Beyond the ABCs—Discovery of Three New Plasmid Types in Rhodobacterales (RepQ, RepY, RepW)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Growth Conditions
2.2. Genome Sequencing, Assembly and Annotation
2.3. Characterization and Comparison of Genomes, Phylogenetic Analyses and Data Handling
2.4. Functionality Test of RepQ, RepY and RepW
3. Results and Discussion
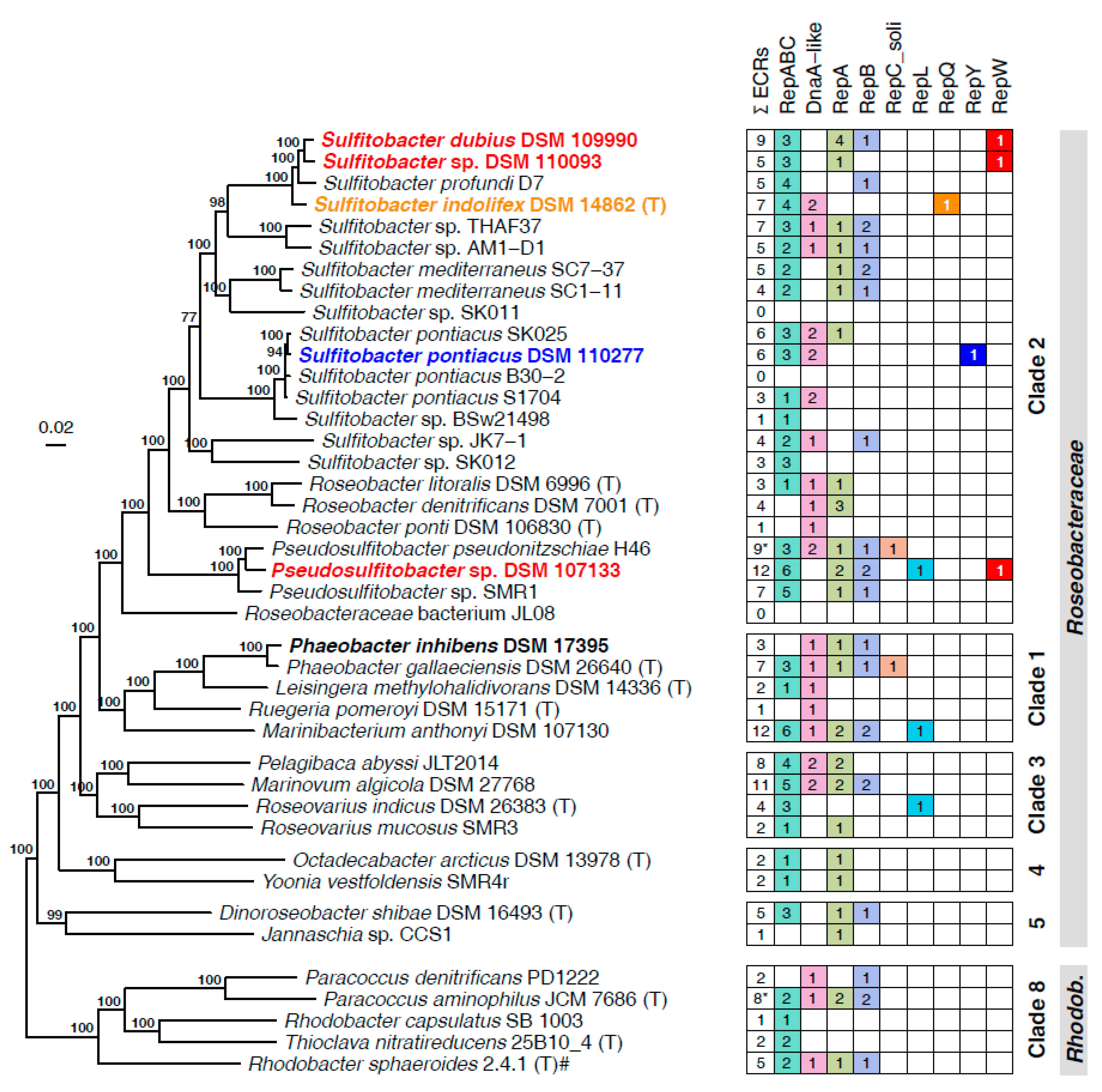
3.1. Genome Sequencing and Classification of Five (Pseudo)Sulfitobacter Strains
3.2. Characterization of Extrachromosomal Replicons
3.3. Identification of Novel Plasmid-Types (RepQ, RepY, RepW)
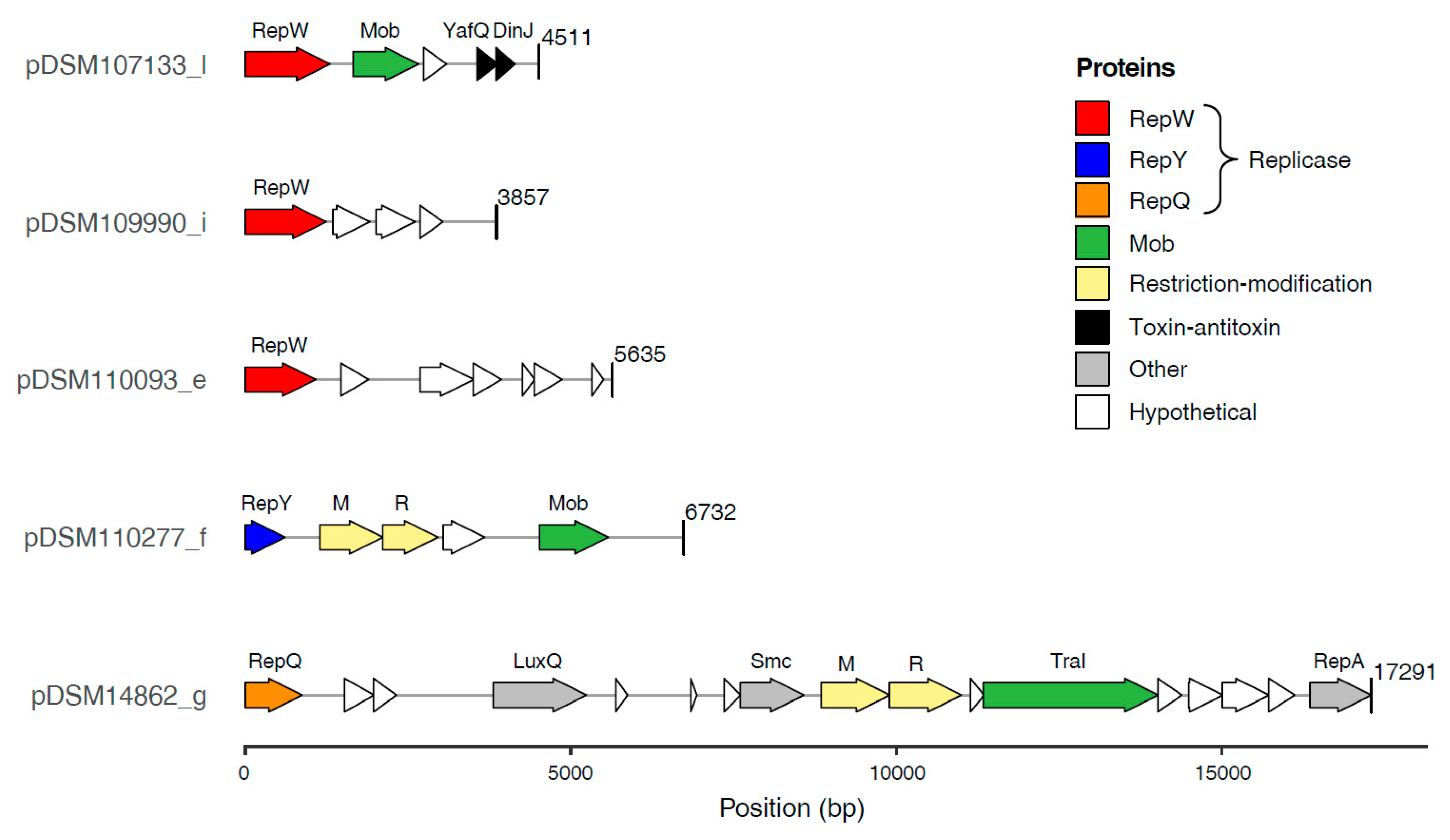
3.4. Characterization of RepQ, RepY and RepW-Type Plasmids
3.4.1. Functionality of the Three Novel Plasmid Replicases
3.4.2. Gene Content of the Novel Plasmids
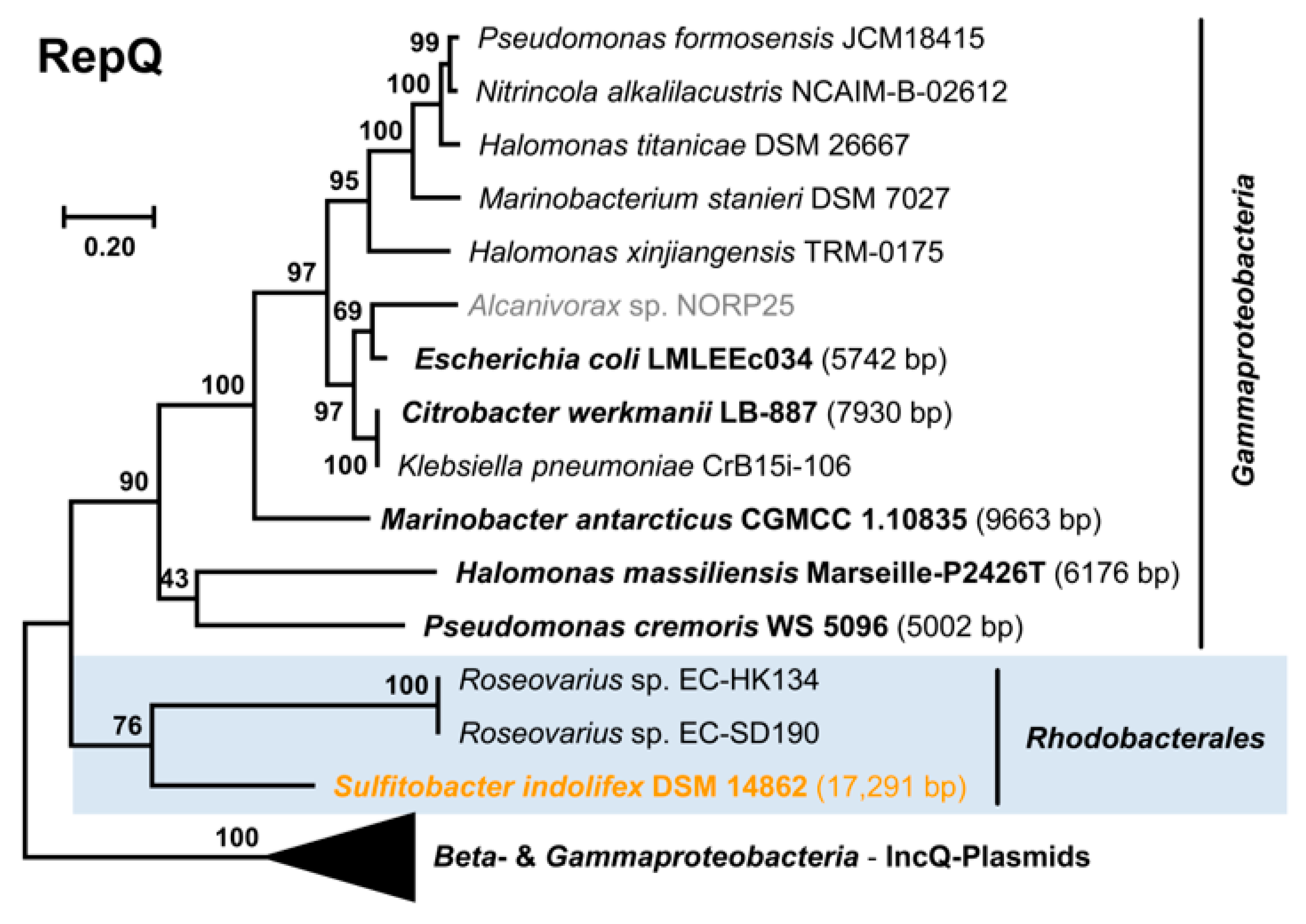
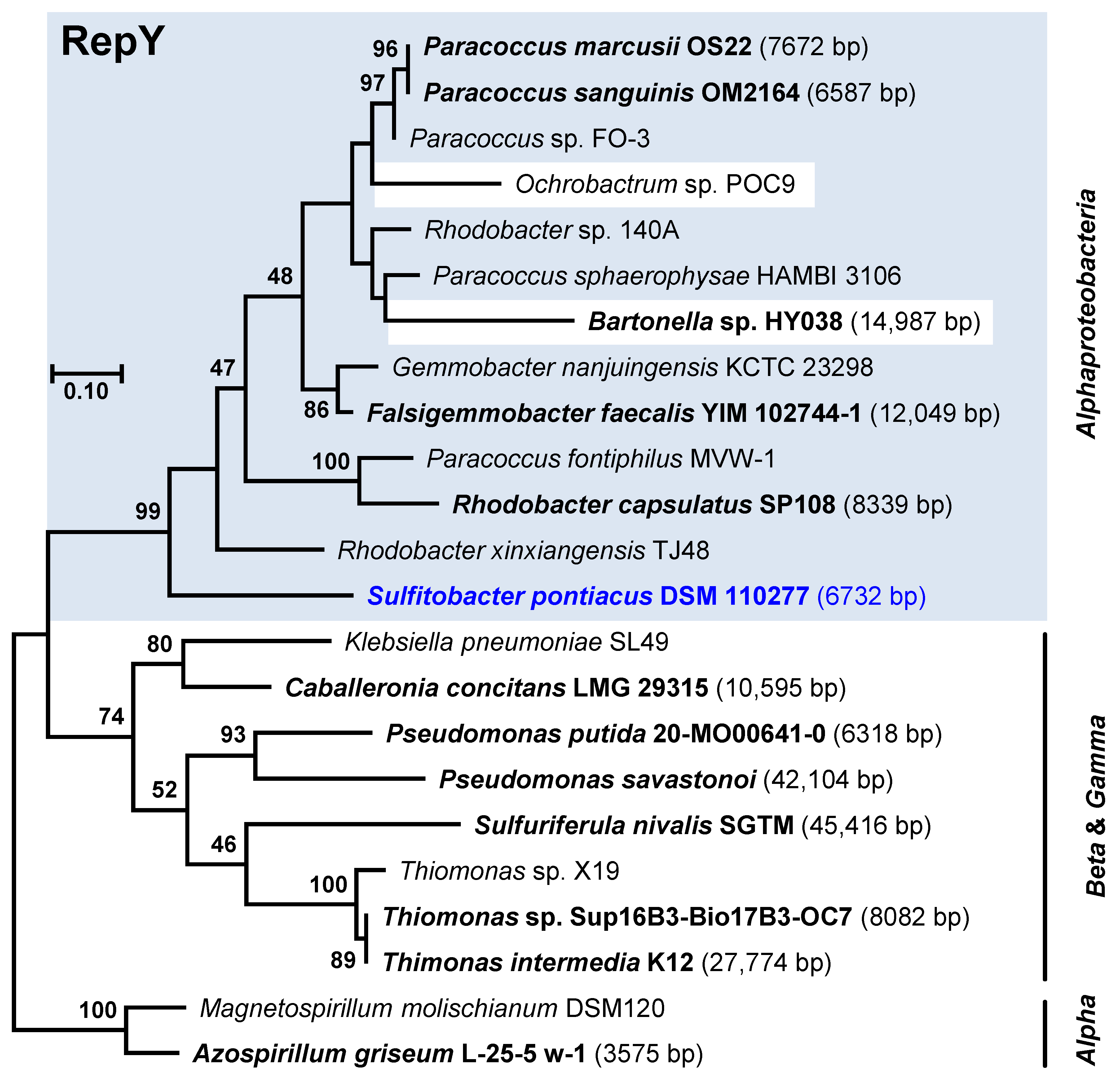
3.5. Distribution, Evolution and Function of the New Plasmid Types
3.5.1. Presence of RepQ, RepY and RepW in Closed Rhodobacterales Genomes
3.5.2. RepQ-Type Plasmids
3.5.3. RepY-Type Plasmids
3.5.4. RepW-Type Plasmids
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Smalla, K.; Jechalke, S.; Top, E.M. Plasmid detection, characterization and ecology. Microbiol. Spectr. 2015, 3, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Elsas, J.D.; Bailey, M.J. The ecology of transfer of mobile genetic elements. FEMS Microbiol. Ecol. 2002, 42, 187–197. [Google Scholar] [CrossRef] [PubMed]
- Heuer, H.; Smalla, K. Plasmids foster diversification and adaptation of bacterial populations in soil. FEMS Microbiol. Rev. 2012, 36, 1083–1104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Birmes, L.; Freese, H.M.; Petersen, J. RepC_soli: A Novel Promiscuous Plasmid Type of Rhodobacteraceae Mediates Horizontal Transfer of Antibiotic Resistances in the Ocean. Environ. Microbiol. 2021, 23, 5395–5411. [Google Scholar] [CrossRef]
- Wen, Y.; Pu, X.; Zheng, W.; Hu, G. High prevalence of plasmid-mediated quinolone resistance and IncQ plasmids carrying qnrS2 gene in bacteria from rivers near hospitals and aquaculture in China. PLoS ONE 2016, 11, e0159418. [Google Scholar] [CrossRef]
- Carattoli, A. Resistance plasmid families in Enterobacteriaceae. Antimicrob. Agents Chemother. 2009, 53, 2227–2238. [Google Scholar] [CrossRef] [Green Version]
- Petersen, J.; Vollmers, J.; Ringel, V.; Brinkmann, H.; Ellebrandt-Sperling, C.; Spröer, C.; Howat, A.M.; Murrell, J.C.; Kaster, A.K. A marine plasmid hitchhiking vast phylogenetic and geographic distances. Proc. Natl. Acad. Sci. USA 2019, 116, 20568–20573. [Google Scholar] [CrossRef] [Green Version]
- Dziewit, L.; Pyzik, A.; Szuplewska, M.; Matlakowska, R.; Mielnicki, S.; Wibberg, D.; Schluter, A.; Puhler, A.; Bartosik, D. Diversity and role of plasmids in adaptation of bacteria inhabiting the Lubin copper mine in Poland, an environment rich in heavy metals. Front. Microbiol. 2015, 6, 152. [Google Scholar] [CrossRef] [Green Version]
- Pérez Carrascal, O.M.; VanInsberghe, D.; Juárez, S.; Polz, M.F.; Vinuesa, P.; González, V. Population genomics of the symbiotic plasmids of sympatric nitrogen-fixing Rhizobium species associated with Phaseolus vulgaris. Environ. Microbiol. 2016, 18, 2660–2676. [Google Scholar] [CrossRef]
- Wang, H.; Tomasch, J.; Michael, V.; Bhuju, S.; Jarek, M.; Petersen, J.; Wagner-Döbler, I. Identification of genetic modules mediating the Jekyll and Hyde interaction of Dinoroseobacter shibae with the dinoflagellate Prorocentrum minimum. Front. Microbiol. 2015, 6, 1262. [Google Scholar] [CrossRef]
- Frank, O.; Michael, V.; Päuker, O.; Boedeker, C.; Jogler, C.; Rohde, M.; Petersen, J. Plasmid curing and the loss of grip—The 65 kb replicon of Phaeobacter inhibens DSM 17395 is required for biofilm formation, motility and the colonization of marine algae. Syst. Appl. Microbiol. 2015, 38, 120–127. [Google Scholar] [CrossRef] [PubMed]
- Czarnecki, J.; Dziewit, L.; Puzyna, M.; Prochwicz, E.; Tudek, A.; Wibberg, D.; Schlüter, A.; Pühler, A.; Bartosik, D. Lifestyle-determining extrachromosomal replicon pAMV1 and its contribution to the carbon metabolism of the methylotrophic bacterium Paracoccus aminovorans JCM 7685. Environ. Microbiol. 2017, 19, 4536–4550. [Google Scholar] [CrossRef] [PubMed]
- Koch, H.; Freese, H.M.; Hahnke, R.L.; Simon, M.; Wietz, M. Adaptations of Alteromonas sp. 76-1 to polysaccharide degradation: A CAZyme plasmid for ulvan degradation and two alginolytic systems. Front. Microbiol. 2019, 10, 504. [Google Scholar] [CrossRef] [PubMed]
- Petersen, J.; Brinkmann, H.; Bunk, B.; Michael, V.; Päuker, O.; Pradella, S. Think pink: Photosynthesis, Plasmids and the Roseobacter Clade. Environ. Microbiol. 2012, 14, 2661–2672. [Google Scholar] [CrossRef]
- Frank, O.; Göker, M.; Pradella, S.; Petersen, J. Ocean’s Twelve: Flagellar and Biofilm Chromids in the Multipartite Genome of Marinovum Algicola DG898 Exemplify Functional Compartmentalization. Environ. Microbiol. 2015, 17, 4019–4034. [Google Scholar] [CrossRef]
- Rodriguez-Beltran, J.; DelaFuente, J.; Leon-Sampedro, R.; MacLean, R.C.; San Millan, A. Beyond horizontal gene transfer: The Role of Plasmids in Bacterial Evolution. Nat. Rev. Microbiol. 2021, 19, 347–359. [Google Scholar] [CrossRef]
- Baxter, J.C.; Funnell, B.E. Plasmid partition mechanisms. Microbiol. Spectr. 2014, 2, PLAS-0023-2014. [Google Scholar] [CrossRef] [Green Version]
- Harms, A.; Brodersen, D.E.; Mitarai, N.; Gerdes, K. Toxins, targets, and triggers: An Overview of Toxin-Antitoxin Biology. Mol. Cell 2018, 70, 768–784. [Google Scholar] [CrossRef] [Green Version]
- Christie, P.J. The Mosaic Type IV Secretion Systems. EcoSal Plus 2016, 7. [Google Scholar] [CrossRef] [Green Version]
- Smillie, C.; Garcillán-Barcia, M.P.; Francia, M.V.; Rocha, E.P.C.; de la Cruz, F. Mobility of plasmids. Microbiol. Mol. Biol. Rev. 2010, 74, 434–452. [Google Scholar] [CrossRef] [Green Version]
- Xue, H.; Cordero, O.X.; Camas, F.M.; Trimble, W.; Meyer, F.; Guglielmini, J.; Rocha, E.P.C.; Polz, M.F. Eco-evolutionary dynamics of episomes among ecologically cohesive bacterial populations. mBio 2015, 6, e00552-15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sevastsyanovich, Y.R.; Krasowiak, R.; Bingle, L.E.H.; Haines, A.S.; Sokolov, S.L.; Kosheleva, I.A.; Leuchuk, A.A.; Titok, M.A.; Smalla, K.; Thomas, C.M. Diversity of IncP-9 plasmids of Pseudomonas. Microbiology 2008, 154, 2929–2941. [Google Scholar] [CrossRef] [Green Version]
- Petersen, J. Phylogeny and compatibility: Plasmid Classification in the Genomics Era. Arch. Microbiol. 2011, 193, 313–321. [Google Scholar] [CrossRef] [PubMed]
- Petersen, J.; Brinkmann, H.; Berger, M.; Brinkhoff, T.; Päuker, O.; Pradella, S. Origin and evolution of a novel DnaA-like plasmid replication type in Rhodobacterales. Mol. Biol. Evol. 2011, 28, 1229–1240. [Google Scholar] [CrossRef] [Green Version]
- Maj, A.; Dziewit, L.; Czarnecki, J.; Wlodarczyk, M.; Baj, J.; Skrzypczyk, G.; Giersz, D.; Bartosik, D. Plasmids of carotenoid-producing Paracoccus spp. (Alphaproteobacteria)—structure, diversity and evolution. PLoS ONE 2013, 8, e80258. [Google Scholar] [CrossRef] [Green Version]
- Simon, M.; Scheuner, C.; Meier-Kolthoff, J.P.; Brinkhoff, T.; Wagner-Döbler, I.; Ulbrich, M.; Klenk, H.P.; Schomburg, D.; Petersen, J.; Göker, M. Phylogenomics of Rhodobacteraceae reveals evolutionary adaptation to marine and non-marine habitats. ISME J. 2017, 11, 1483–1499. [Google Scholar] [CrossRef] [PubMed]
- Newton, R.J.; Griffin, L.E.; Bowles, K.M.; Meile, C.; Gifford, S.; Givens, C.E.; Howard, E.C.; King, E.; Oakley, C.A.; Reisch, C.R.; et al. Genome characteristics of a generalist marine bacterial lineage. ISME J. 2010, 4, 784–798. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fadeev, E.; De Pascale, F.; Vezzi, A.; Hubner, S.; Aharonovich, D.; Sher, D. Why close a bacterial genome? The plasmid of Alteromonas macleodii HOT1A3 is a vector for inter-specific transfer of a flexible genomic island. Front. Microbiol. 2016, 7, 248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, K.Y.H.; Orata, F.D.; Boucher, Y.F.; Case, R.J. Roseobacters in a sea of poly- and paraphyly: Whole Genome-Based Taxonomy of the Family Rhodobacteraceae and the Proposal for the Split of the “Roseobacter Clade” into a Novel Family, Roseobacteraceae fam. nov. Front. Microbiol. 2021, 12, 683109. [Google Scholar] [CrossRef]
- Yang, Q.; Ge, Y.-M.; Iqbal, N.M.; Yang, X.; Zhang, X.-L. Sulfitobacter alexandrii sp. nov., a new microalgae growth-promoting bacterium with exopolysaccharides bioflocculanting potential isolated from marine phycosphere. Antonie Leeuwenhoek 2021, 114, 1091–1106. [Google Scholar] [CrossRef]
- Pohlner, M.; Degenhardt, J.; von Hoyningen-Huene, A.J.E.; Wemheuer, B.; Erlmann, N.; Schnetger, B.; Badewien, T.H.; Engelen, B. The biogeographical distribution of benthic Roseobacter group members along a Pacific transect is structured by nutrient availability within the sediments and primary production in different oceanic provinces. Front. Microbiol. 2017, 8, 2550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.; Kang, S.-H.; Yang, E.J.; Macdonald, A.M.; Joo, H.M.; Park, J.; Kim, K.; Lee, G.S.; Kim, J.-H.; Yoon, J.-E.; et al. Latitudinal distributions and controls of bacterial community composition during the summer of 2017 in western Arctic surface waters (from the Bering Strait to the Chukchi Borderland). Sci. Rep. 2019, 9, 16822. [Google Scholar] [CrossRef] [PubMed]
- Allgaier, M.; Uphoff, H.; Felske, A.; Wagner-Döbler, I. Aerobic anoxygenic photosynthesis in Roseobacter clade bacteria from diverse marine habitats. Appl. Environ. Microbiol. 2003, 69, 5051–5059. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baym, M.; Kryazhimskiy, S.; Lieberman, T.D.; Chung, H.; Desai, M.M.; Kishony, R. Inexpensive multiplexed library preparation for megabases-sized genomes. PLoS ONE 2015, 10, e0128036. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [Green Version]
- Koboldt, D.C.; Zhang, Q.; Larson, D.E.; Shen, D.; McLellan, M.D.; Lin, L.; Miller, C.A.; Mardis, E.R.; Ding, L.; Wilson, R.K. VarScan 2: Somatic Mutation and Copy Number Alteration Discovery in Cancer by Exome Sequencing. Genome Res. 2012, 22, 568–576. [Google Scholar] [CrossRef] [Green Version]
- Zerbino, D.R.; Birney, E. Velvet: Algorithms for de Novo Short Read Assembly Using de Bruijn Graphs. Genome Res. 2008, 18, 821–829. [Google Scholar] [CrossRef] [Green Version]
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [Green Version]
- Harrison, P.W.; Lower, R.P.J.; Kim, N.K.D.; Young, J.P.W. Introducing the bacterial ‘chromid’: Not a Chromosome, not a Plasmid. Trends Microbiol. 2010, 18, 141–148. [Google Scholar] [CrossRef]
- Petersen, J.; Frank, O.; Göker, M.; Pradella, S. Extrachromosomal, extraordinary and essential—The plasmids of the Roseobacter clade. Appl. Microbiol. Biotechnol. 2013, 97, 2805–2815. [Google Scholar] [CrossRef] [PubMed]
- Garcillán-Barcia, M.P.; Redondo-Salvo, S.; Vielva, L.; de la Cruz, F. MOBscan: Automated Annotation of MOB Relaxases. Methods Mol. Biol. 2020, 2075, 295–308. [Google Scholar] [CrossRef]
- Parte, A.C.; Sardà Carbasse, J.; Meier-Kolthoff, J.P.; Reimer, L.C.; Göker, M. List of Prokaryotic names with Standing in Nomenclature (LPSN) moves to the DSMZ. Int. J. Syst. Evol. Microbiol. 2020, 70, 5607–5612. [Google Scholar] [CrossRef]
- Na, S.I.; Kim, Y.O.; Yoon, S.H.; Ha, S.M.; Baek, I.; Chun, J. UBCG: Up-to-date bacterial core gene set and pipeline for phylogenomic tree reconstruction. J. Microbiol. 2018, 56, 280–285. [Google Scholar] [CrossRef] [PubMed]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2—Approximately Maximum-Likelihood trees for large alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Göker, M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 2019, 10, 2182. [Google Scholar] [CrossRef] [PubMed]
- Petersen, J.; Brinkmann, H.; Pradella, S. Diversity and evolution of repABC type plasmids in Rhodobacterales. Environ. Microbiol. 2009, 11, 2627–2638. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The Protein Families Database in 2021. Nucleic Acids Res. 2020, 49, D412–D419. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple Sequence Alignment with High Accuracy and High Throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [Green Version]
- Jones, D.T.; Taylor, W.R.; Thornton, J.M. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 1992, 8, 275–282. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Yu, G. Using ggtree to Visualize Data on Tree-Like Structures. Curr. Protoc. Bioinform. 2020, 69, e96. [Google Scholar] [CrossRef] [PubMed]
- Wilkins, D. gggenes: Draw Gene Arrow Maps in ‘ggplot2’, R package version 0.4.1; 2020; Available online: https://wilkox.org/gggenes/ (accessed on 28 February 2022).
- Bartling, P.; Brinkmann, H.; Bunk, B.; Overmann, J.; Göker, M.; Petersen, J. The composite 259-kb plasmid of Martelella mediterranea DSM 17316(T)—A natural replicon with functional RepABC modules from Rhodobacteraceae and Rhizobiaceae. Front. Microbiol. 2017, 8, 1787. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bartling, P.; Vollmers, J.; Petersen, J. The first world swimming championships of roseobacters—Phylogenomic insights into an exceptional motility phenotype. Syst. Appl. Microbiol. 2018, 41, 544–554. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.P.; Göker, M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef] [Green Version]
- Luo, H.; Swan, B.K.; Stepanauskas, R.; Hughes, A.L.; Moran, M.A. Evolutionary analysis of a streamlined lineage of surface ocean Roseobacters. ISME J. 2014, 8, 1428–1439. [Google Scholar] [CrossRef] [Green Version]
- Galardini, M.; Pini, F.; Bazzicalupo, M.; Biondi, E.G.; Mengoni, A. Replicon-dependent bacterial genome evolution: The Case of Sinorhizobium Meliloti. Genome Biol. Evol. 2013, 5, 542–558. [Google Scholar] [CrossRef]
- Sonnenschein, E.C.; Nielsen, K.F.; D’Alvise, P.; Porsby, C.H.; Melchiorsen, J.; Heilmann, J.; Kalatzis, P.G.; Lopez-Perez, M.; Bunk, B.; Sproer, C.; et al. Global occurrence and heterogeneity of the Roseobacter-clade species Ruegeria mobilis. ISME J. 2017, 11, 569–583. [Google Scholar] [CrossRef] [Green Version]
- Tomasch, J.; Ringel, V.; Wang, H.; Freese, H.M.; Bartling, P.; Brinkmann, H.; Vollmers, J.; Jarek, M.; Wagner-Döbler, I.; Petersen, J. Fatal Affairs—Conjugational transfer of a Dinoflagellate-killing plasmid between marine Rhodobacterales. Microb. Genom. 2022, 8, 000787. [Google Scholar] [CrossRef]
- Manageiro, V.; Romao, R.; Moura, I.B.; Sampaio, D.A.; Vieira, L.; Ferreira, E.; Canica, M.; Network Eu, S.P. Molecular epidemiology and risk factors of carbapenemase-producing Enterobacteriaceae isolates in Portuguese hospitals: Results from European survey on carbapenemase-producing Enterobacteriaceae (EuSCAPE). Front. Microbiol. 2018, 9, 8. [Google Scholar] [CrossRef] [Green Version]
- Kaplan, E.; Sela, N.; Doron-Faigenboim, A.; Navon-Venezia, S.; Jurkevitch, E.; Cytryn, E. Genomic and functional characterization of qnr-encoding plasmids from municipal wastewater biosolid Klebsiella pneumoniae isolates. Front. Microbiol. 2015, 6, 12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marsin, S.; Forterre, P. A rolling circle replication initiator protein with a nucleotidyl-transferase activity encoded by the plasmid pGT5 from the hyperthermophilic archaeon Pyrococcus abyssi. Mol. Microbiol. 1998, 27, 1183–1192. [Google Scholar] [CrossRef] [PubMed]
- Rocha, E.P.C.; Danchin, A. Base composition bias might result from competition for metabolic resources. Trends Genet. 2002, 18, 291–294. [Google Scholar] [CrossRef]
- Dietel, A.-K.; Merker, H.; Kaltenpoth, M.; Kost, C. Selective advantages favour high genomic AT-contents in intracellular elements. PLoS Genet. 2019, 15, e1007778. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, M.; Zhu, X.; Zhang, C.; Zhao, Z. LuxQ-LuxU-LuxO pathway regulates biofilm formation by Vibrio parahaemolyticus. Microbiol. Res. 2021, 250, 126791. [Google Scholar] [CrossRef]
- Miller, M.B.; Skorupski, K.; Lenz, D.H.; Taylor, R.K.; Bassler, B.L. Parallel quorum sensing systems converge to regulate virulence in Vibrio cholerae. Cell 2002, 110, 303–314. [Google Scholar] [CrossRef] [Green Version]
- Byrd, D.R.; Matson, S.W. Nicking by transesterification: The Reaction Catalysed by a Relaxase. Mol. Microbiol. 1997, 25, 1011–1022. [Google Scholar] [CrossRef] [Green Version]
- Jagura-Burdzy, G.; Thomas, C.M. kfrA gene of broad host range plasmid RK2 encodes a novel DNA-binding protein. J. Mol. Biol. 1992, 225, 651–660. [Google Scholar] [CrossRef]
- Greene, P.J.; Gupta, M.; Boyer, H.W.; Brown, W.E.; Rosenberg, J.M. Sequence analysis of the DNA encoding the Eco RI endonuclease and methylase. J. Biol. Chem. 1981, 256, 2143–2153. [Google Scholar] [CrossRef]
- Roberts, R.J.; Vincze, T.; Posfai, J.; Macelis, D. REBASE—A database for DNA restriction and modification: Enzymes, Genes and Genomes. Nucleic Acids Res. 2015, 43, D298–D299. [Google Scholar] [CrossRef]
- Ichige, A.; Kobayashi, I. Stability of EcoRI restriction-modification enzymes in vivo differentiates the EcoRI restriction-modification system from other postsegregational cell killing systems. J. Bacteriol. 2005, 187, 6612–6621. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dziewit, L.; Kuczkowska, K.; Adamczuk, M.; Radlinska, M.; Bartosik, D. Functional characterization of the type II PamI restriction-modification system derived from plasmid pAMI7 of Paracoccus aminophilus JCM 7686. FEMS Microbiol. Lett. 2011, 324, 56–63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Motiejūnaite, R.; Armalyte, J.; Markuckas, A.; Suziedeliene, E. Escherichia coli dinJ-yafQ genes act as a toxin-antitoxin module. FEMS Microbiol. Lett. 2007, 268, 112–119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prysak, M.H.; Mozdzierz, C.J.; Cook, A.M.; Zhu, L.; Zhang, Y.; Inouye, M.; Woychik, N.A. Bacterial toxin YafQ is an endoribonuclease that associates with the ribosome and blocks translation elongation through sequence-specific and frame-dependent mRNA cleavage. Mol. Microbiol. 2009, 71, 1071–1087. [Google Scholar] [CrossRef] [PubMed]
- Ni, S.W.; Li, B.Y.; Tang, K.H.; Yao, J.Y.; Wood, T.K.; Wang, P.X.; Wang, X.X. Conjugative plasmid-encoded toxin-antitoxin system PrpT/PrpA directly controls plasmid copy number. Proc. Natl. Acad. Sci. USA 2021, 118, 10. [Google Scholar] [CrossRef]
- Bönemann, G.; Stiens, M.; Pühler, A.; Schlüter, A. Mobilizable IncQ-related plasmid carrying a new quinolone resistance gene, qnrS2, isolated from the bacterial community of a wastewater treatment plant. Antimicrob. Agents Chemother. 2006, 50, 3075–3080. [Google Scholar] [CrossRef] [Green Version]
- Bardaji, L.; Pérez-Martínez, I.; Rodríguez-Moreno, L.; Rodríguez-Palenzuela, P.; Sundin, G.W.; Ramos, C.; Murillo, J. Sequence and role in virulence of the three plasmid complement of the model tumor-inducing bacterium Pseudomonas savastanoi pv. savastanoi NCPPB 3335. PLoS ONE 2011, 6, e25705. [Google Scholar] [CrossRef] [Green Version]
- Blankenfeldt, W.; Kuzin, A.P.; Skarina, T.; Korniyenko, Y.; Tong, L.; Bayer, P.; Janning, P.; Thomashow, L.S.; Mavrodi, D.V. Structure and function of the phenazine biosynthetic protein PhzF from Pseudomonas fluorescens. Proc. Natl. Acad. Sci. USA 2004, 101, 16431–16436. [Google Scholar] [CrossRef] [Green Version]
- Jørgensen, T.S.; Hansen, M.A.; Xu, Z.; Tabak, M.A.; Sørensen, S.J.; Hansen, L.H. Plasmids, viruses, and other circular elements in rat gut. bioRxiv 2017, 143420. [Google Scholar] [CrossRef] [Green Version]
- Jørgensen, T.S.; Xu, Z.; Hansen, M.A.; Sørensen, S.J.; Hansen, L.H. Hundreds of circular novel plasmids and DNA elements identified in a rat cecum metamobilome. PLoS ONE 2014, 9, e87924. [Google Scholar] [CrossRef] [Green Version]
- Sheu, S.Y.; Chen, W.M.; Lin, G.H. Characterization and application of a rolling-circle-type plasmid from Cupriavidus taiwanensis. Plasmid 2007, 57, 275–285. [Google Scholar] [CrossRef] [PubMed]
- Okamura, Y.; Takeyama, H.; Sekine, T.; Sakaguchi, T.; Wahyudi, A.T.; Sato, R.; Kamiya, S.; Matsunaga, T. Design and application of a new cryptic-plasmid-based shuttle vector for Magnetospirillum magneticum. Appl. Environ. Microbiol. 2003, 69, 4274–4277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Attéré, S.A.; Vincent, A.T.; Paccaud, M.; Frenette, M.; Charette, S.J. The role for the small cryptic plasmids as moldable vectors for genetic innovation in Aeromonas salmonicida subsp. salmonicida. Front. Genet. 2017, 8, 211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodriguez-Beltran, J.; Hernandez-Beltran, J.C.R.; DelaFuente, J.; Escudero, J.A.; Fuentes-Hernandez, A.; MacLean, R.C.; Pena-Miller, R.; Millan, A.S. Multicopy plasmids allow bacteria to escape from fitness trade-offs during evolutionary innovation. Nat. Ecol. Evol. 2018, 2, 873–881. [Google Scholar] [CrossRef] [PubMed]
- Kovach, M.E.; Elzer, P.H.; Steven Hill, D.; Robertson, G.T.; Farris, M.A.; Roop, R.M.; Peterson, K.M. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 1995, 166, 175–176. [Google Scholar] [CrossRef]
- Patzelt, D.; Wang, H.; Buchholz, I.; Rohde, M.; Gröbe, L.; Pradella, S.; Neumann, A.; Schulz, S.; Heyber, S.; Münch, K.; et al. You are what you talk: Quorum sensing induces individual morphologies and cell division modes in Dinoroseobacter shibae. ISME J. 2013, 7, 2274–2286. [Google Scholar] [CrossRef] [Green Version]
- Döhlemann, J.; Wagner, M.; Happel, C.; Carrillo, M.; Sobetzko, P.; Erb, T.J.; Thanbichler, M.; Becker, A. A family of single copy repABC-type shuttle vectors stably maintained in the alpha-proteobacterium Sinorhizobium meliloti. ACS Synth. Biol. 2017, 6, 968–984. [Google Scholar] [CrossRef]

| Replicon Id | Size (bp) | Replicon Type | GC | Copy No. | Replication Module | Partitioning System | Mobility | NCBI Accession |
|---|---|---|---|---|---|---|---|---|
| Sulfitobacter indolifexDSM 14862 (HEL-45) | ||||||||
| cDSM14862 | 3,271,523 | chromosome | 60.0 | 1.0 | DnaA | yes | no | CP084951 |
| pDSM14862_a | 313,826 | plasmid | 55.4 | 0.5 | RepABC-20 | yes | no | CP084952 |
| pDSM14862_b | 307,297 | plasmid | 56.1 | 0.8 | DnaA-like-I | yes | no | CP084953 |
| pDSM14862_c | 200,719 | chromid | 60.3 | 0.8 | RepABC-8 | yes | no | CP084954 |
| pDSM14862_d | 160,516 | plasmid | 56.5 | 0.8 | DnaA-like-II | yes | no | CP084955 |
| pDSM14862_e | 123,234 | plasmid | 57.1 | 0.6 | RepABC-4 | yes | T4SS | CP084956 |
| pDSM14862_f | 104,164 | plasmid | 59.8 | 0.7 | RepABC-7 | yes | T4SS | CP084957 |
| pDSM14862_g | 17,291 | plasmid | 54.9 | 5.3 | RepQ | no | MOBP | CP084958 |
| Sulfitobacter pontiacusDSM 110277 (SO248Ex84) | ||||||||
| cDSM110277 | 3,012,962 | chromosome | 60.5 | 1.0 | DnaA | yes | no | CP084959 |
| pDSM110277_a | 239,416 | plasmid | 60.2 | 0.7 | RepABC-10 | yes | T4SS | CP084960 |
| pDSM110277_b | 230,345 | chromid | 59.7 | 0.7 | DnaA-like-II | yes | no | CP084961 |
| pDSM110277_c | 177,750 | chromid | 60.3 | 0.6 | DnaA-like-I | yes | no | CP084962 |
| pDSM110277_d | 128,813 | plasmid | 59.6 | 0.5 | RepABC-9-1 | yes | T4SS | CP084963 |
| pDSM110277_e | 53,945 | plasmid | 57.0 | 1.8 | RepABC-8 | yes | no | CP084964 |
| pDSM110277_f | 6732 | plasmid | 52.1 | 124.3 | RepY | no | MOBQ | CP084965 |
| Sulfitobacter sp. DSM 110093 (2RS2_G6) | ||||||||
| cDSM110093 | 3,434,207 | chromosome | 59.7 | 1.0 | DnaA | yes | MOBV | CP085167 |
| pDSM110093_a | 336,544 | plasmid | 55.3 | 1.1 | RepABC-21 | yes | no | CP085168 |
| pDSM110093_b | 273,772 | plasmid | 54.9 | 1.1 | RepABC-20 | yes | no | CP085169 |
| pDSM110093_c | 268,356 | chromid | 59.7 | 1.0 | RepABC-8 | yes | no | CP085170 |
| pDSM110093_d | 21,520 | plasmid | 52.6 | 4.3 | RepA | no | MOBP | CP085171 |
| pDSM110093_e | 5635 | plasmid | 53.9 | 23.4 | RepW | no | no | CP085172 |
| Sulfitobacter dubius DSM 109990 (3RS2_G4b) | ||||||||
| cDSM109990 | 3,274,709 | chromosome | 60.2 | 1.0 | DnaA | yes | no | CP085144 |
| pDSM109990_a | 284,454 | plasmid | 55.4 | 1.2 | RepB-I | yes | MOBP | CP085145 |
| pDSM109990_b | 247,035 | chromid | 60.5 | 0.9 | RepABC-8 | yes | no | CP085146 |
| pDSM109990_c | 183,486 | plasmid | 57.6 | 0.9 | RepABC-2 | yes | no | CP085147 |
| pDSM109990_d | 108,277 | plasmid | 59.9 | 0.6 | RepABC-1 | yes | T4SS | CP085148 |
| pDSM109990_e | 47,721 | plasmid | 58.7 | 2.6 | RepA_a | no | MOBQ | CP085149 |
| pDSM109990_f | 6286 | plasmid | 59.7 | 9.5 | RepA_b | no | MOBQ | CP085150 |
| pDSM109990_g | 4878 | plasmid | 52.3 | 12.5 | RepA_c | no | no | CP085151 |
| pDSM109990_h | 4609 | plasmid | 58.4 | 12.4 | RepA_d | no | MOBV | CP085152 |
| pDSM109990_i | 3857 | plasmid | 55.0 | 15.3 | RepW | no | no | CP085153 |
| Pseudosulfitobacter sp. DSM 107133 (PIC-76) | ||||||||
| cDSM107133 | 3,635,847 | chromosome | 61.1 | 1.0 | DnaA | yes | no | CP085154 |
| pDSM107133_a | 571,401 | chromid | 61.0 | 1.1 | RepABC-3 | yes | no | CP085155 |
| pDSM107133_b | 246,683 | chromid | 61.1 | 0.6 | RepB-I | yes | no | CP085156 |
| pDSM107133_c | 158,961 | plasmid | 60.3 | 0.5 | RepABC-11 | yes | T4SS | CP085157 |
| pDSM107133_d | 135,500 | plasmid | 58.0 | 0.4 | RepABC-9-1 | yes | MOBP | CP085158 |
| pDSM107133_e | 129,047 | plasmid | 58.4 | 0.4 | RepABC-5 | yes | T4SS | CP085159 |
| pDSM107133_f | 128,511 | plasmid | 60.2 | 0.4 | RepABC-1 | yes | T4SS | CP085160 |
| pDSM107133_g | 105,694 | plasmid | 60.4 | 0.4 | RepABC-9-2 | yes | T4SS | CP085161 |
| pDSM107133_h | 65,426 | chromid | 63.6 | 0.6 | RepB-III | yes | no | CP085162 |
| pDSM107133_i | 17,449 | plasmid | 53.1 | 3.8 | RepA_a | no | no | CP085163 |
| pDSM107133_j | 7190 | plasmid | 59.6 | 9.1 | RepA_b | no | MOBQ | CP085164 |
| pDSM107133_k | 5670 | plasmid | 59.0 | 8.6 | RepL | no | MOBQ | CP085165 |
| pDSM107133_l | 4511 | plasmid | 53.5 | 21.3 | RepW | no | MOBV | CP085166 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Freese, H.M.; Ringel, V.; Overmann, J.; Petersen, J. Beyond the ABCs—Discovery of Three New Plasmid Types in Rhodobacterales (RepQ, RepY, RepW). Microorganisms 2022, 10, 738. https://doi.org/10.3390/microorganisms10040738
Freese HM, Ringel V, Overmann J, Petersen J. Beyond the ABCs—Discovery of Three New Plasmid Types in Rhodobacterales (RepQ, RepY, RepW). Microorganisms. 2022; 10(4):738. https://doi.org/10.3390/microorganisms10040738
Chicago/Turabian StyleFreese, Heike M., Victoria Ringel, Jörg Overmann, and Jörn Petersen. 2022. "Beyond the ABCs—Discovery of Three New Plasmid Types in Rhodobacterales (RepQ, RepY, RepW)" Microorganisms 10, no. 4: 738. https://doi.org/10.3390/microorganisms10040738
APA StyleFreese, H. M., Ringel, V., Overmann, J., & Petersen, J. (2022). Beyond the ABCs—Discovery of Three New Plasmid Types in Rhodobacterales (RepQ, RepY, RepW). Microorganisms, 10(4), 738. https://doi.org/10.3390/microorganisms10040738






