Abstract
This study aims to explore the intra-species distribution of genetic characteristics that favor the persistence in the gastrointestinal tract (GIT) and host interaction of bacteria belonging to species of the Lacticaseibacillus genus. These bacterial species comprise commercial probiotics with the widest use among consumers and strains naturally occurring in GIT and in fermented food. Since little is known about the distribution of genetic traits for adhesion capacity, polysaccharide production, biofilm formation, and utilization of substrates critically important for survival in GIT, which influence probiotic characteristics, a list of genetic determinants possibly involved in such functions was created by a search for specific genes involved in the above aspects in the genome of the extensively characterized probiotic L. rhamnosus GG. Eighty-two gene loci were retrieved and their presence and variability in other Lacticaseibacillus spp. genomes were assessed by alignment with the publicly available fully annotated genome sequences of L. casei, L. paracasei, L. rhamnosus, and L. zeae. Forty-nine of these genes were found to be absent in some strains or species. The remaining genes were conserved and covered almost all the functions considered, indicating that all strains of the genus may exert some probiotic effects. Among the variable loci, a taurine utilization operon and a α-L-fucosidase were examined for the presence/absence in 26 strains isolated from infant feces by PCR-based tests. Results were variable among the isolates, though their common origin indicated the capacity to survive in the intestinal niche. This study indicated that the capacity to exert probiotic actions of Lacticaseibacillus spp. depends on a conserved set of genes but variable genetic factors, whose role is only in part elucidated, are more numerous and can explain the enhanced probiotic characteristics for some strains. The selection of the most promising probiotic candidates to be used in food is feasible by analyzing the presence/absence of a set of variable traits.
1. Introduction
Bacteria of the Lacticaseibacillus genus, in particular the species Lacticaseibacillus casei, paracasei, rhamnosus, and zeae, comprise strains able to colonize the human gastrointestinal tract (GIT) and exert probiotic effects. These species are also adapted to fermented food environments, primarily dairy products, but also fermented foods of plant origin. Many studies have dealt with the demonstration of the beneficial effects of these bacteria and some of them exert probiotic effects able to ameliorate or prevent different medical conditions [1,2,3]. Among these, L. rhamnosus GG (ATCC 53103) is the most extensively studied [4]. These bacteria are commercially available as food supplements or in probiotic food products, but they also constitute the spontaneous microbiota of fermented foods of large consumption, including traditional products. In particular, L. paracasei strains constitute one of the main microbial components in traditional cheeses during and at the end of ripening. These foods can be a source of Lacticaseibacillus strains in numbers sufficient to influence host health [5]. However, strains that are part of the natural dairy microbiota could have a variable capacity to exert beneficial effects, so that it could be useful to select those with enhanced probiotic potential. Indeed, the selection, among the autochthonous microorganisms, of Lacticaseibacillus bacteria with traits associated to probiotic functions and use of these as added cultures, could confer health-promoting properties to the dairy products and also prevent the development of adventitious bacteria with undesirable characteristics.
Therefore, this study was focused on identifying genetic traits that, if present, can increase the potential of a bacterial strain belonging to the Lacticaseibacillus genus to behave as a probiotic. To this aim, the genome of the most-studied Lacticaseibacillus probiotic, L. rhamnosus GG, was used as a reference for identifying gene loci encoding for characteristics involved in survival in GIT and colonization capacity, adhesion to mucus or other host molecules, and production of cell-surface-associated macromolecules, including exopolysaccharides (EPS) involved in adhesion and immune modulation [6]. All the gene loci to which any of the above functions could be assigned, based on the existing annotation or on protein databases and scientific literature consultation, were searched by Blastn in the other Lacticaseibacillus spp. completely annotated genomes. The variability in the presence/absence of each gene, or gene cluster where appropriate, is presented. In addition, the presence/absence analysis of variable loci encoding taurine uptake and fucose utilization, as well as physiological features such as tolerance to bile salts and biofilm formation, were determined for 26 Lacticaseibacillus fecal isolates. These properties were selected on the basis of previous evidence on their involvement in survival in the intestinal environment. Indeed, taurine utilization capacity conferred by the tauBAC gene cluster was suggested to increase bile tolerance and persistence in GIT [7], while utilization of L-fucose, one of the most common monosaccharides in glycans on mammalian cell surfaces and intestinal mucus, allowed by the presence of α-L-fucosidases, is advantageous for use of this sugar as a carbon source in GIT [8].
2. Materials and Methods
2.1. Analysis of Lacticaseibacillus spp. Genomes for Genes Involved in Survival in GIT and Adhesion
The whole genome sequence of L. rhamnosus GG (GenBank acc. n. FM179322.1) was examined visually for the presence of genes involved in survival in GIT, polysaccharide production, and adhesion on the basis of the predicted function indicated for each gene locus. Membrane- and cell-surface-associated proteins without an assigned function were analyzed by Blastp (https://blast.ncbi.nlm.nih.gov/, accessed on 2 May 2022), Interpro (https://ebi.ac.uk/interpro/, accessed on 29 March 2022), and UniProt (https://www.uniprot.org, accessed on 29 March 2022) searches to derive a putative functional role. Only genes encoding proteins with a function assigned on the basis of the above analyses, or experimentally proven on the basis of scientific literature, were retained for the search of homologs of the encoding genes in the available complete and fully annotated genomes of L. casei, L. paracasei, L. rhamnosus, and L. zeae by Blastn. The latter species was considered for its close relatedness with L. casei and for its probiotic potential [9]. The cut-off values fixed for the definition of homology were a minimum of 30% query coverage and sequence identity above 60%.
Polysaccharide production gene clusters were graphically represented by using the https://katlabs.cc/genegraphics/app, accessed on 1 March 2022.
2.2. Bacterial Strains and Culture Conditions
Bacterial strains examined were isolated from the feces of children in a previous study [10] and assigned to the species L. casei, L. paracasei, L. rhamnosus, and L. zeae according to the highest Blastn scores of their 16S rRNA gene sequences. L. rhamnosus GG ATCC 53103 was used as positive control in PCR-amplification tests for genes tauB and the α-L-fucosidase gene LGG_02652. Lactobacilli were subcultured in an MRS broth or agar (Biolife Italiana, Milan, Italy) at 37 °C in aerobiosis for 48 h.
2.3. Molecular Techniques
DNA was extracted from 1 mL of fresh bacterial culture according to Amadoro et al. (2018). PCR tests for the screening of relevant genetic traits were carried out with primers 27f (5′-AGAGTTTGATCCTGGCTCAG-3′)/1492r (5′- GGTTACCTTGTTACGACTT-3′) targeted on the 16S rRNA gene, TauBF (5′-AGG(C/G)TC(G/T)GCATAGGC-3′)/TauBR (5′-CATGT(A/G)G(A/C)(C/T)TA(C/T)TGTTAC-3′) targeted on the taurine uptake gene tauB, locus LGG_00172, and FucF (5′-(G/T)AAC(C/G)ACCCAGTCACT-3′)/FucR (5′-G(A/T)CAGAACCA(C/T)TACCG) targeted on the α-fucosidase gene, locus LGG_02652. The latter two degenerate primer pairs were designed on consensus nucleotide positions in the genes tauB and LGG_02652 after alignment of the homologous gene sequences by Clustal Ω (https://www.ebi.ac.uk/Tools/msa/clustalo/, accessed on 10 September 2021) for L. paracasei and L. rhamnosus and all the Lacticaseibacillus species, respectively. Primer specificity was checked by Blastn and their melting temperature and tendency to form dimers were optimized by the Eurofins Genomics (Ebersberg, Germany) Oligo Calculator (https://www.google.com/search?client=firefox-b-d&q=eurofins+oligo+calculator, accessed on 12 September 2021).
The expected length of the amplification product was 649 for L. paracasei and 665 bp for L. rhamnosus for TauBF/TauBR primers and 1220 bp for FucF/FucR primers for both species. In the PCR reactions, primers were used in 0.5 µM final concentration carried out with the Takara Bio EmeraldAmp GT PCR Master Mix (Diatech, Jesi, AN, Italy). PCR programs comprised an initial denaturation at 94 °C for 5 min, 40 cycles of denaturation at 94 °C for 30 s, annealing for 30 s, and elongation at 72 °C for 1 min. A final elongation at 72 °C for 5 min was executed. The annealing temperatures were 55 °C for primer pair 27f/1492r and 50 °C for the other primer pairs.
The amplification products were separated on a 1.5% (w/v) agarose gel prepared in 1× TAE buffer (80 mM Tris/acetate, 2 mM EDTA, pH 8.0), stained with GelRed (Biotium, Fremont, CA, USA, DiaTech, Jesi, Italy) in the recommended amount, and run at 120 V in 1× TAE buffer.
Sequencing of the amplification products was carried out after purification with the Wizard® SV Gel and PCR Clean-Up System (Promega Italia Srl, Milan, Italy) at Eurofins Genomics and the same primers used for amplification were used as sequencing primers.
2.4. Biofilm Forming Capacity
Two hundred µL of a 48 h culture in an MRS broth of each strain were transferred in triplicate in a microtiter plate well and incubated at 37 °C for 24 h. After the incubation the well contents were aspirated and the well was washed thrice with sterile saline. The well was filled with 200 µL of 99% methanol and kept for 5 min at room temperature. Methanol was aspirated and the well was let dry before adding 200 ul of a 2% (w/v) aqueous solution of crystal violet (Merck Life Science S.r.l., Milan, Italy). This solution was left in contact for 5 min. The colorant solution was removed; the wells were let dry and washed several times with water. Finally, 160 µL of 33% (w/v) acetic acid solution were added and the optical density (OD) of the wells was read at 620 nm in a 1420 multilabel counter Victor 3 v plate reader (PerkinElmer Italia, Milan, Italy).
2.5. Bile Salt Tolerance Test
Bile tolerance was assayed as described by Bustos et al., 2006 [11] by determination of the transmembrane electrical potential (Δψ) dissipation of energized cells, after the addition of bile salts to a 1.5% (w/v) final concentration. The method of Bustos et al., 2006 [11] was modified for use of safranin O (λ excitation 520 nm, λ emission 570 nm) 1.25 µM as a cell uptake probe and the use of 1 µM of protonophore carbonyl cyanide-p-trifluoromethoxy-phenyl hydrazine (FCCP) to completely depolarize cell membranes. The experiments were carried out in triplicate. Changes in fluorescence were measured with a LS50B spectrofluorimeter (PerkinElmer). The Δψ dissipation level caused by bile salts was expressed as a percentage of the increase in fluorescence after bile salt addition on the increase in fluorescence measured after total (100%) Δψ dissipation with FCCP.
2.6. Statistical Analyses
The data series of bile salt tolerance and biofilm formation were compared among strains by the Student’s t-test in Microsoft Excel. These were considered to be statistically distinct for p ≤ 0.05.
3. Results
3.1. Distribution of Genetic Traits Required for Probiotic Activity in Lacticaseibacillus Species
The selection of genetic traits to be compared among strains in the genome of L. rhamnosus GG, after identification of some proteins with unassigned function by search in protein databases, resulted in the list reported in Table 1.

Table 1.
Gene loci in the genome of L. rhamnosus GG encoding traits relevant for the capacity to exert probiotic properties separated on the basis of their functional role.
It can be stated that at least one representative of each functional group of genes was found to be present in all genomes, except the bile salt hydrolase homologous to LGG_00501, genes for taurine utilization, sortases and pili, the anti-inflammatory protein PrtR encoded by locus LGG_02734, and fibrinogen-binding functions.
The role of each gene and the percentage of the analyzed genomes in which it is found is detailed in Table S1. Each gene locus in the list was aligned by Blastn to the completely annotated genomes of the species Lacticaseibacillus casei, L. paracasei, L. rhamnosus, and L. zeae, which were represented by 5, 54, 38, and 2 entries, respectively, in NCBI at the time of analysis. Strain 12A was included in the analyses as a member of the L. paracasei species because it was found to be erroneously identified as L. casei [12].
Traits that are present in all genomes include the bile salt hydrolase (bsh) gene. This indicates the common ability to detoxify bile salts conferred by this enzyme in Lacticaseibacillus species [13]. An exception is L. paracasei ATCC 334, in which a truncated copy of the gene is present. Surface antigens p40, p75, p60, MetQ, and LGG_00503 are also conserved. However, the MetQ-encoding gene appears to be truncated in L. paracasei N1115, and therefore it is possibly not functional in this strain. Among these surface antigens, p40 and p75 have a role in the protection of inflammation and integrity of the intestinal epithelium [14] and in L. paracasei BL23 have cell-wall hydrolase activity and are secreted in microvesicles [15,16]. Antigen p60 has an immunomodulatory function [17] and the myosin-cross-reactive antigen encoded by locus LGG_00503 may be also involved in the production of conjugated linoleic acid [18]. Other conserved genes are 11 glycosyltransferases for the biosynthesis of polysaccharides or lipopolysaccharides, a flippase-like protein LGG_00827, a lipotheicoic acid (LTA) synthase LGG_00830, a polysaccharide biosynthesis transport protein LGG_00851, the LiaX daptomycin-sensing surface protein LGG_00914, a PspC domain-containing protein that in Staphylococcus mutans mediates biofilm formation in vivo [19], a toxin immunity protein LGG_01002, a lipopolysaccharide assembly protein LGG_01366, a fibronectin binding protein FbpA, a AI-2E family transporter possibly involved in biofilm formation, a mucus-binding protein MucBP, the first proteins of two gene clusters for polysaccharide production LGG_01990 and LGG_02036, a teichoic acid glycosylation protein LGG_02144, two PsaA putative adhesion lipoproteins and a polysaccharide transport protein LGG_02520.
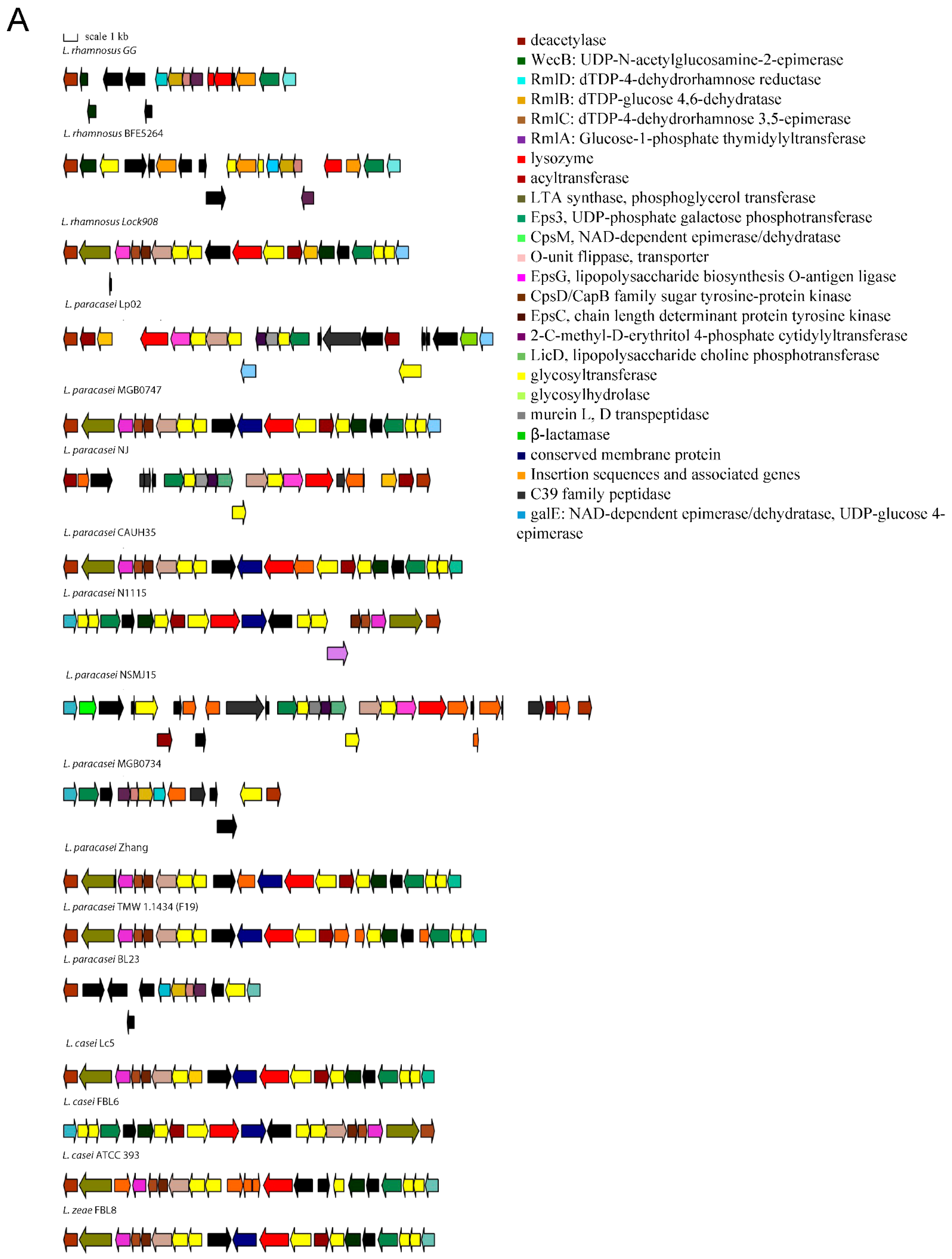
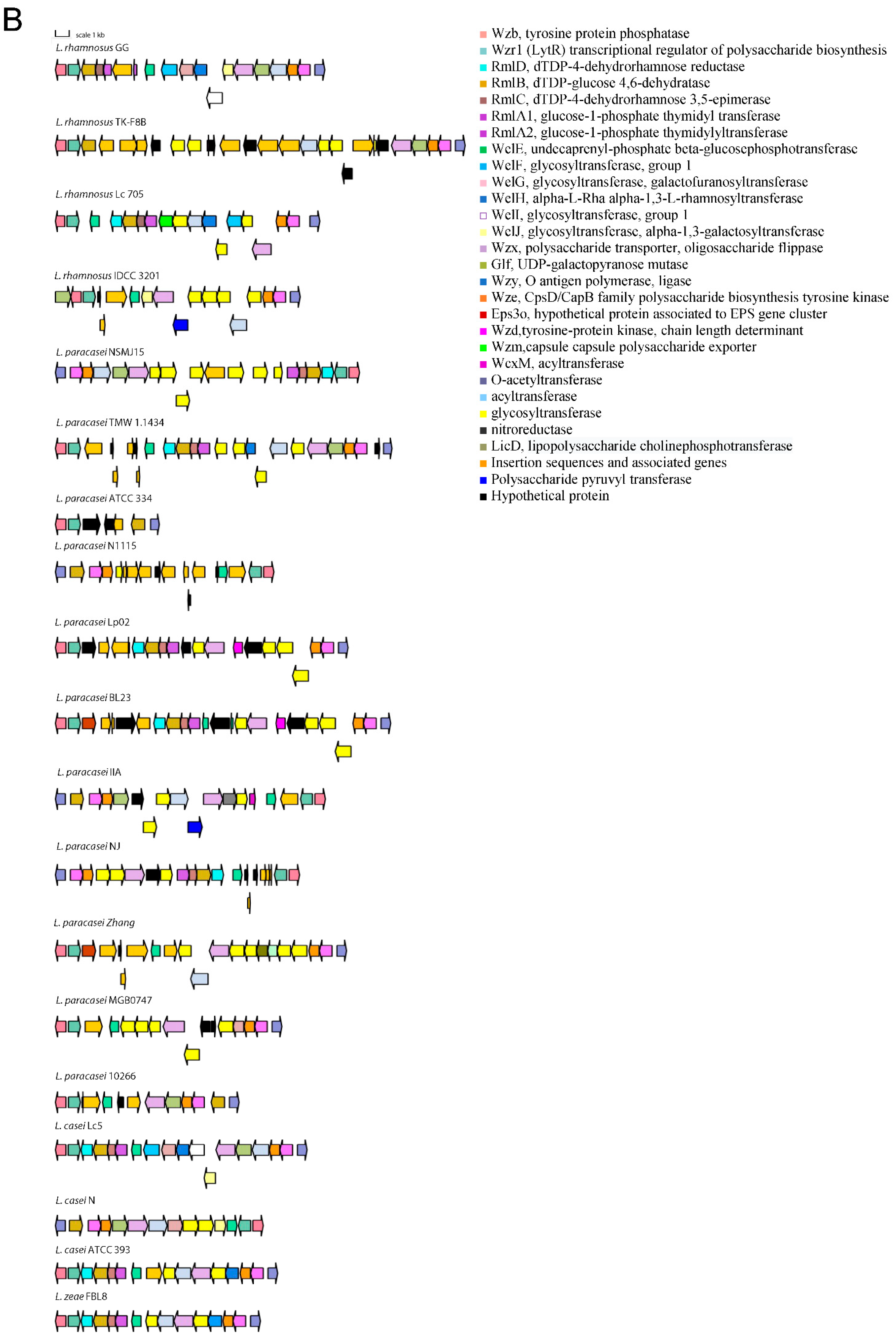
The variable genes follow a species-specific or a strain-specific distribution. Namely, some strains of L. casei do not have a FeoB for Fe(II) uptake-encoding gene that confers increased gut colonization ability [20]. The L. paracasei species lacks a β-N-acetylhexosaminidase that is variable in L. rhamnosus strains, the TauE protein involved in taurine metabolism, the lectin-like protein LGG_00579, the cell surface protein LGG_00584, the MabA extracellular matrix binding protein, a modulator of adhesion and biofilm formation [21], the cell envelope-associated proteinase LGG_02734, lactocepin PrtR, able to selectively degrade pro-inflammatory chemokines and reduce inflammation in experimental IBD models [22], and the fibrinogen-binding protein LGG_02282. Some L. paracasei strains lack the InlJ internalin LGG_02337. L. casei and L. zeae lack genes encoding the SpaCBA and SpaFED pili, a pilin subunit LGG_00422, the adhesion exoprotein LGG_02923, and five proteins containing the WxL domain [23]. These proteins are involved in single-species biofilm formation and for some a lectin function was proven [24]. Other proteins not encoded in the genomes of L. casei and L. zeae are the extracellular complex proteins SpcA and B, the adhesin LGG_01590, one glycosyltransferase, and the cell surface protein LGG_00578. The cell-surface-docked proteins encoded by the gene cluster LGG_01589 to LGG_01592 that might be involved in adhesion, are highly conserved among S-layer-forming lactobacilli, are expressed constitutively, and are specific to vertebrate-adapted species, suggesting a role in adaptation to these hosts [25]. Worthy of note is that the SpaCBA pilus has a variable presence in L. rhamnosus, while it was found to be present in all analyzed L. paracasei strains, with six of them having an additional plasmid-encoded copy. In L. paracasei, an additional pilin subunit D1, SpaA (LGG_00422), present in all genomes, is also found on plasmid in six strains. The remaining genes with an intra-species varying distribution are the SpaFED pili, the taurine metabolism system, and proteins associated with EPS production. The role of the SpaFED pilus is not well-defined and it was reported not to be expressed in L. rhamnosus GG [26,27]. The EPS production genes in L. rhamnosus GG are arranged in three gene clusters, namely loci from LGG_00278 to LGG_00283, loci from LGG_01990 to LGG_02005 and loci from LGG_02036 to LGG_02054. The first cluster comprises genes present only in some L. paracasei and L. rhamnosus strains. These are three ramnosyltransferases, a polysaccharide transporter Eps1, and an Eps2 protein involved in polysaccharide biosynthesis that in some strains are dislocated in different EPS gene clusters. Given the high variability in EPS production gene arrangement, the remaining two clusters are shown graphically in Figure 1 for strains representative of the different gene content and order observed.

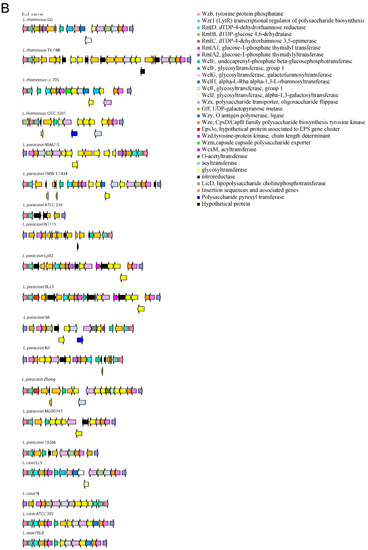
Figure 1.
Gene arrangement in the EPS production gene clusters starting from proteins orthologous to LGG_01990 (A) and LGG_02036 (B).
3.2. Testing of Genetic and Physiological Features in Lacticaseibacillus Strains from Feces
As relevant features for adaptation to the intestinal environment, taurine- and fucose-utilization capacity were tested in 26 strains identified as Lacticaseibacillus species by 16S rRNA gene sequencing among lactobacilli from the feces of children obtained in a previous study [10]. Moreover, bile salt resistance was determined by a fluorimetric method and biofilm formation capacity in microtiter plates was also examined. Results are summarized in Table 2.

Table 2.
Distribution of variable genes involved in survival in GIT in intestinal isolates of L. casei, L. paracasei, L. rhamnosus, and L. zeae, percent dissipation of membrane potential in presence of 1.5% bile salts and biofilm formation extent.
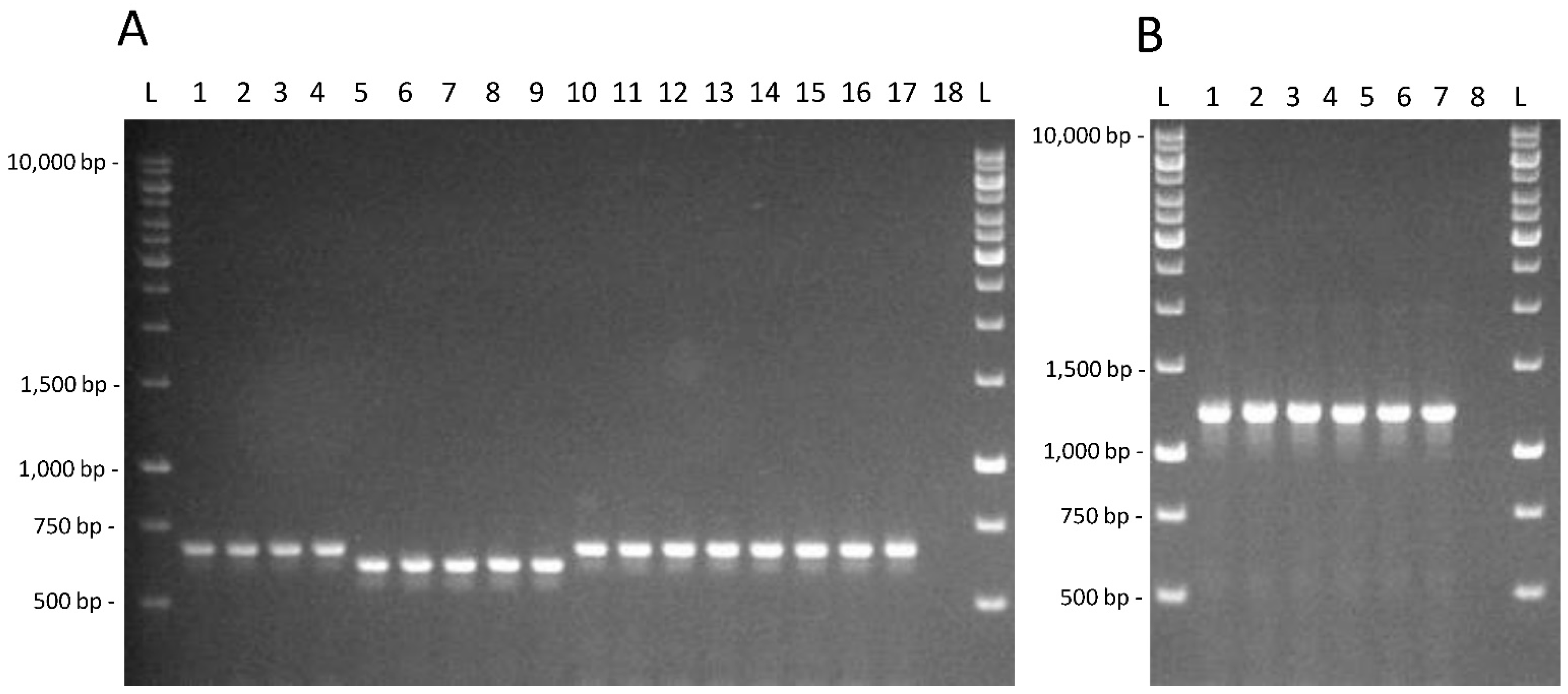
It is possible to observe that taurine utilization and fucosidase activity presence are neither shared by all the intestinal isolates nor present in most of them. The amplification products for the PCR assays targeted on the tauB gene and on the α-fucosidase LGG_02652, whose identity was confirmed by the sequencing of the amplicons obtained from L. rhamnosus GG, are shown in Figure 2.
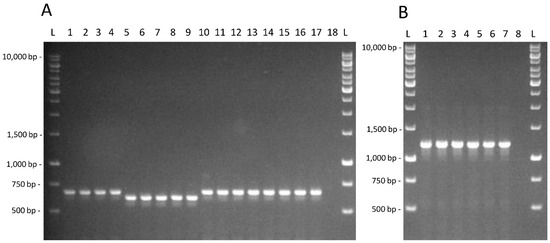
Figure 2.
(A) Amplification products obtained for the tauB gene for strains L. casei AB-15-6 (1); C-15-1 (2); C-15-1b (3); G-0-6 (4); L. paracasei AN-15-2 (5); AN-15-3 (6); J-7-1 (7); J-15-4 (8); P-7-13 (9); L. rhamnosus AN-7-4 (10); AN-21-1 (11); D-0-5 (12); G-7-14 (13); G-7-16 (14); J-7-4 (15); Z-15-4 (16); GG (17); negative control (18). (B). Amplification products obtained for the α-fucosidase LGG_02652 targeted PCR test for strains L. casei C-15-1b (1); L. paracasei P-7-13 (2); L. rhamnosus AN-21-2 (3); D-0-5 (4); Z-15-4 (5); GG (6); negative control (7). L; GeneRuler 1 kb DNA Ladder (ThermoFisher Scientific, Monza, Italy).
The strains formed four statistically distinct groups (a–d) on the basis of tolerance to bile salts and three strains, L. paracasei 6-15-1, L. rhamnosus Z-15-4, and L. zeae J-7-2, were not included in any group. Strain L. zeae J-7-2 was the most sensitive to bile salts based on the 95% extent of membrane depolarization.
According to biofilm-forming capacity, the strains were distributed in six statistically distinct groups (a–f) with strains L. rhamnosus G-7-14 and SA-7-6 not included in any of those groups. Four isolates, L. casei C-15-1b, L. paracasei P-7-13, L. rhamnosus AN-0-1, and G-7-16, showed a biofilm-forming capacity comparable to that of L. rhamnosus GG, while strains L. paracasei AN-15-1 and J-7-3 and L. rhamnosus G-7-14 and SA-7-6 exhibited a biofilm-forming capacity stronger than L. rhamnosus GG.
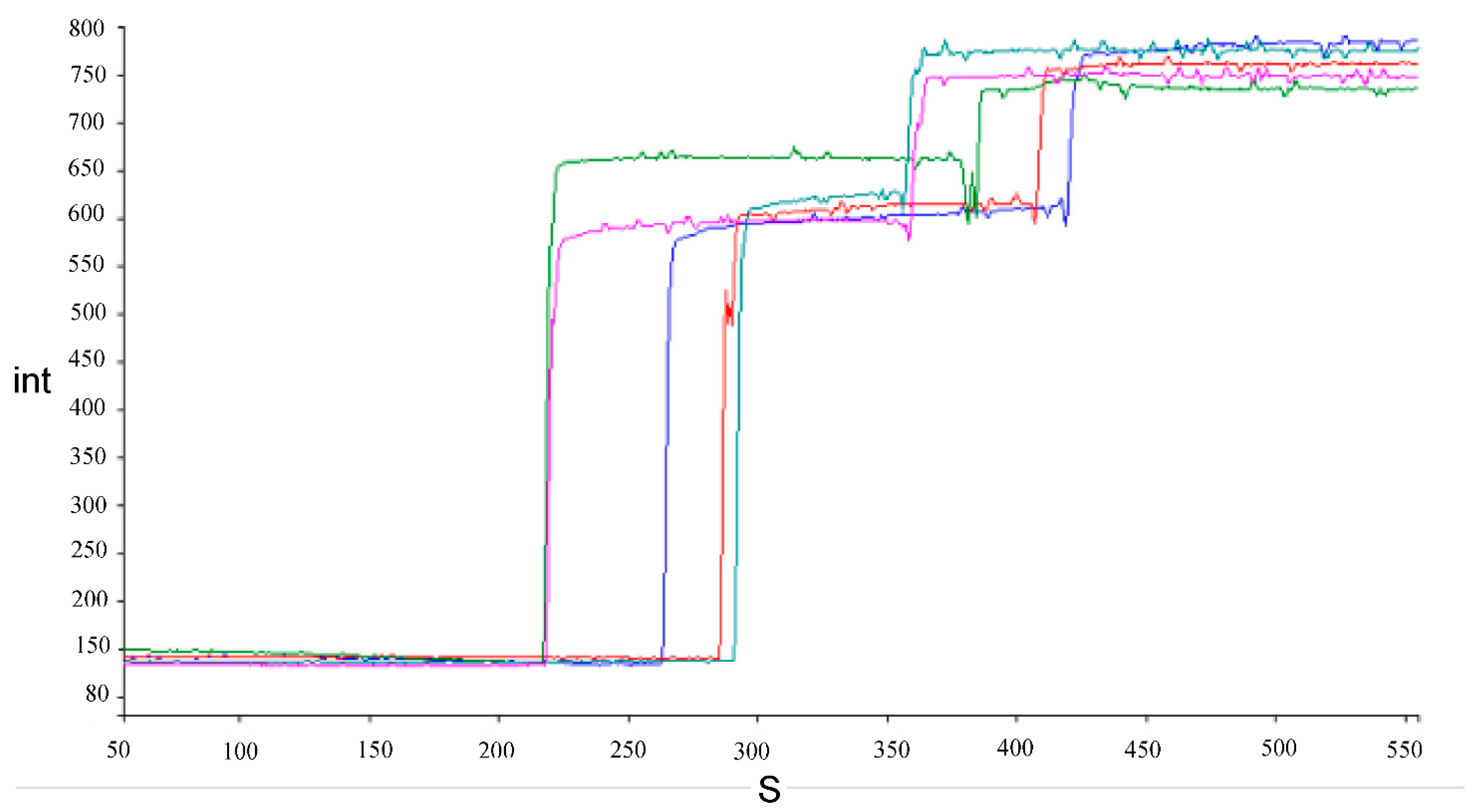
Examples of traces showing the increase in fluorescence intensity caused by the release of safranin O after the addition of bile salts to energized cells and complete depolarization after addition of FCCP are shown in Figure 3.
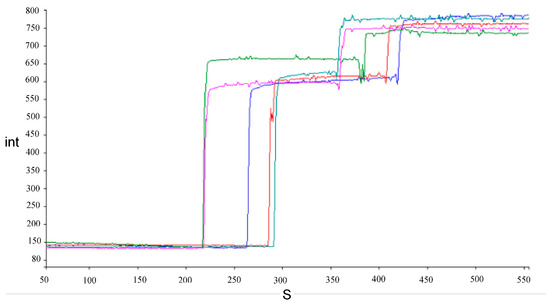
Figure 3.
Examples of traces of fluorescence intensity (int) increase during time in seconds (S) for energized Lacticaseibacillus cells after addition of 1.5% (w/v) bile salts and FCCP in succession. Each color represents a different strain, namely, L. paracasei AN-15-3 (blue), J-7-1 (olive green) and J-15-4 (sea green), L. rhamnosus AN-21-2 (purple), and G-7-14 (red).
4. Discussion
This study highlighted that the genetic characteristics that influence survival and persistence in GIT and the probiotic effects exerted by bacteria of the Lacticaseibacillus genus are very complex. Genetic loci common to all the analyzed genomes encode for functions such as adhesion, bile resistance, polysaccharide production, fibronectin binding, and cell–cell signaling and represent a constant endowment of genes that allow coping with the GIT environment by all bacteria of the species considered. An exception among the analyzed genomes is represented by the dairy strain L. paracasei ATCC 334 (Acc. N. NC_008526.1), in which a nonfunctional bsh gene is present and one of the main EPS-production gene clusters is almost completely deleted (Figure 1B). Among the conserved genes the one encoding of the MetQ protein was also found to be truncated in one out of 54 L. paracasei genomes analyzed, thus indicating that in a minority of strains some of the most conserved traits may also be lacking.
Among the common traits, the bsh gene product has particular relevance for the probiotic function, since it has been shown to efficiently lower total and low-density lipoprotein cholesterol [28].
Another common trait that favors host colonization is the presence of the fibronectin-binding protein (FnBP) FbpA. This protein type binds fibronectin, a multidomain glycoprotein found in the human body fluids, extracellular matrices, and intestinal epithelial cells that are common targets for bacterial adhesins in GIT. The FbpA from L. paracasei BL23 has been characterized and found to exhibit a strong affinity for immobilized fibronectin [29]. In L. acidophilus, a mutant with inactivated fbpA, exhibited a significant decrease in adhesion to epithelial cells in vitro. While in pathogens, some FnBPs contribute to virulence, FnBPs in commensal and probiotic strains these proteins are essential for persistence in their ecological niches and might exert competition against pathogens for binding to fibronectin [30].
In most of the genomes, except for two L. casei strains, a FeoB protein, essential for the uptake of ferrous iron and gut colonization, is encoded [20].
Proteins p75 and p40 present in all Lacticaseibacillus genomes were found to mitigate intestinal inflammation through activation of the epidermal growth factor receptor [31] and upregulation of a proliferation-inducing ligand in the epithelium that stimulates the secretion of immunoglobulin A and relieves cytokine-induced apoptosis in the intestinal epithelial cells [32]. In addition, p75 and p40 stimulate epithelial cells to activate pathways that enhance their survival and barrier function to prevent bacterial translocation and the invasion by toxins [33].
The majority of the genetic determinants with a role in adaptation in GIT considered in this study was variable in the Lacticaseibacillus genomes. Among these, the sortase-dependent pili SpaCBA and SpaFEG, with the first having a proven role in adhesion to mucus, collagen, biofilm formation, and immune cell response stimulation, are included [26]. Similarly to what observed in this study, Douillard et al. [7] found that all the L. paracasei strains that they examined contained a SpaCBA pilus cluster. However, only the L. rhamnosus strains produced a functional SpaCBA pilus since the insertion of an IS 30 element upstream of the pilus gene cluster had constituted a strong promoter that allowed pilus expression. On the other hand, they observed that some strains displayed a mucus-binding capacity also in absence of SpaCBA pili, suggesting the existence of alternative mucus-binding mechanisms.
The relevance of the SpaCBA pilus in L. paracasei physiology should be better elucidated. This is present in two copies in some L. paracasei strains, among which strain LP10266 (Acc. n. NZ_CP031785.1 and n. NZ_CP031786.1) was recently shown to exhibit increased adhesion capacity. This strain was isolated from a patient with endocarditis and it was hypothesized that increased adherence can represent a virulence trait [34]. High adherence determined by a plasmid-encoded SpaCBA pilus was reported also for a L. paracasei strain isolated from raw cow’s milk [35].
The SpaCBA pilus was absent in the L. casei and L. zeae genomes analyzed, but this can be attributed to their low number, that is five genomes and two genomes, respectively. This might explain why the tauB gene, absent in the five L. casei genomes analyzed, was instead detected in the fecal isolates.
However, in general, the strains of these two species analyzed in this study are defective of many important adaptive traits. This might explain their infrequent occurrence in different ecological niches compared to L. paracasei and L. rhamnosus.
The taurine uptake system is another variable trait that, based on the genome analysis of 19 L. casei/paracasei strains, was proposed to be lost by strains adapted to dairy niches [36]. However, in this study, it appeared to be absent also in strains of intestinal origin, indicating that it may not be essential, at least in the short term, for survival in GIT. It can be noted that this gene, predicted to be absent in the L. casei genomes, was instead found to be present in the strains screened by PCR, thus indicating that the few genomes available for this species do not allow an exhaustive analysis of the distribution of genetic determinants relevant for probiotic action.
The α-L-fucosidase found to be absent in some genomes in this study and corresponding to the experimentally characterized LCABL_28270 in L. paracasei BL23 (GenBank acc. n. FM177140) (Rodríguez-Díaz et al., 2011 [8]) was also proposed to be lost by dairy strains [7]. However, it appeared to be present only in a minority of fecal isolates.
A great variability was displayed by the EPS-production gene clusters. This observation explains the diverse EPS types produced by Lacticaseibacillus strains and experimentally characterized [37,38]. Given the multiple actions exerted by these macromolecules, such as immunomodulation, antioxidant properties, enhancement of the hydrophobicity of bacterial cell surface that increases binding to the intestinal mucosa [37,38], variability in composition of these macromolecules can create also a variability of the effects that these bacteria can exert, that deserve to be defined to the strain level.
MucBP proteins were found to be present in most genomes. One of these, namely the LGG_02337, an adhesin distributed on the whole cell surface, participates in the adhesion of L. rhamnosus GG to mucus, and was recognized to be involved in pilus-mediated mucosal adhesion [25].
Finally, proteins with a WxL C-terminal domain, shown to possibly form a cell-surface protein complex involved in the degradation of plant polysaccharides in other lactobacilli, have a lectin-like function [24]. One protein of this group was found to be responsible for the adherence of L. rhamnosus GR-1 to the vaginal epithelium [39].
5. Conclusions
This study highlighted that all the Lacticaseibacillus spp. genomes analyzed comprise a common set of genes that could favor probiotic functions to be exerted by most of the microorganisms belonging to this genus. However, another set of gene loci is variable and can confer increased colonization capacity and beneficial interaction with the host to some strains. The presence of genetic traits relevant for survival in GIT, namely taurine and fucose utilization, as well as bile salt tolerance and biofilm formation, were found to be variable in fecal isolates, showing that a complex of features, not single traits, play a role in adaptation to the intestinal niche. Numerous traits emerged whose functional role is still little explored so far. This study resulted in the identification of variable genetic traits to be analyzed in the preliminary selection of the natural Lacticaseibacillus strain to be used to increase the beneficial properties of fermented products and probiotic candidates to be further characterized by whole-genome sequencing, in accordance with the current European food safety guidelines [40].
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/microorganisms10051023/s1: Table S1: Genes encoding functions required for survival/persistence in GIT, adhesion and exopolysaccharides production in the genome of L. rhamnosus GG (GenBank Acc. n. NC_013198.1/FM179322.1) and their presence/absence, identity and coverage percentages in other fully annotated Lacticaseibacillus genomes.
Author Contributions
F.R. and G.C. conceptualized the study, F.R. contributed to methodology, wrote the manuscript draft and made corrections, C.A. and M.L.P. equally contributed to methodology, interpreting data and editing, G.C. supervised and coordinated the study and critically revised the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Jacouton, E.; Chain, F.; Sokol, H.; Langella, P.; Bermúdez-Humarán, L.G. Probiotic Strain Lactobacillus casei BL23 Prevents Colitis-Associated Colorectal Cancer. Front. Immunol. 2017, 8, 1553. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Cao, C.; Wu, Z.; Zhang, H.; Sun, Z.; Wang, M.; Xu, H.; Zhao, Z.; Wang, Y.; Pei, G.; et al. The probiotic L. casei Zhang slows the progression of acute and chronic kidney disease. Cell Metab. 2021, 33, 1926–1942.e8. [Google Scholar] [CrossRef] [PubMed]
- Colombo Pimentel, T.; Ramalho Brandão, L.; Pereira de Oliveira, M.; Almeida da Costa, W.K.; Magnani, M. Health benefits and technological effects of Lacticaseibacillus casei-01: An overview of the scientific literature. Trends Food Sci. Technol. 2021, 114, 722–737. [Google Scholar] [CrossRef]
- Capurso, L. Thirty Years of Lactobacillus rhamnosus GG: A Review. J. Clin. Gastroenterol. 2019, 53 (Suppl. 1), S1–S41. [Google Scholar] [CrossRef] [PubMed]
- Del Matto, I.; Rossi, F.; Iannitto, G.; Petrone, D.; Mastrodomenico, M.T.; Alessiani, A.; Sacchini, L.; Amadoro, C.; Tucci, P.; Marino, L. Variability of the microbiota in traditional Caciocavallo, Scamorza and Caciotta cheeses manufactured with raw milk and natural cultures. Int. J. Dairy Technol. 2021, 74, 564–574. [Google Scholar] [CrossRef]
- Lu, Y.; Han, S.; Zhang, S.; Wang, K.; Lv, L.; McClements, D.J.; Xiao, H.; Berglund, B.; Yao, M.; Li, L. The role of probiotic exopolysaccharides in adhesion to mucin in different gastrointestinal conditions. Curr. Res. Food Sci. 2022, 5, 581–589. [Google Scholar] [CrossRef]
- Douillard, F.P.; Ribbera, A.; Kant, R.; Pietilä, T.E.; Järvinen, H.M.; Messing, M.; Randazzo, C.L.; Paulin, L.; Laine, P.; Ritari, J.; et al. Comparative genomic and functional analysis of 100 Lactobacillus rhamnosus strains and their comparison with strain GG. PLoS Gen. 2013, 9, e1003683. [Google Scholar] [CrossRef]
- Rodríguez-Díaz, J.; Monedero, V.; Yebra, M.J. Utilization of Natural Fucosylated Oligosaccharides by Three Novel α-l-Fucosidases from a Probiotic Lactobacillus casei Strain. Appl. Environ. Microbiol. 2011, 77, 703–705. [Google Scholar] [CrossRef] [Green Version]
- Kim, E.; Yang, S.M.; Kim, D.; Kim, H.Y. Complete Genome Sequencing and Comparative Genomics of Three Potential Probiotic Strains, Lacticaseibacillus casei FBL6, Lacticaseibacillus chiayiensis FBL7, and Lacticaseibacillus zeae FBL8. Front. Microbiol. 2022, 12, 794315. [Google Scholar] [CrossRef]
- Amadoro, C.; Rossi, F.; Pallotta, M.L.; Gasperi, M.; Colavita, G. Traditional dairy products can supply beneficial microorganisms able to survive in the gastrointestinal tract. LWT 2018, 93, 376–383. [Google Scholar] [CrossRef]
- Bustos, A.Y.; Raya, R.; Bru, E.; de Valdez, G.F.; Taranto, M.P. Application of fluorescent techniques to evaluate the survival of probiotic lactobacilli to bile acid. Biotechnol. Lett. 2011, 33, 1389–1394. [Google Scholar] [CrossRef] [PubMed]
- Frolova, M.; Yudin, S.; Makarov, V.; Glazunova, O.; Alikina, O.; Markelova, N.; Kolzhetsov, N.; Dzhelyadin, T.; Shcherbakova, V.; Trubitsyn, V.; et al. Lacticaseibacillus paracasei: Occurrence in the Human Gut Microbiota and K-Mer-Based Assessment of Intraspecies Diversity. Life 2021, 11, 1246. [Google Scholar] [CrossRef] [PubMed]
- Begley, M.; Hill, C.; Gahan, C.G. Bile salt hydrolase activity in probiotics. Appl. Environ. Microbiol. 2006, 72, 1729–1738. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Baarlen, P.; Wells, J.M.; Kleerebezem, M. Regulation of intestinal homeostasis and immunity with probiotic lactobacilli. Trends Immunol. 2013, 34, 208–215. [Google Scholar] [CrossRef] [PubMed]
- Bäuerl, C.; Pérez-Martínez, G.; Yan, F.; Polk, D.B.; Monedero, V. Functional analysis of the p40 and p75 proteins from Lactobacillus casei BL23. J. Mol. Microbiol. Biotechnol. 2010, 19, 231–241. [Google Scholar] [CrossRef] [Green Version]
- Domínguez Rubio, A.P.; Martínez, J.H.; Martínez Casillas, D.C.; Coluccio Leskow, F.; Piuri, M.; Pérez, O.E. Lactobacillus casei BL23 Produces Microvesicles Carrying Proteins That Have Been Associated with Its Probiotic Effect. Front. Microbiol. 2017, 8, 1783. [Google Scholar] [CrossRef] [Green Version]
- Karlsson, M.; Scherbak, N.; Khalaf, H.; Olsson, P.E.; Jass, J. Substances released from probiotic Lactobacillus rhamnosus GR-1 potentiate NF-κB activity in Escherichia coli-stimulated urinary bladder cells. FEMS Immunol. Med. Microbiol. 2012, 66, 147–156. [Google Scholar] [CrossRef] [Green Version]
- Yang, B.; Chen, H.; Song, Y.; Chen, Y.Q.; Zhang, H.; Chen, W. Myosin-cross-reactive antigens from four different lactic acid bacteria are fatty acid hydratases. Biotechnol. Lett. 2013, 35, 75–81. [Google Scholar] [CrossRef]
- Jung, C.J.; Hsu, C.C.; Chen, J.W.; Cheng, H.W.; Yuan, C.T.; Kuo, Y.M.; Hsu, R.B.; Chia, J.S. PspC domain-containing protein (PCP) determines Streptococcus mutans biofilm formation through bacterial extracellular DNA release and platelet adhesion in experimental endocarditis. PLoS Pathog. 2021, 17, e1009289. [Google Scholar] [CrossRef]
- Naikare, H.; Palyada, K.; Panciera, R.; Marlow, D.; Stintzi, A. Major role for FeoB in Campylobacter jejuni ferrous iron acquisition, gut colonization, and intracellular survival. Infect. Immun. 2006, 74, 5433–5444. [Google Scholar] [CrossRef] [Green Version]
- Vélez, M.P.; Petrova, M.I.; Lebeer, S.; Verhoeven, T.L.; Claes, I.; Lambrichts, I.; Tynkkynen, S.; Vanderleyden, J.; De Keersmaecker, S.C. Characterization of MabA, a modulator of Lactobacillus rhamnosus GG adhesion and biofilm formation. FEMS Immunol. Med. Microbiol. 2010, 59, 386–398. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hörmannsperger, G.; von Schillde, M.A.; Haller, D. Lactocepin as a protective microbial structure in the context of IBD. Gut Microbes 2013, 4, 152–157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brinster, S.; Furlan, S.; Serror, P. C-terminal WxL domain mediates cell wall binding in Enterococcus faecalis and other gram-positive bacteria. J. Bacteriol. 2007, 189, 1244–1253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petrova, M.I.; Lievens, E.; Verhoeven, T.L.; Macklaim, J.M.; Gloor, G.; Schols, D.; Vanderleyden, J.; Reid, G.; Lebeer, S. The lectin-like protein 1 in Lactobacillus rhamnosus GR-1 mediates tissue-specific adherence to vaginal epithelium and inhibits urogenital pathogens. Sci. Rep. 2016, 6, 37437. [Google Scholar] [CrossRef]
- Gagic, D.; Wen, W.; Collett, M.A.; Rakonjac, J. Unique secreted-surface protein complex of Lactobacillus rhamnosus, identified by phage display. Microbiology 2013, 2, 1–17. [Google Scholar] [CrossRef]
- Chaurasia, P.; Pratap, S.; von Ossowski, I.; Palva, A.; Krishnan, V. New insights about pilus formation in gut-adapted Lactobacillus rhamnosus GG from the crystal structure of the SpaA backbone-pilin subunit. Sci. Rep. 2016, 6, 28664. [Google Scholar] [CrossRef] [Green Version]
- von Ossowski, I.; Reunanen, J.; Satokari, R.; Vesterlund, S.; Kankainen, M.; Huhtinen, H.; Tynkkynen, S.; Salminen, S.; de Vos, W.M.; Palva, A. Mucosal adhesion properties of the probiotic Lactobacillus rhamnosus GG SpaCBA and SpaFED pilin subunits. Appl. Environ. Microbiol. 2010, 76, 2049–2057. [Google Scholar] [CrossRef] [Green Version]
- Jones, M.L.; Tomaro-Duchesneau, C.; Martoni, C.J.; Prakash, S. Cholesterol lowering with bile salt hydrolase-active probiotic bacteria, mechanism of action, clinical evidence, and future direction for heart health applications. Expert Opin. Biol. Ther. 2013, 13, 631–642. [Google Scholar] [CrossRef]
- Munoz-Provencio, D.; Perez-Martinez, G.; Monedero, V. Characterization of a fibronectin-binding protein from Lactobacillus casei BL23. J. Appl. Microbiol. 2010, 108, 1050–1059. [Google Scholar] [CrossRef] [Green Version]
- Hymes, J.P.; Klaenhammer, T.R. Stuck in the Middle: Fibronectin-Binding Proteins in Gram-Positive Bacteria. Front. Microbiol. 2016, 7, 1504. [Google Scholar] [CrossRef] [Green Version]
- Yan, F.; Cao, H.; Cover, T.L.; Washington, M.K.; Shi, Y.; Liu, L.; Chaturvedi, R.; Peek, R.M., Jr.; Wilson, K.T.; Polk, D.B. Colon-specific delivery of a probiotic-derived soluble protein ameliorates intestinal inflammation in mice through an EGFR-dependent mechanism. J. Clin. Investig. 2011, 121, 2242–2253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Liu, L.; Moore, D.J.; Shen, X.; Peek, R.M.; Acra, S.A.; Li, H.; Ren, X.; Polk, D.B.; Yan, F. An LGG-derived protein promotes IgA production through upregulation of APRIL expression in intestinal epithelial cells. Mucosal. Immunol. 2017, 10, 373–384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, F.; Cao, H.; Cover, T.L.; Whitehead, R.; Washington, M.K.; Polk, D.B. Soluble proteins produced by probiotic bacteria regulate intestinal epithelial cell survival and growth. Gastroenterology 2007, 132, 562–575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, Q.; Hao, Y.; Wang, L.; Lu, C.; Li, M.; Si, Z.; Wu, X.; Lu, Z. Characterization of a bacterial strain Lactobacillus paracasei LP10266 recovered from an endocarditis patient in Shandong, China. BMC Microbiol. 2021, 21, 183. [Google Scholar] [CrossRef] [PubMed]
- Koryszewska-Bagińska, A.; Gawor, J.; Nowak, A.; Grynberg, M.; Aleksandrzak-Piekarczyk, T. Comparative genomics and functional analysis of a highly adhesive dairy Lactobacillus paracasei subsp. paracasei IBB3423 strain. Appl. Microbiol. Biotechnol. 2019, 103, 7617–7634. [Google Scholar] [CrossRef] [Green Version]
- Fontana, A.; Zacconi, C.; Morelli, L. Genetic Signatures of Dairy Lactobacillus casei Group. Front. Microbiol. 2018, 9, 2611. [Google Scholar] [CrossRef]
- Liu, C.F.; Tseng, K.C.; Chiang, S.S.; Lee, B.H.; Hsu, W.H.; Pan, T.M. Immunomodulatory and antioxidant potential of Lactobacillus exopolysaccharides. J. Sci. Food Agric. 2011, 91, 2284–2291. [Google Scholar] [CrossRef]
- Balzaretti, S.; Taverniti, V.; Guglielmetti, S.; Fiore, W.; Minuzzo, M.; Ngo, H.N.; Ngere, J.B.; Sadiq, S.; Humphreys, P.N.; Laws, A.P. A Novel Rhamnose-Rich Hetero-exopolysaccharide Isolated from Lactobacillus paracasei DG Activates THP-1 Human Monocytic Cells. Appl. Environ. Microbiol. 2017, 83, e02702-16. [Google Scholar] [CrossRef] [Green Version]
- Siezen, R.; Boekhorst, J.; Muscariello, L.; Molenaar, D.; Renckens, B.; Kleerebezem, M. Lactobacillus plantarum gene clusters encoding putative cell-surface protein complexes for carbohydrate utilization are conserved in specific gram-positive bacteria. BMC Genom. 2006, 7, 126. [Google Scholar] [CrossRef] [Green Version]
- European Food Safety Authority. EFSA statement on the requirements for whole genome sequence analysis of microorganisms intentionally used in the food chain. EFSA J. 2021, 19, 6506. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).