.; Guo, J.; Wang, X.; Li, J.; Zhang, X.; Cai, X. Listeria monocytogenes in RNase III rncS. Kafkas Univ. Vet. Fak. Derg. 2019, 26, 269–277. [Google Scholar] [CrossRef]
- Freire, P.; Amaral, J.D.; Santos, J.M.; Arraiano, C.M. Adaptation to carbon starvation: RNase III ensures normal expression levels of bolA1p mRNA and σS. Biochimie 2006, 88, 341–346. [Google Scholar] [CrossRef] [PubMed]
- Basineni, S.R.; Madhugiri, R.; Kolmsee, T.; Hengge, R.; Klug, G. The influence of Hfq and ribonucleases on the stability of the small non-coding RNA OxyS and its target rpoS in E. coli is growth phase dependent. RNA Biol. 2009, 6, 584–594. [Google Scholar] [CrossRef] [Green Version]
- Resch, A.; Afonyushkin, T.; Lombo, T.B.; McDowall, K.J.; Bläsi, U.; Kaberdin, V.R. Translational activation by the noncoding RNA DsrA involves alternative RNase III processing in the rpoS 5′-leader. RNA 2008, 14, 454–459. [Google Scholar] [CrossRef] [Green Version]
- Lioliou, E.; Sharma, C.M.; Caldelari, I.; Helfer, A.C.; Fechter, P.; Vandenesch, F.; Vogel, J.; Romby, P. Global regulatory functions of the Staphylococcus aureus endoribonuclease III in gene expression. PLoS Genet. 2012, 8, e1002782. [Google Scholar] [CrossRef]
- Viegas, S.C.; Mil-Homens, D.; Fialho, A.M.; Arraiano, C.M. The Virulence of Salmonella enterica Serovar Typhimurium in the Insect Model Galleria mellonella Is Impaired by Mutations in RNase E and RNase III. Appl. Environ. Microbiol. 2013, 79, 6124–6133. [Google Scholar] [CrossRef] [Green Version]
- Salze, M.; Muller, C.; Bernay, B.; Hartke, A.; Clamens, T.; Lesouhaitier, O.; Rincé, A. Study of key RNA metabolism proteins in Enterococcus faecalis. RNA Biol. 2020, 17, 794–804. [Google Scholar] [CrossRef]
- Goldblum, K.; Apirion, D. Inactivation of the ribonucleic acid-processing enzyme ribonuclease E blocks cell division. J. Bacteriol. 1981, 146, 128–132. [Google Scholar]
- Regnier, P.; Hajnsdorf, E. Decay of mRNA encoding ribosomal protein S15 of Escherichia coli is initiated by an RNase E-dependent endonucleolytic cleavage that removes the 3' stabilizing stem and loop structure. J. Mol. Biol. 1991, 217, 283–292. [Google Scholar] [PubMed]
- Reuven, N.B.; Deutscher, M.P. Multiple exoribonucleases are required for the 3′ processing of Escherichia coli tRNA precursors in vivo. Faseb J. 1993, 7, 143–148. [Google Scholar] [PubMed]
- Korshunov, S.; Imlay, J.A. Two sources of endogenous hydrogen peroxide in Escherichia coli. Mol. Microbiol. 2010, 75, 1389–1401. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lorenz, R.; Bernhart, S.H.; Höner zu Siederdissen, C.; Tafer, H.; Flamm, C.; Stadler, P.F.; Hofacker, I.L. ViennaRNA Package 2.0. Algorithms Mol. Biol. 2011, 6, 26. [Google Scholar] [CrossRef]







| Transcripts | Temperature | wt | rnc | pnp | rnc pnp |
|---|---|---|---|---|---|
| rpoH | 45 °C | 4.01 ± 0.04 | 3.39 ± 0.08 | 3.14 ± 0.05 | 2.06 ± 0.09 |
| sodA | 37 °C | 4.72 ± 0.09 | 2.57 ± 0.05 | 2.02 ± 0.10 | 1.50 ± 0.13 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lejars, M.; Hajnsdorf, E. RNase III Participates in the Adaptation to Temperature Shock and Oxidative Stress in Escherichia coli. Microorganisms 2022, 10, 699. https://doi.org/10.3390/microorganisms10040699
Lejars M, Hajnsdorf E. RNase III Participates in the Adaptation to Temperature Shock and Oxidative Stress in Escherichia coli. Microorganisms. 2022; 10(4):699. https://doi.org/10.3390/microorganisms10040699
Chicago/Turabian StyleLejars, Maxence, and Eliane Hajnsdorf. 2022. "RNase III Participates in the Adaptation to Temperature Shock and Oxidative Stress in Escherichia coli" Microorganisms 10, no. 4: 699. https://doi.org/10.3390/microorganisms10040699
APA StyleLejars, M., & Hajnsdorf, E. (2022). RNase III Participates in the Adaptation to Temperature Shock and Oxidative Stress in Escherichia coli. Microorganisms, 10(4), 699. https://doi.org/10.3390/microorganisms10040699






