A New ICEclc Subfamily Integrative and Conjugative Element Responsible for Horizontal Transfer of Biphenyl and Salicylic Acid Catabolic Pathway in the PCB-Degrading Strain Pseudomonas stutzeri KF716
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Culture Conditions
2.2. Sequence Annotation and Computational Analysis
2.3. Detection of Target DNA by Polymerase Chain Reaction
2.4. Conjugal Transfer of ICE
3. Results and Discussion
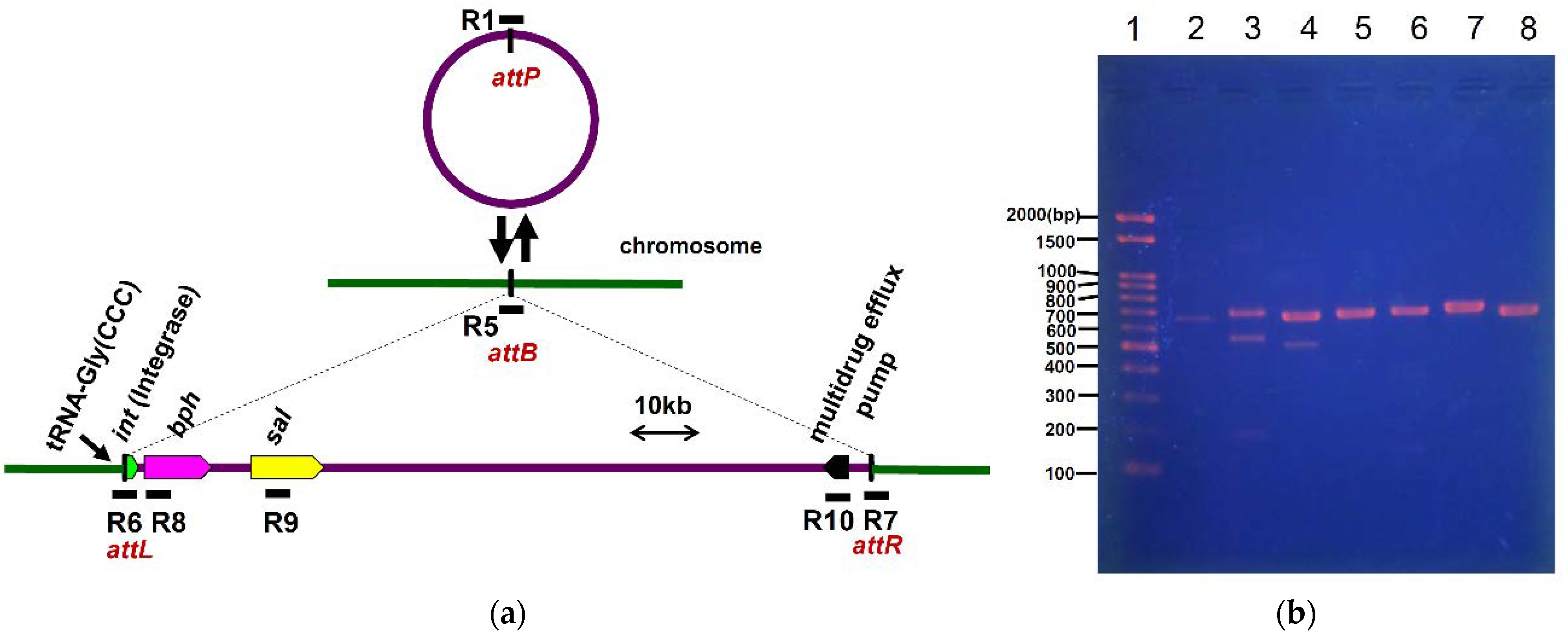
3.1. Detection of ICEbph-salKF716
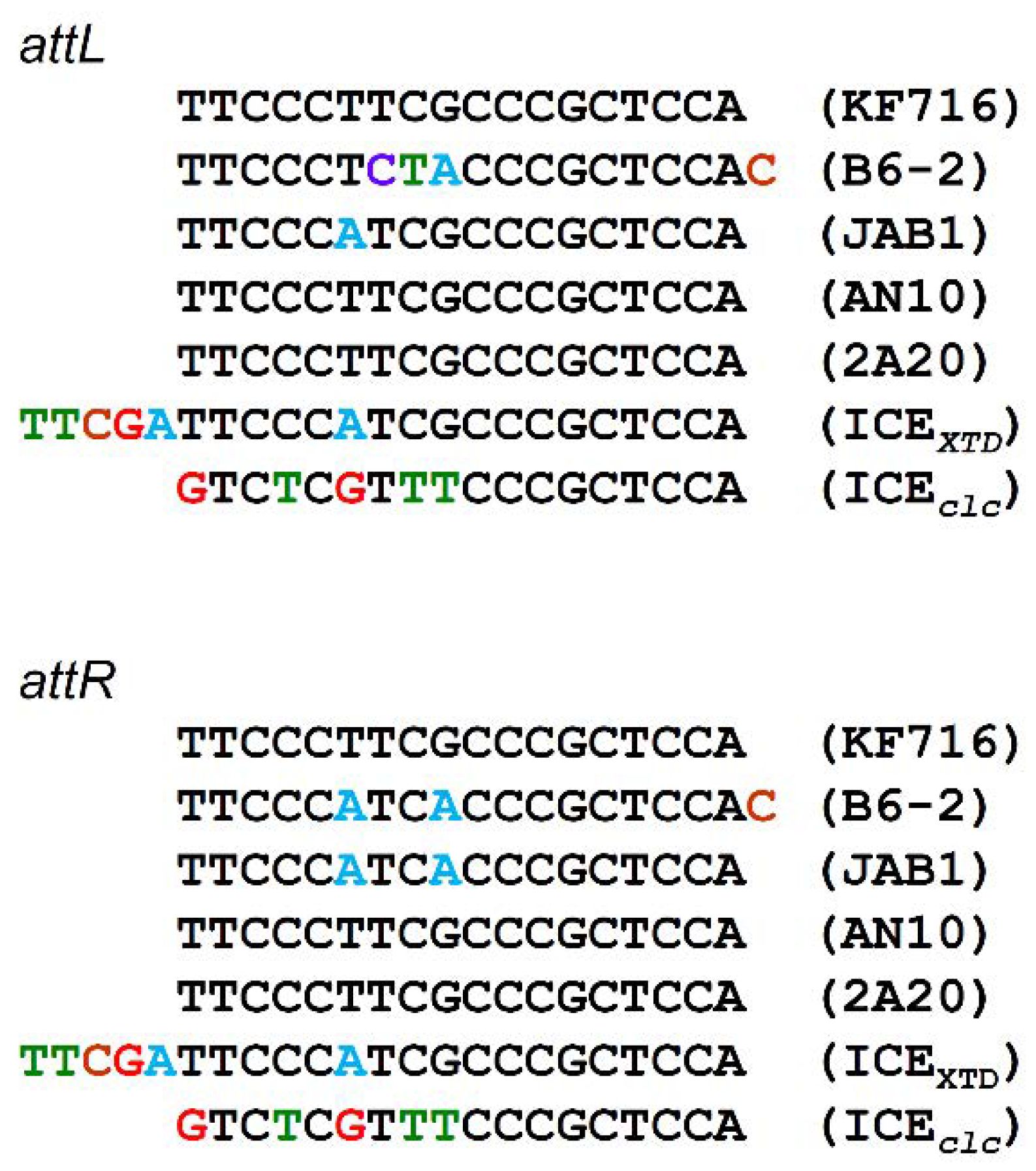
3.2. Core Region of ICEbph-salKF716
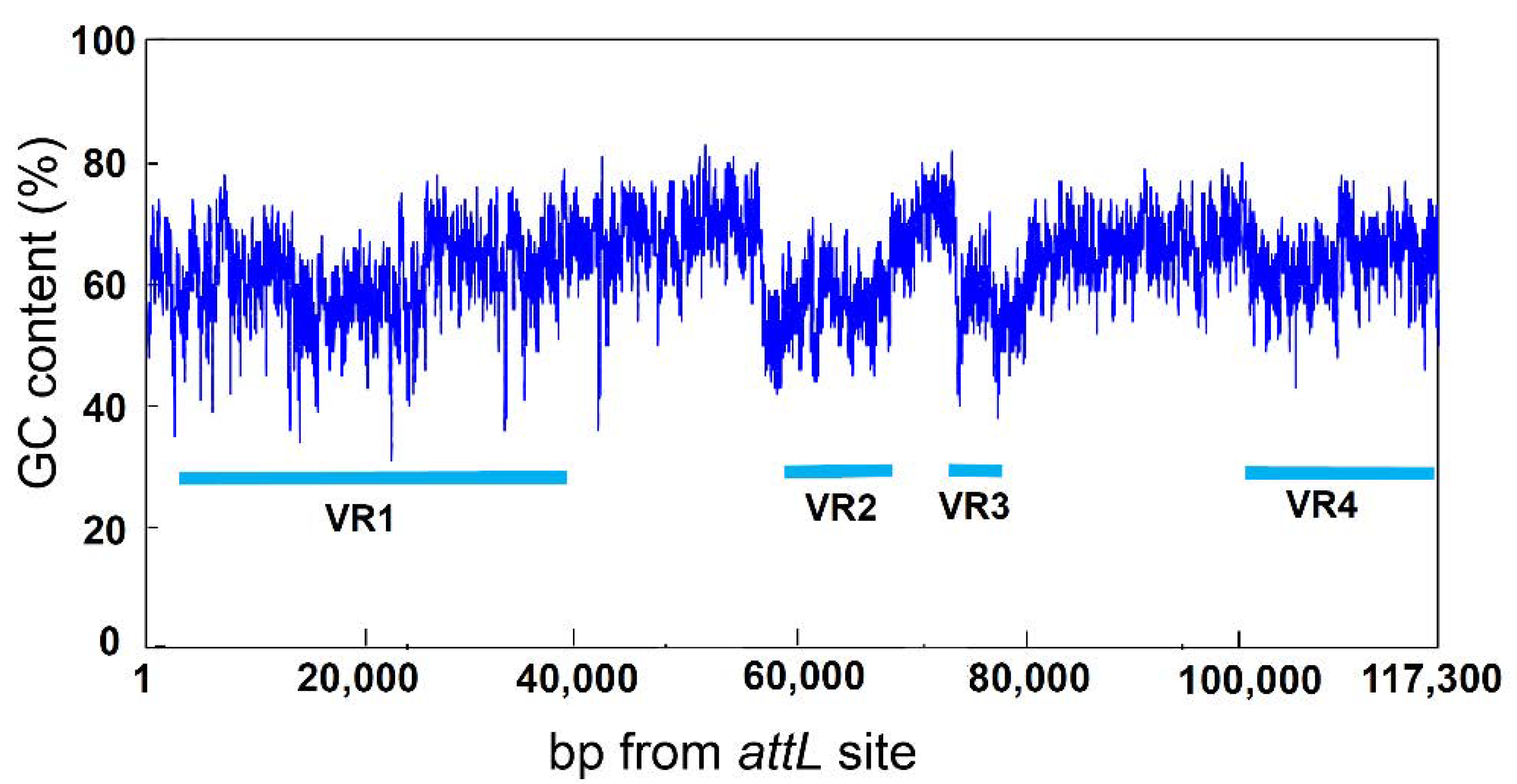
3.3. Variable Region of ICEbph-salKF716
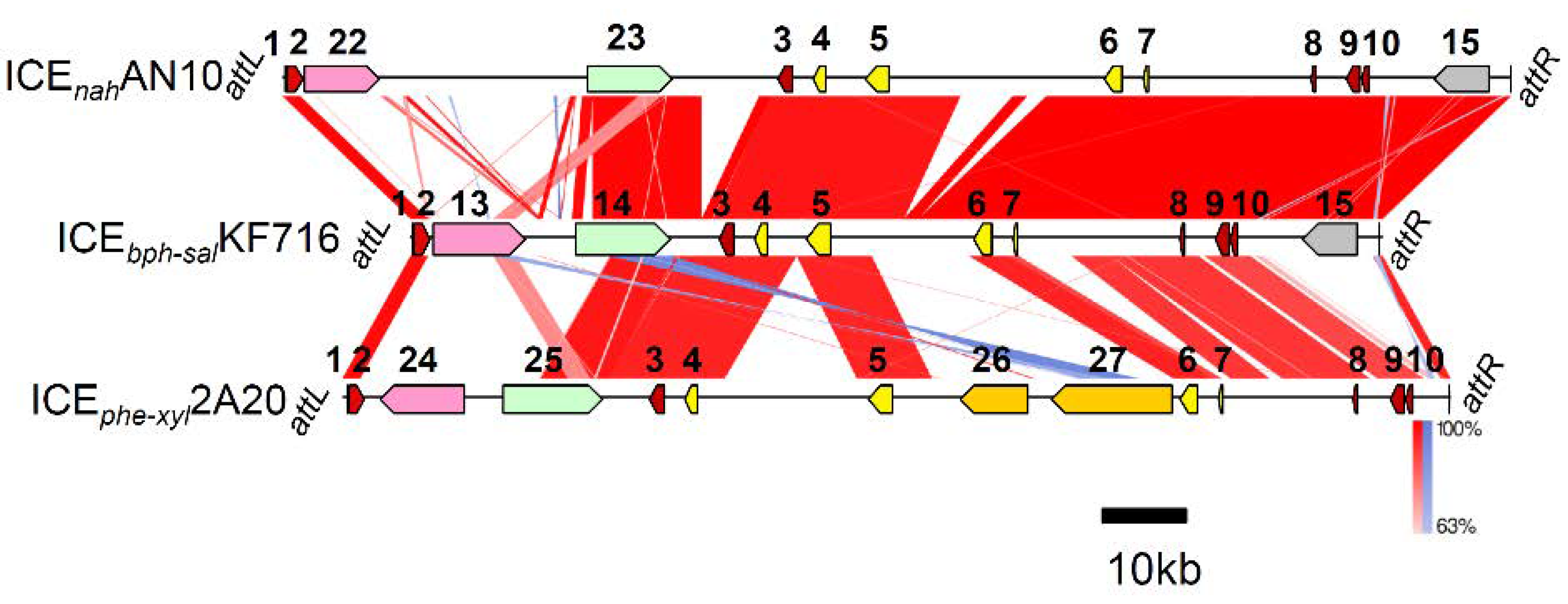
3.4. Comparison of ICEbph-salKF716 and Other Putative ICEs
3.5. Excision and Formation of Circular Intermediate Form of ICEbph-salKF716
3.6. Transfer and Integration of ICEbph-salKF716
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bellanger, X.; Payot, S.; Leblond-Bourget, N.; Guédon, G. Conjugative and mobilizable genomic islands in bacteria: Evolution and diversity. FEMS Microbiol. Rev. 2014, 38, 720–760. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gaillard, M.; Vallaeys, T.; Vorhölter, F.J.; Minoia, M.; Werlen, C.; Sentchilo, V.; Pühler, A.; van der Meer, J.R. The clc element of Pseudomonas sp. strain B13, a genomic island with various catabolic properties. J. Bacteriol. 2006, 188, 1999–2013. [Google Scholar] [CrossRef] [Green Version]
- Chain, P.S.G.; Denef, V.J.; Konstantinidis, K.T.; Vergez, L.M.; Agulló, L.; Reyes, V.L.; Hauser, L.; Córdova, M.; Gómez, L.; González, M.; et al. Burkholderia xenovorans LB400 harbors a multi-replicon, 9.73-Mbp genome shaped for versatility. Proc. Natl. Acad. Sci. USA 2006, 103, 15280–15287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Obi, C.C.; Vayla, S.; de Gannes, V.; Berres, M.E.; Walker, J.; Pavelec, D.; Hyman, J.; Hickey, W.J. The integrative conjugative element clc (ICEclc) of Pseudomonas aeruginosa JB2. Front. Microbiol. 2018, 9, 1532. [Google Scholar] [CrossRef] [Green Version]
- Miyazaki, R.; Bertelli, C.; Benaglio, P.; Canton, J.; De Coi, N.; Gharib, W.H.; Gjoksi, B.; Goesmann, A.; Greub, G.; Harshman, K.; et al. Comparative genome analysis of Pseudomonas knackmussii B13, the first bacterium known to degrade chloroaromatic compounds. Environ. Microbiol. 2015, 17, 91–104. [Google Scholar] [CrossRef] [Green Version]
- Zamarro, M.T.; Martín-Moldes, Z.; Díaz, E. The ICEXTD of Azoarcus sp. CIB, an integrative and conjugative element with aerobic and anaerobic catabolic properties. Environ. Microbiol. 2016, 18, 5018–5031. [Google Scholar] [CrossRef]
- Xiang, Y.; Xing, Z.; Liu, J.; Qin, W.; Huang, X. Recent advances in the biodegradation of polychlorinated biphenyls. World J. Microbiol. Biotechnol. 2020, 36, 145. [Google Scholar] [CrossRef]
- Toussaint, A.; Merlin, C.; Monchy, S.; Benotmane, M.A.; Leplae, R.; Mergeay, M.; Springael, D. The biphenyl- and 4-chlorobiphenyl-catabolic transposon Tn4371, a member of a new family of genomic islands related to IncP and Ti plasmids. Appl. Environ. Microbiol. 2003, 69, 4837–4845. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohtsubo, Y.; Ishibashi, Y.; Naganawa, H.; Hirokawa, S.; Atobe, S.; Nagata, Y.; Tsuda, M. Conjugal transfer of polychlorinated biphenyl/biphenyl degradation genes in Acidovorax sp. strain KKS102, which are located on an integrative and conjugative element. J. Bacteriol. 2012, 194, 4237–4248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirose, J.; Fujihara, H.; Watanabe, T.; Kimura, N.; Suenaga, H.; Futagami, T.; Goto, M.; Suyama, A.; Furukawa, K. Biphenyl/PCB degrading bph genes of ten bacterial strains isolated from biphenyl-contaminated soil in Kitakyushu, Japan: Comparative and dynamic features as integrative conjugative elements (ICEs). Genes 2019, 10, 404. [Google Scholar] [CrossRef] [Green Version]
- Suenaga, H.; Fujihara, H.; Kimura, N.; Hirose, J.; Watanabe, T.; Futagami, T.; Goto, M.; Shimodaira, J.; Furukawa, K. Insights into the genomic plasticity of Pseudomonas putida KF715, a strain with unique biphenyl-utilizing activity and genome instability properties. Environ. Microbiol. Rep. 2017, 9, 589–598. [Google Scholar] [CrossRef] [PubMed]
- Hirose, J.; Yamazoe, A.; Hosoyama, A.; Kimura, N.; Suenaga, H.; Watanabe, T.; Fujihara, H.; Futagami, T.; Goto, M.; Furukawa, K. Draft genome sequence of the polychlorinated biphenyl-degrading bacterium Pseudomonas stutzeri KF716 (NBRC 110668). Genome Announc. 2015, 3, e01215-15. [Google Scholar] [CrossRef] [Green Version]
- Tatusova, T.; DiCuccio, M.; Badretdin, A.; Chetvernin, V.; Nawrocki, E.P.; Zaslavsky, L.; Lomsadze, A.; Pruitt, K.D.; Borodovsky, M.; Ostell, J. NCBI Prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016, 44, 6614–6624. [Google Scholar] [CrossRef]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef] [Green Version]
- Johnson, M.; Zaretskaya, I.; Raytselis, Y.; Merezhuk, Y.; McGinnis, S.; Madden, T.L. NCBI BLAST: A better web interface. Nucleic Acids Res. 2008, 36, W5–W9. [Google Scholar] [CrossRef]
- Sullivan, M.J.; Petty, N.K.; Beatson, S.A. Easyfig: A genome comparison visualizer. Bioinformatics 2011, 27, 1009–1010. [Google Scholar] [CrossRef]
- Alvarez-Martinez, C.E.; Christie, P.J. Biological diversity of prokaryotic Type IV secretion systems. Microbiol. Mol. Biol. Rev. 2009, 73, 775–808. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Christie, P.J.; Whitaker, N.; González-Rivera, C. Mechanism and structure of the bacterial Type IV secretion systems. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 2014, 1843, 1578–1591. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bignell, C.; Thomas, C.M. The bacterial parA-parB partitioning proteins. J. Biotechnol. 2001, 91, 1–34. [Google Scholar] [CrossRef]
- Minoia, M.; Gaillard, M.; Reinhard, F.; Stojanov, M.; Sentchilo, V.; van der Meer, J.R. Stochasticity and bistability in horizontal transfer control of a genomic island in Pseudomonas. Proc. Natl. Acad. Sci. USA 2008, 105, 20792–20797. [Google Scholar] [CrossRef] [Green Version]
- De La Cruz, F.; Frost, L.S.; Meyer, R.J.; Zechner, E.L. Conjugative DNA metabolism in gram-negative bacteria. FEMS Microbiol. Rev. 2010, 34, 18–40. [Google Scholar] [CrossRef]
- Marrero, J.; Waldor, M.K. Determinants of entry exclusion within Eex and TraG are cytoplasmic. J. Bacteriol. 2007, 189, 6469–6473. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taira, K.; Hirose, J.; Hayashida, S.; Furukawa, K. Analysis of bph operon from the polychlorinated biphenyl-degrading strain of Pseudomonas pseudoalcaligenes KF707. J. Biol. Chem. 1992, 267, 4844–4853. [Google Scholar] [CrossRef]
- Tang, H.; Yu, H.; Li, Q.; Wang, X.; Gai, Z.; Yin, G.; Su, F.; Tao, F.; Ma, C.; Xu, P. Genome sequence of Pseudomonas putida strain B6-2, a superdegrader of polycyclic aromatic hydrocarbons and dioxin-like compounds. J. Bacteriol. 2011, 193, 6789–6790. [Google Scholar] [CrossRef]
- Wang, W.; Li, Q.; Zhang, L.; Cui, J.; Yu, H.; Wang, X.; Ouyang, X.; Tao, F.; Xu, P.; Tang, H. Genetic mapping of highly versatile and solvent-tolerant Pseudomonas putida B6-2 (ATCC BAA-2545) as a ‘superstar’ for mineralization of PAHs and dioxin-like compounds. Environ. Microbiol. 2021, 23, 4309–4325. [Google Scholar] [CrossRef]
- Ridl, J.; Suman, J.; Fraraccio, S.; Hradilova, M.; Strejcek, M.; Cajthaml, T.; Zubrova, A.; Macek, T.; Strnad, H.; Uhlik, O. Complete genome sequence of Pseudomonas alcaliphila JAB1 (=DSM 26533), a versatile degrader of organic pollutants. Stand. Genom. Sci. 2018, 13, 3. [Google Scholar] [CrossRef] [PubMed]
- Brunet-Galmés, I.; Busquets, A.; Peña, A.; Gomila, M.; Nogales, B.; García-Valdés, E.; Lalucat, J.; Bennasar, A.; Bosch, R. Complete genome sequence of the naphthalene-degrading bacterium Pseudomonas stutzeri AN10 (CCUG 29243). J. Bacteriol. 2012, 194, 6642–6643. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heinaru, E.; Naanuri, E.; Grünbach, M.; Jõesaar, M.; Heinaru, A. functional redundancy in phenol and toluene degradation in Pseudomonas stutzeri strains isolated from the Baltic Sea. Gene 2016, 589, 90–98. [Google Scholar] [CrossRef]
- Bosch, R.; Garcıía-Valdés, E.; Moore, E.R.B. Genetic characterization and evolutionary implications of a chromosomally encoded naphthalene-degradation upper pathway from Pseudomonas stutzeri AN10. Gene 1999, 236, 149–157. [Google Scholar] [CrossRef]
- Bosch, R.; Garcıía-Valdés, E.; Moore, E.R.B. Complete nucleotide sequence and evolutionary significance of a chromosomally encoded naphthalene-degradation lower pathway from Pseudomonas stutzeri AN10. Gene 2000, 245, 65–74. [Google Scholar] [CrossRef]
- Fujihara, H.; Yoshida, H.; Matsunaga, T.; Goto, M.; Furukawa, K. Cross-regulation of biphenyl- and salicylate-catabolic genes by two regulatory systems in Pseudomonas pseudoalcaligenes KF707. J. Bacteriol. 2006, 188, 4690–4697. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Sousa, L.P. Mobile genetic elements in Pseudomonas stutzeri. Curr. Microbiol. 2020, 77, 179–184. [Google Scholar] [CrossRef]
- Stover, C.K.; Pham, X.Q.; Erwin, A.L.; Mizoguchi, S.D.; Warrener, P.; Hickey, M.J.; Brinkman, F.S.; Hufnagle, W.O.; Kowalik, D.J.; Lagrou, M.; et al. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 2000, 406, 959–964. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hirose, J.; Watanabe, T.; Futagami, T.; Fujihara, H.; Kimura, N.; Suenaga, H.; Goto, M.; Suyama, A.; Furukawa, K. A New ICEclc Subfamily Integrative and Conjugative Element Responsible for Horizontal Transfer of Biphenyl and Salicylic Acid Catabolic Pathway in the PCB-Degrading Strain Pseudomonas stutzeri KF716. Microorganisms 2021, 9, 2462. https://doi.org/10.3390/microorganisms9122462
Hirose J, Watanabe T, Futagami T, Fujihara H, Kimura N, Suenaga H, Goto M, Suyama A, Furukawa K. A New ICEclc Subfamily Integrative and Conjugative Element Responsible for Horizontal Transfer of Biphenyl and Salicylic Acid Catabolic Pathway in the PCB-Degrading Strain Pseudomonas stutzeri KF716. Microorganisms. 2021; 9(12):2462. https://doi.org/10.3390/microorganisms9122462
Chicago/Turabian StyleHirose, Jun, Takahito Watanabe, Taiki Futagami, Hidehiko Fujihara, Nobutada Kimura, Hikaru Suenaga, Masatoshi Goto, Akiko Suyama, and Kensuke Furukawa. 2021. "A New ICEclc Subfamily Integrative and Conjugative Element Responsible for Horizontal Transfer of Biphenyl and Salicylic Acid Catabolic Pathway in the PCB-Degrading Strain Pseudomonas stutzeri KF716" Microorganisms 9, no. 12: 2462. https://doi.org/10.3390/microorganisms9122462
APA StyleHirose, J., Watanabe, T., Futagami, T., Fujihara, H., Kimura, N., Suenaga, H., Goto, M., Suyama, A., & Furukawa, K. (2021). A New ICEclc Subfamily Integrative and Conjugative Element Responsible for Horizontal Transfer of Biphenyl and Salicylic Acid Catabolic Pathway in the PCB-Degrading Strain Pseudomonas stutzeri KF716. Microorganisms, 9(12), 2462. https://doi.org/10.3390/microorganisms9122462





