Putrescine Production by Latilactobacillus curvatus KP 3-4 Isolated from Fermented Foods
Abstract
:1. Introduction
2. Materials and Methods
2.1. Mice
2.2. Bacterial Strains
2.3. Gnotobiotic Mouse Experiment
2.4. Isolation of Lactic Acid Bacteria from Food
2.5. Screening for Polyamine-Producing Bacteria
2.6. Culture of Representative Probiotic Lactic Acid Bacteria and L. curvatus
2.7. Culturing of L. curvatus KP 3-4 in Medium Supplemented with Ornithine
2.8. Measurement of Polyamine Concentration in Culture Supernatant by HPLC
2.9. Measurement of Concentration of Intracellular Polyamines by HPLC
2.10. Whole Genome Sequencing and De Novo Assembly of L. curvatus KP 3-4
2.11. Conservation Analysis of Polyamine Biosynthetic Proteins and Transporters
2.12. Measurement of Polyamine Concentration in Mice Feces
2.13. Statistical Analysis
3. Results
3.1. Latilactobacillus curvatus KP 3-4 Derived from Kabura-Zushi Produces Putrescine
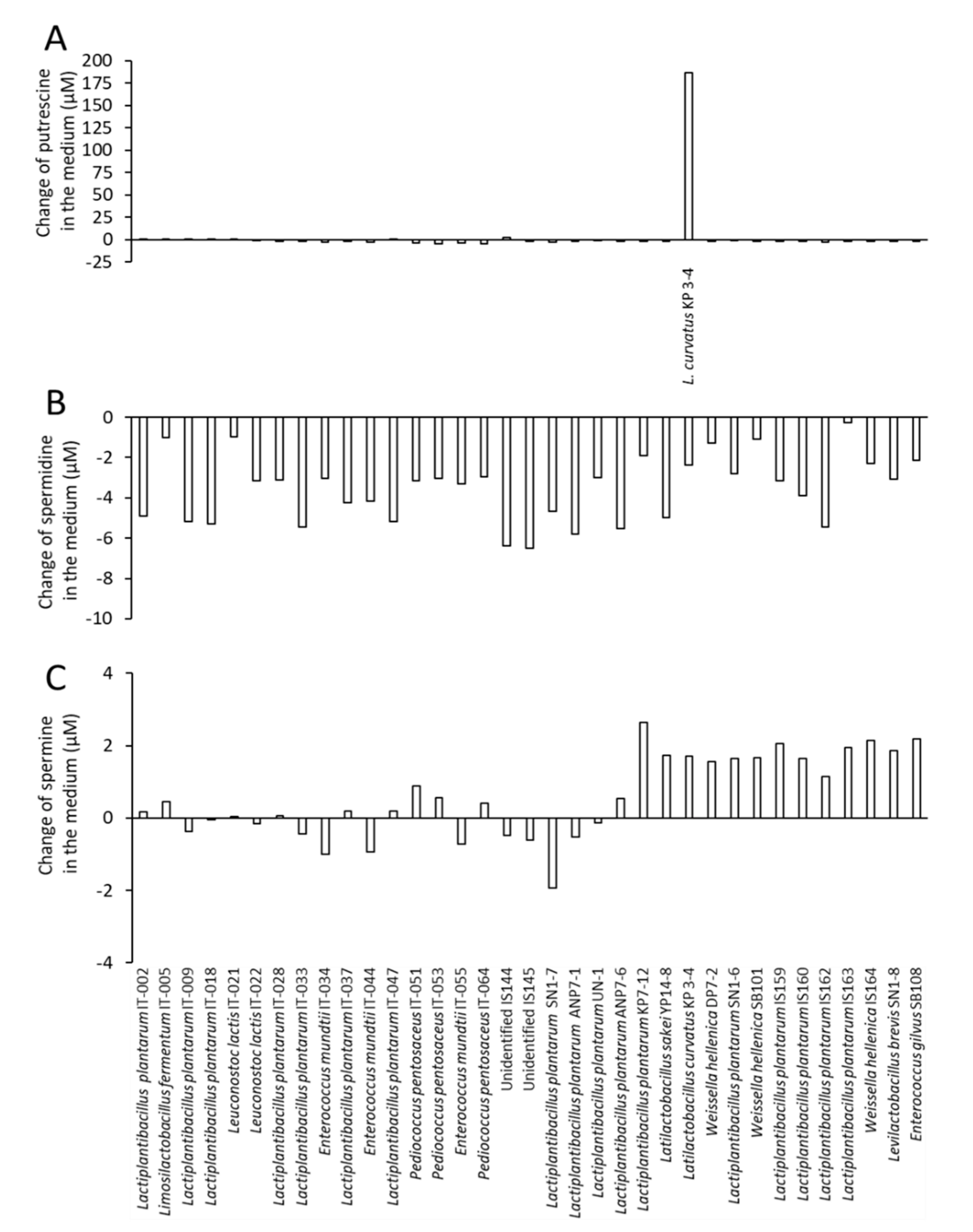
3.2. Comparison of Putrescine Productivity between L. curvatus KP 3-4 and Probiotic Lactic Acid Bacteria



3.3. Analyses of the Mechanism of Putrescine Production by L. curvatus KP 3-4

3.4. Oral Administration of L. curvatus KP 3-4 Increases Polyamine Levels in Feces of Mice

4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Soda, K.; Dobashi, Y.; Kano, Y.; Tsujinaka, S.; Konishi, F. Polyamine-rich food decreases age-associated pathology and mortality in aged mice. Exp. Gerontol. 2009, 44, 727–732. [Google Scholar] [CrossRef] [PubMed]
- Eisenberg, T.; Knauer, H.; Schauer, A.; Büttner, S.; Ruckenstuhl, C.; Carmona-Gutierrez, D.; Ring, J.; Schroeder, S.; Magnes, C.; Antonacci, L.; et al. Induction of autophagy by spermidine promotes longevity. Nat. Cell Biol. 2009, 11, 1305–1314. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, M.; Kurihara, S.; Kibe, R.; Ashida, H.; Benno, Y. Longevity in Mice Is Promoted by Probiotic-Induced Suppression of Colonic Senescence Dependent on Upregulation of Gut Bacterial Polyamine Production. PLoS ONE 2011, 6, e23652. [Google Scholar] [CrossRef] [Green Version]
- Soda, K.; Kano, Y.; Chiba, F.; Koizumi, K.; Miyaki, Y. Increased Polyamine Intake Inhibits Age-Associated Alteration in Global DNA Methylation and 1,2-Dimethylhydrazine-Induced Tumorigenesis. PLoS ONE 2013, 8, e64357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kibe, R.; Kurihara, S.; Sakai, Y.; Suzuki, H.; Ooga, T.; Sawaki, E.; Muramatsu, K.; Nakamura, A.; Yamashita, A.; Kitada, Y.; et al. Upregulation of colonic luminal polyamines produced by intestinal microbiota delays senescence in mice. Sci. Rep. 2014, 4, 4548. [Google Scholar] [CrossRef] [PubMed]
- Gupta, V.K.; Scheunemann, L.; Eisenberg, T.; Mertel, S.; Bhukel, A.; Koemans, T.S.; Kramer, J.M.; Liu, K.S.Y.; Schroeder, S.; Stunnenberg, H.G.; et al. Restoring polyamines protects from age-induced memory impairment in an autophagy-dependent manner. Nat. Neurosci. 2013, 16, 1453–1460. [Google Scholar] [CrossRef]
- Eisenberg, T.; Abdellatif, M.; Schroeder, S.; Primessnig, U.; Stekovic, S.; Pendl, T.; Harger, A.; Schipke, J.; Zimmermann, A.; Schmidt, A.; et al. Cardioprotection and lifespan extension by the natural polyamine spermidine. Nat. Med. 2016, 22, 1428–1438. [Google Scholar] [CrossRef]
- Casero, R.A.; Murray Stewart, T.; Pegg, A.E. Polyamine metabolism and cancer: Treatments, challenges and opportunities. Nat. Rev. Cancer 2018, 18, 681–695. [Google Scholar] [CrossRef]
- Sturgill, G.; Rather, P.N. Evidence that putrescine acts as an extracellular signal required for swarming in Proteus mirabilis. Mol. Microbiol. 2004, 51, 437–446. [Google Scholar] [CrossRef]
- Nishimura, K.; Shiina, R.; Kashiwagi, K.; Igarashi, K. Decrease in Polyamines with Aging and Their Ingestion from Food and Drink. J. Biochem. 2006, 139, 81–90. [Google Scholar] [CrossRef]
- Wirth, M.; Benson, G.; Schwarz, C.; Köbe, T.; Grittner, U.; Schmitz, D.; Sigrist, S.J.; Bohlken, J.; Stekovic, S.; Madeo, F.; et al. The effect of spermidine on memory performance in older adults at risk for dementia: A randomized controlled trial. Cortex 2018, 109, 181–188. [Google Scholar] [CrossRef] [PubMed]
- Ralph, A.E.K.; Bardocz, S. Polyamine content of the human diet. In Polyamines in Health and Nutrition; Kluwer Academic Publishers: London, UK, 1999; p. 15. [Google Scholar]
- Zoumas-Morse, C.; Rock, C.L.; Quintana, E.L.; Neuhouser, M.L.; Gerner, E.W.; Meyskens, F.L. Development of a Polyamine Database for Assessing Dietary Intake. J. Am. Diet. Assoc. 2007, 107, 1024–1027. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buyukuslu, N.; Hizli, H.; Esin, K.; Garipagaoglu, M. A Cross-Sectional Study: Nutritional Polyamines in Frequently Consumed Foods of the Turkish Population. Foods 2014, 3, 541–557. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nishibori, N.; Fujihara, S.; Akatuki, T. Amounts of polyamines in foods in Japan and intake by Japanese. Food Chem. 2007, 100, 491–497. [Google Scholar] [CrossRef]
- Ali, M.A.; Poortvliet, E.; Strömberg, R.; Yngve, A. Polyamines: Total daily intake in adolescents compared to the intake estimated from the Swedish Nutrition Recommendations Objectified (SNO). Food Nutr. Res. 2011, 55, 5455. [Google Scholar] [CrossRef] [Green Version]
- Novella-Rodríguez, S.; Veciana-Nogués, M.T.; Izquierdo-Pulido, M.; Vidal-Carou, M.C. Distribution of Biogenic Amines and Polyamines in Cheese. J. Food Sci. 2003, 68, 750–756. [Google Scholar] [CrossRef]
- Landete, J.M.; Ferrer, S.; Polo, L.; Pardo, I. Biogenic Amines in Wines from Three Spanish Regions. J. Agric. Food Chem. 2005, 53, 1119–1124. [Google Scholar] [CrossRef]
- Sugiyama, Y.; Nara, M.; Sakanaka, M.; Kitakata, A.; Okuda, S.; Kurihara, S. Analysis of polyamine biosynthetic- and transport ability of human indigenous Bifidobacterium. Biosci. Biotechnol. Biochem. 2018, 82, 1606–1614. [Google Scholar] [CrossRef]
- Bover-Cid, S.; Holzapfel, W.H. Improved screening procedure for biogenic amine production by lactic acid bacteria. Int. J. Food Microbiol. 1999, 53, 33–41. [Google Scholar] [CrossRef]
- Li, L.; Zou, D.; Ruan, L.; Wen, Z.; Chen, S.; Xu, L.; Wei, X. Evaluation of the Biogenic Amines and Microbial Contribution in Traditional Chinese Sausages. Front. Microbiol. 2019, 10, 872. [Google Scholar] [CrossRef] [Green Version]
- Arena, M.E.; Manca de Nadra, M.C. Biogenic amine production by Lactobacillus. J. Appl. Microbiol. 2001, 90, 158–162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barbieri, F.; Montanari, C.; Gardini, F.; Tabanelli, G. Biogenic Amine Production by Lactic Acid Bacteria: A Review. Foods 2019, 8, 17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lonvaud-Funel, A. Biogenic amines in wines: Role of lactic acid bacteria. FEMS Microbiol. Lett. 2001, 199, 9–13. [Google Scholar] [CrossRef] [PubMed]
- Shortt, C. The probiotic century: Historical and current perspectives. Trends Food Sci. Technol. 1999, 10, 411–417. [Google Scholar] [CrossRef]
- Kiyohara, M.; Koyanagi, T.; Matsui, H.; Yamamoto, K.; Take, H.; Katsuyama, Y.; Tsuji, A.; Miyamae, H.; Kondo, T.; Nakamura, S.; et al. Changes in Microbiota Population during Fermentation of Narezushi as Revealed by Pyrosequencing Analysis. Biosci. Biotechnol. Biochem. 2012, 76, 48–52. [Google Scholar] [CrossRef] [Green Version]
- Barla, F.; Koyanagi, T.; Tokuda, N.; Matsui, H.; Katayama, T.; Kumagai, H.; Michihata, T.; Sasaki, T.; Tsuji, A.; Enomoto, T. The γ-aminobutyric acid-producing ability under low pH conditions of lactic acid bacteria isolated from traditional fermented foods of Ishikawa Prefecture, Japan, with a strong ability to produce ACE-inhibitory peptides. Biotechnol. Rep. 2016, 10, 105–110. [Google Scholar] [CrossRef] [Green Version]
- Gotoh, A.; Nara, M.; Sugiyama, Y.; Sakanaka, M.; Yachi, H.; Kitakata, A.; Nakagawa, A.; Minami, H.; Okuda, S.; Katoh, T.; et al. Use of Gifu Anaerobic Medium for culturing 32 dominant species of human gut microbes and its evaluation based on short-chain fatty acids fermentation profiles. Biosci. Biotechnol. Biochem. 2017, 81, 2009–2017. [Google Scholar] [CrossRef] [Green Version]
- Sakanaka, M.; Sugiyama, Y.; Kitakata, A.; Katayama, T.; Kurihara, S. Carboxyspermidine decarboxylase of the prominent intestinal microbiota species Bacteroides thetaiotaomicron is required for spermidine biosynthesis and contributes to normal growth. Amino Acids 2016, 48, 2443–2451. [Google Scholar] [CrossRef]
- Koren, S.; Walenz, B.P.; Berlin, K.; Miller, J.R.; Bergman, N.H.; Phillippy, A.M. Canu: Scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017, 27, 722–736. [Google Scholar] [CrossRef] [Green Version]
- Sugiyama, Y.; Nara, M.; Sakanaka, M.; Gotoh, A.; Kitakata, A.; Okuda, S.; Kurihara, S. Comprehensive analysis of polyamine transport and biosynthesis in the dominant human gut bacteria: Potential presence of novel polyamine metabolism and transport genes. Int. J. Biochem. Cell Biol. 2017, 93, 52–61. [Google Scholar] [CrossRef]
- Nakamura, A.; Kurihara, S.; Takahashi, D.; Ohashi, W.; Nakamura, Y.; Kimura, S.; Onuki, M.; Kume, A.; Sasazawa, Y.; Furusawa, Y.; et al. Symbiotic polyamine metabolism regulates epithelial proliferation and macrophage differentiation in the colon. Nat. Commun. 2021, 12, 2105. [Google Scholar] [CrossRef] [PubMed]
| Strains | Isolated Source | Isolation Method (A–C), GenBank Accession Number, or Reference |
|---|---|---|
| Lactiplantibacillus plantarum IT-002 | cabbage | A |
| Limosilactobacillus fermentum IT-005 | spinach | A |
| Lactiplantibacillus plantarum IT-009 | mandarin orange | A |
| Lactiplantibacillus plantarum IT-018 | cabbage | A |
| Leuconostoc lactis IT-021 | spinach | A |
| Leuconostoc lactis IT-022 | spinach | A |
| Lactiplantibacillus plantarum IT-028 | mandarin orange | A |
| Lactiplantibacillus plantarum IT-033 | lettuce | A |
| Enterococcus mundtii IT-034 | lettuce | A |
| Lactiplantibacillus plantarum IT-037 | onion | A |
| Enterococcus mundtii IT-044 | lettuce | A |
| Lactiplantibacillus plantarum IT-047 | onion | A |
| Pediococcus pentosaceus IT-051 | mandarin orange | A |
| Pediococcus pentosaceus IT-053 | broccoli | A |
| Enterococcus mundtii IT-055 | broccoli | A |
| Pediococcus pentosaceus IT-064 | mandarin orange | A |
| Unidentified IS144 | Japanese Greens, Mibuna | A |
| Unidentified IS145 | Japanese Greens, Mibuna | A |
| Lactiplantibacillus plantarum SN1-7 | Nare-zushi using mackerel | B |
| Lactiplantibacillus plantarum ANP7-1 | Nare-zushi using horse-mackerel | AB666313, [26] |
| Lactiplantibacillus plantarum UN-1 | Nare-zushi using Japanese dace | B |
| Lactiplantibacillus plantarum ANP7-6 | Nare-zushi using horse-mackerel | AB666315, [26,27] |
| Lactiplantibacillus plantarum KP7-12 | Kabura-zushi | B |
| Latilactobacillus sakei YP14-8 | Yamahai (yeast starter) | B |
| Latilactobacillus curvatus KP 3-4 | Kabura-zushi | B |
| Weissella hellenica DP7-2 | Radish sushi | B |
| Lactiplantibacillus plantarum SN1-6 | Nare-zushi using mackerel | B |
| Weissella hellenica SB101 | Squid pickled in malted rice | LC136890, [27] |
| Lactiplantibacillus plantarum IS159 | Nare-zushi using mackerel | C |
| Lactiplantibacillus plantarum IS160 | Nare-zushi using mackerel | C |
| Lactiplantibacillus plantarum IS162 | Nare-zushi using mackerel | C |
| Lactiplantibacillus plantarum IS163 | Nare-zushi using mackerel | C |
| Weissella hellenica IS164 | Nare-zushi using mackerel | C |
| Levilactobacillus brevis SN1-8 | Nare-zushi using mackerel | B |
| Enterococcus gilvus SB108 | Nare-zushi using horse-mackerel | B |
| Species | Reference |
|---|---|
| Lacticaseibacillus casei JCM1134T | GCF_000829055.1_ASM82905v1_genomic.fna |
| Lacticaseibacillus rhamnosus ATCC 7469T | GCF_007990855.1_ASM799085v1_genomic.fna |
| Limosilactobacillus reuteri subsp. reuteri JCM 1112T | GCF_000010005.1_ASM1000v1_genomic.fna |
| Lactiplantibacillus plantarum subsp. plantarum JCM 1149T | GCF_014131735.1_ASM1413173v1_genomic.fna |
| Ligilactobacillus salivarius JCM 1231T | GCF_001435955.1_ASM143595v1_genomic.fna |
| Lactobacillus johnsonii JCM 2012T | GCF_001433975.1_ASM143397v1_genomic.fna |
| Lactobacillus acidophilus JCM 1132T | GCF_001591845.1_ASM159184v1_genomic.fna |
| Lactobacillus crispatus JCM 1185T | GCA_001311685.1_ASM131168v1_genomic.fna |
| Latilactobacillus curvatus JCM1096T | GCF_004101845.1_ASM410184v1_genomic.fna |
| Latilactobacillus curvatus JCM1091 | No assembly |
| Strains | Source |
|---|---|
| Lacticaseibacillus casei JCM1134T | JCM |
| Lacticaseibacillus rhamnosus ATCC 7469T | ATCC |
| Limosilactobacillus reuteri subsp. reuteri JCM 1112T | JCM |
| Lactiplantibacillus plantarum subsp. plantarum JCM 1149T | JCM |
| Lactobacillus acidophilus JCM 1132T | JCM |
| Lactobacillus crispatus JCM 1185T | JCM |
| Ligilactobacillus salivarius JCM 1231T | JCM |
| Lactobacillus johnsonii JCM 2012T | JCM |
| Latilactobacillus curvatus JCM1096T | JCM |
| Latilactobacillus curvatus JCM1091 | JCM |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hirano, R.; Kume, A.; Nishiyama, C.; Honda, R.; Shirasawa, H.; Ling, Y.; Sugiyama, Y.; Nara, M.; Shimokawa, H.; Kawada, H.; et al. Putrescine Production by Latilactobacillus curvatus KP 3-4 Isolated from Fermented Foods. Microorganisms 2022, 10, 697. https://doi.org/10.3390/microorganisms10040697
Hirano R, Kume A, Nishiyama C, Honda R, Shirasawa H, Ling Y, Sugiyama Y, Nara M, Shimokawa H, Kawada H, et al. Putrescine Production by Latilactobacillus curvatus KP 3-4 Isolated from Fermented Foods. Microorganisms. 2022; 10(4):697. https://doi.org/10.3390/microorganisms10040697
Chicago/Turabian StyleHirano, Rika, Aiko Kume, Chisato Nishiyama, Ryosuke Honda, Hideto Shirasawa, Yiwei Ling, Yuta Sugiyama, Misaki Nara, Hiromi Shimokawa, Hiroki Kawada, and et al. 2022. "Putrescine Production by Latilactobacillus curvatus KP 3-4 Isolated from Fermented Foods" Microorganisms 10, no. 4: 697. https://doi.org/10.3390/microorganisms10040697






