Proteome and Physiological Characterization of Halotolerant Nodule Endophytes: The Case of Rahnella aquatilis and Serratia plymuthica
Abstract
:1. Introduction
2. Materials and Methods
2.1. Isolation of Bacterial Strains from Vicia fava and Pisum sativum Nodules
2.2. Identification of Bacterial Endophytes
2.3. Physiological Characterization of Plant Beneficial Traits
2.4. Tolerance to Temperature and Antibiotic Resistance Profile
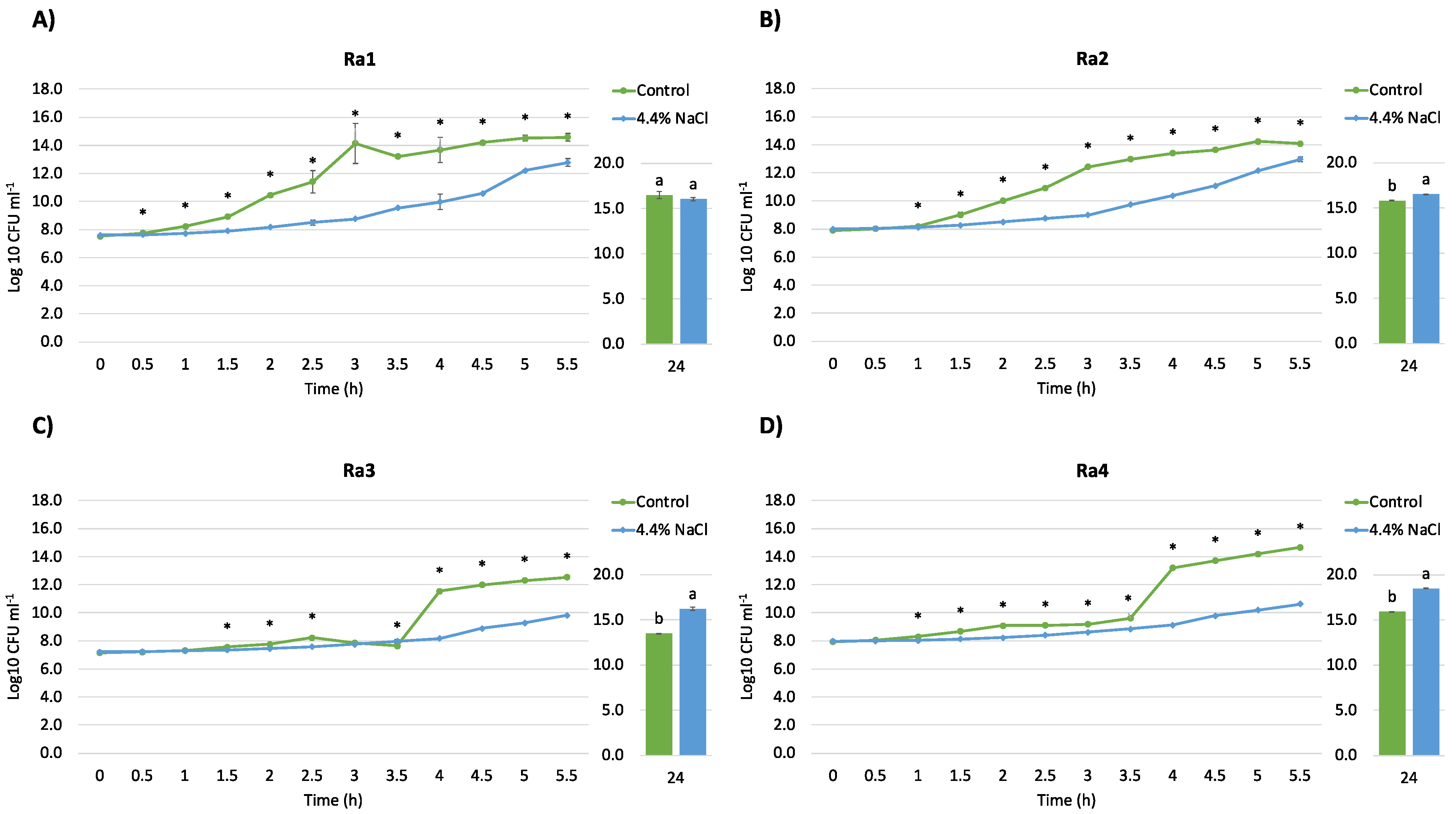
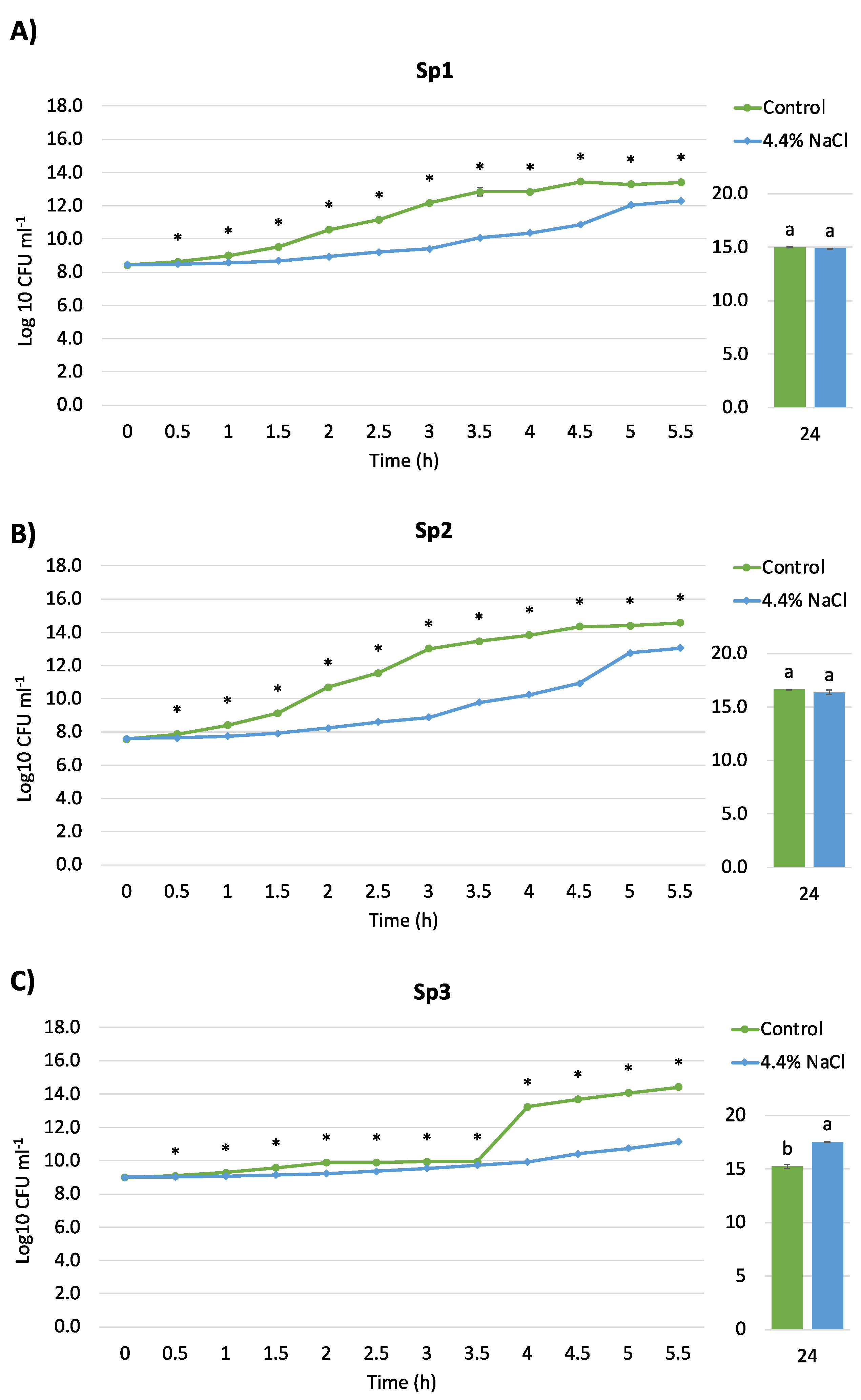
2.5. Salt Tolerance and Growth Curve
2.6. 1-Aminocyclopropane-1-carboxylic Acid (ACC) Deaminase Assay
2.7. Proteome Analysis
2.8. Bioinformatic Analysis
2.9. Statistical Analysis
3. Results and Discussion
3.1. Isolation, Identification, and Physiological Characterization of Bacterial Strains
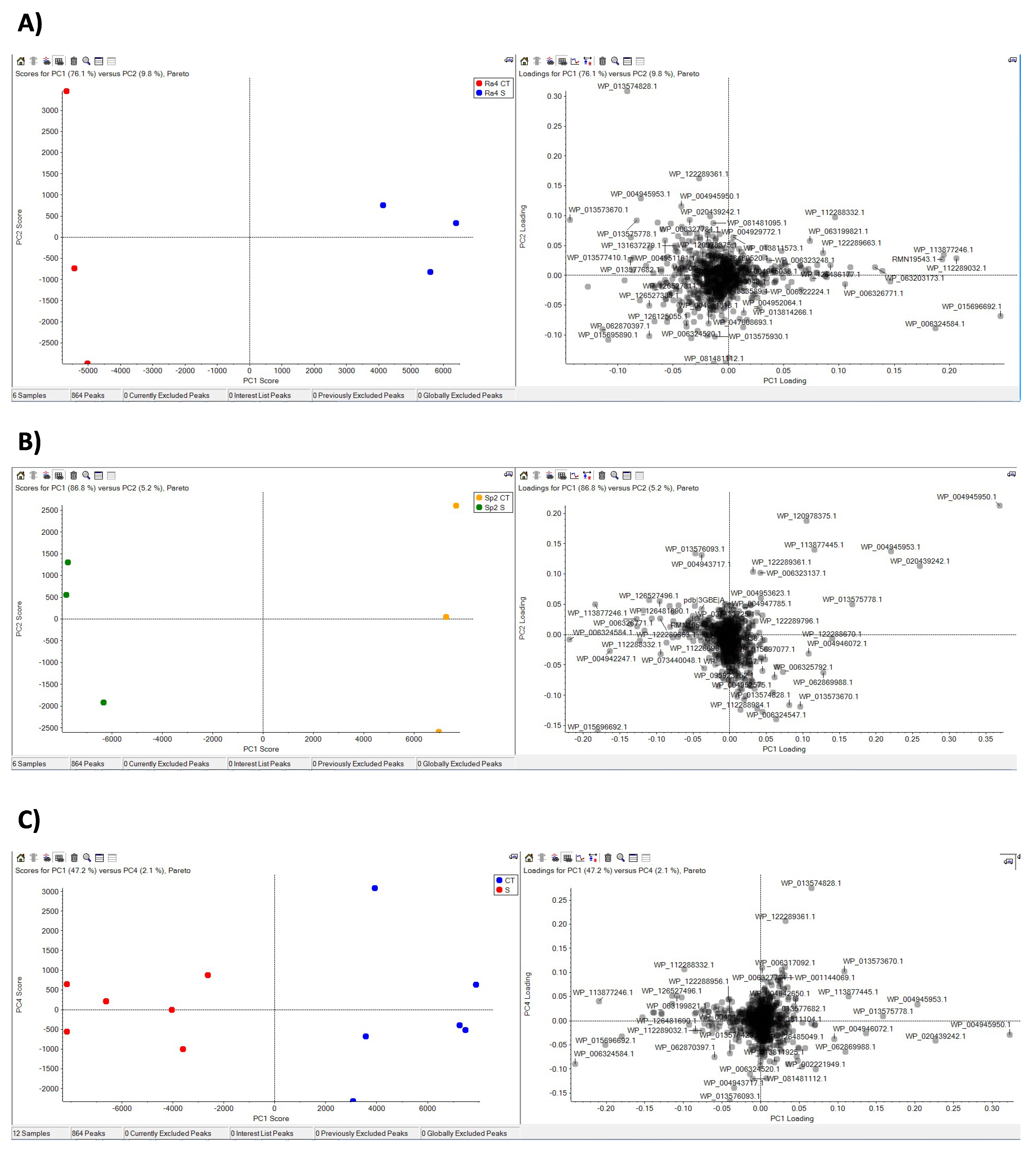
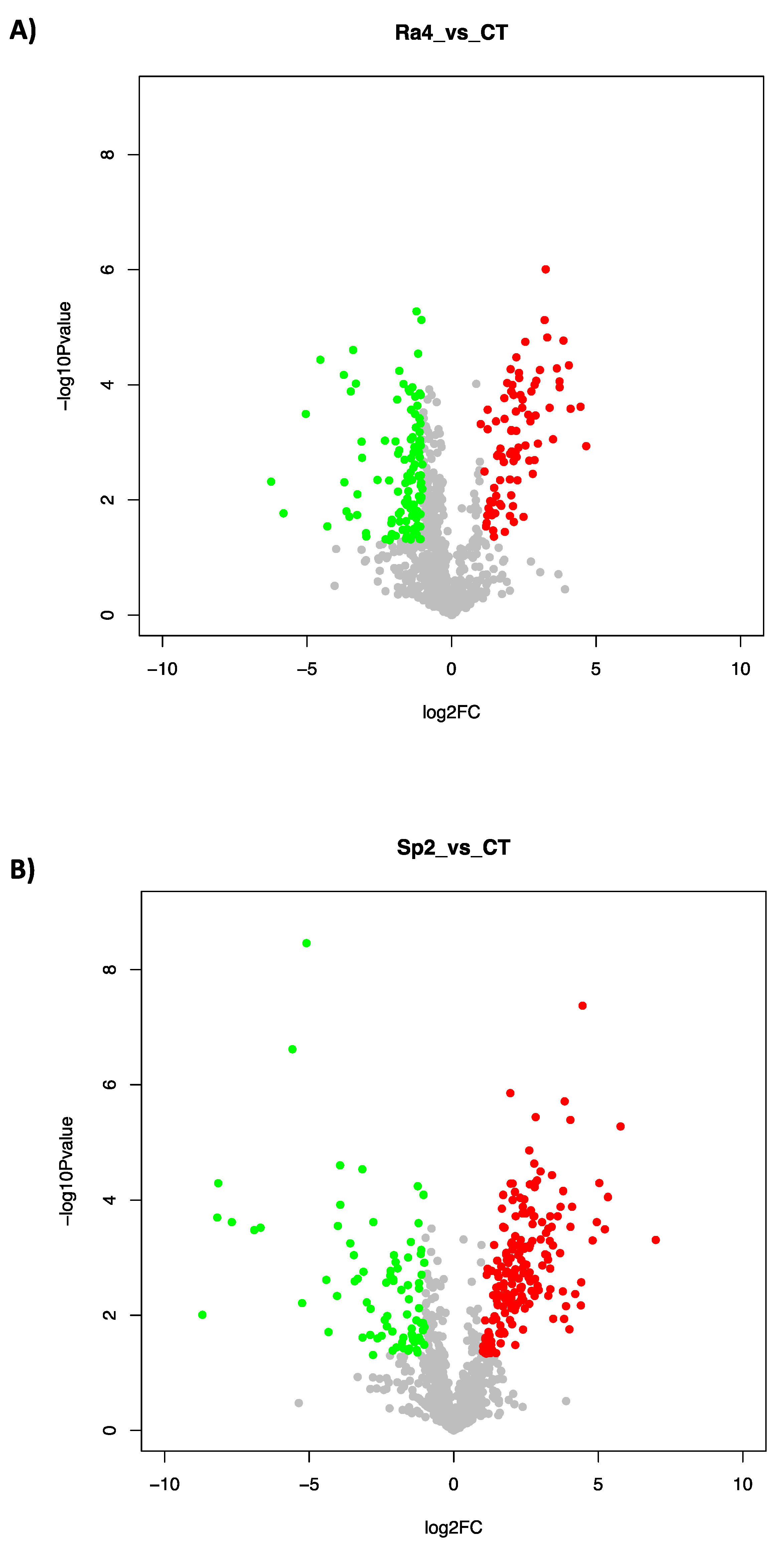
3.2. Proteome Response to Salt Stress
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Velazquez, E.; Martinez-Hidalgo, P.; Carro, L.; Alonso, P.; Peix, A. Nodular Endophytes: Untapped Diversity. In Beneficial Plant-Microbial Interactions; CRC Press: Boca Raton, FL, USA, 2013. [Google Scholar]
- De Meyer, S.E.; De Beuf, K.; Vekeman, B.; Willems, A. A Large Diversity of Non-Rhizobial Endophytes Found in Legume Root Nodules in Flanders (Belgium). Soil Biol. Biochem. 2015, 83, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Zgadzaj, R.; James, E.K.; Kelly, S.; Kawaharada, Y.; de Jonge, N.; Jensen, D.B.; Madsen, L.H.; Radutoiu, S. A Legume Genetic Framework Controls Infection of Nodules by Symbiotic and Endophytic Bacteria. PLoS Genet. 2015, 11, e1005280. [Google Scholar] [CrossRef] [Green Version]
- Pandya, M.; Rajput, M.; Rajkumar, S. Exploring Plant Growth Promoting Potential of Non Rhizobial Root Nodules Endophytes of Vigna radiata. Microbiology 2015, 84, 80–89. [Google Scholar] [CrossRef]
- Liu, H.; Carvalhais, L.C.; Crawford, M.; Singh, E.; Dennis, P.G.; Pieterse, C.M.J.; Schenk, P.M. Inner Plant Values: Diversity, Colonization and Benefits from Endophytic Bacteria. Front. Microbiol. 2017, 8, 2552. [Google Scholar] [CrossRef] [PubMed]
- Ibáñez, F.; Wall, L.; Fabra, A. Starting Points in Plant-Bacteria Nitrogen-Fixing Symbioses: Intercellular Invasion of the Roots. J. Exp. Bot. 2017, 68, 1905–1918. [Google Scholar] [CrossRef]
- Muresu, R.; Porceddu, A.; Sulas, L.; Squartini, A. Nodule-Associated Microbiome Diversity in Wild Populations of Sulla coronaria Reveals Clues on the Relative Importance of Culturable Rhizobial Symbionts and Co-Infecting Endophytes. Microbiol. Res. 2019, 221, 10–14. [Google Scholar] [CrossRef]
- Soares, R.; Trejo, J.; Lorite, M.J.; Figueira, E.; Sanjuán, J.; Videira e Castro, I. Diversity, Phylogeny and Plant Growth Promotion Traits of Nodule Associated Bacteria Isolated from Lotus parviflorus. Microorganisms 2020, 8, 499. [Google Scholar] [CrossRef] [Green Version]
- Leite, J.; Fischer, D.; Rouws, L.F.M.; Fernandes-Júnior, P.I.; Hofmann, A.; Kublik, S.; Schloter, M.; Xavier, G.R.; Radl, V. Cowpea Nodules Harbor Non-Rhizobial Bacterial Communities That Are Shaped by Soil Type Rather than Plant Genotype. Front. Plant. Sci. 2017, 7. [Google Scholar] [CrossRef]
- Estévez, J.; Dardanelli, M.S.; Megías, M.; Rodríguez-Navarro, D.N. Symbiotic Performance of Common Bean and Soybean Co-Inoculated with Rhizobia and Chryseobacterium balustinum Aur9 under Moderate Saline Conditions. Symbiosis 2009, 49, 29–36. [Google Scholar] [CrossRef]
- Rajendran, G.; Sing, F.; Desai, A.J.; Archana, G. Enhanced Growth and Nodulation of Pigeon Pea by Co-Inoculation of Bacillus Strains with Rhizobium Spp. Bioresour. Technol. 2008, 99, 4544–4550. [Google Scholar] [CrossRef]
- Bai, Y.; Zhou, X.; Smith, D.L. Enhanced Soybean Plant Growth Resulting from Coinoculation of Bacillus Strains with Bradyrhizobium japonicum. Crop. Sci. 2003, 43, 1774–1781. [Google Scholar] [CrossRef]
- Tariq, M.; Hameed, S.; Yasmeen, T.; Zahid, M.; Zafar, M. Molecular Characterization and Identification of Plant Growth Promoting Endophytic Bacteria Isolated from the Root Nodules of Pea (Pisum sativum L.). World, J. Microbiol. Biotechnol. 2014, 30, 719–725. [Google Scholar] [CrossRef] [PubMed]
- Egamberdieva, D.; Wirth, S.J.; Shurigin, V.V.; Hashem, A.; Abd_Allah, E.F. Endophytic Bacteria Improve Plant Growth, Symbiotic Performance of Chickpea (Cicer arietinum L.) and Induce Suppression of Root Rot Caused by Fusarium solani under Salt Stress. Front. Microbiol. 2017, 8, 1887. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Wang, S.; Zhu, X.; Wang, Y.; Liu, X.; Duan, Y.; Fan, H.; Chen, L. Isolation and Characterization of Nodules Endophytic Bacteria Pseudomonas protegens Sneb1997 and Serratia plymuthica Sneb2001 for the Biological Control of Root-Knot Nematode. Appl. Soil Ecol. 2021, 164, 103924. [Google Scholar] [CrossRef]
- Peix, A.; Ramírez-Bahena, M.H.; Velázquez, E.; Bedmar, E.J. Bacterial Associations with Legumes. Crit. Rev. Plant. Sci. 2015, 34, 17–42. [Google Scholar] [CrossRef]
- Andrews, M.; Hodge, S.; Raven, J.A. Positive Plant Microbial Interactions. Ann. Appl. Biol. 2010, 157, 317–320. [Google Scholar] [CrossRef]
- Saïdi, S.; Chebil, S.; Gtari, M.; Mhamdi, R. Characterization of Root-Nodule Bacteria Isolated from Vicia faba and Selection of Plant Growth Promoting Isolates. World J. Microbiol. Biotechnol. 2013, 29, 1099–1106. [Google Scholar] [CrossRef]
- Maheshwari, R.; Bhutani, N.; Suneja, P. Isolation and Characterization of ACC Deaminase Producing Endophytic Bacillus mojavensis PRN2 from Pisum sativum. IRAN J. Biotech. 2020, 18, e2308. [Google Scholar] [CrossRef]
- Maas, E.V.; Hoffman, G.J. Crop Salt Tolerance—Current Assessment. J. Irrig. Drain. Div. 1977, 103, 115–134. [Google Scholar] [CrossRef]
- Bruning, B.; Rozema, J. Symbiotic Nitrogen Fixation in Legumes: Perspectives for Saline Agriculture. Environ. Exp. Bot. 2013, 92, 134–143. [Google Scholar] [CrossRef]
- Hashem, F.M.; Swelim, D.M.; Kuykendall, L.D.; Mohamed, A.I.; Abdel-Wahab, S.M.; Hegazi, N.I. Identification and Characterization of Salt- and Thermo-Tolerant Leucaena -Nodulating Rhizobium Strains. Biol. Fertil. Soils 1998, 27, 335–341. [Google Scholar] [CrossRef]
- Saxena, A.K.; Rewari, R.B. Differential Responses of Chick Pea (Cicer arietinum L.)- Rhizobium Combination to Saline Soil Conditions. Biol. Fertil. Soils 1992, 13, 31–34. [Google Scholar] [CrossRef]
- Massa, N.; Cesaro, P.; Todeschini, V.; Capraro, J.; Scarafoni, A.; Cantamessa, S.; Copetta, A.; Anastasia, F.; Gamalero, E.; Lingua, G.; et al. Selected Autochthonous Rhizobia, Applied in Combination with AM Fungi, Improve Seed Quality of Common Bean Cultivated in Reduced Fertilization Condition. Appl. Soil Ecol. 2020, 148, 103507. [Google Scholar] [CrossRef]
- Weisburg, W.G.; Barns, S.M.; Pelletier, D.A.; Lane, D.J. 16S Ribosomal DNA Amplification for Phylogenetic Study. J. Bacteriol. 1991, 173, 697–703. [Google Scholar] [CrossRef] [Green Version]
- Gamalero, E.; Favale, N.; Bona, E.; Novello, G.; Cesaro, P.; Massa, N.; Glick, B.; Orozco-Mosqueda, M.; Berta, G.; Lingua, G. Screening of Bacterial Endophytes Able to Promote Plant Growth and Increase Salinity Tolerance. Appl. Sci. 2020, 10, 5767. [Google Scholar] [CrossRef]
- Schwyn, B.; Neilands, J. Universal Chemical Assay for the Detection and Determination of Siderophore. Anal. Biochem. 1987, 160, 47–56. [Google Scholar] [CrossRef]
- De Brito, A.M.; Gagne, S.; Antoun, H. Effect of Compost on Rhizosphere Microflora of the Tomato and on the Incidence of Plant Growth-Promoting Rhizobacteria. Appl. Environ. Microbiol. 1995, 61, 194–199. [Google Scholar] [CrossRef] [Green Version]
- Stock, I.; Gruger, T.; Wiedemann, B. Natural Antibiotic Susceptibility of Rahnella aquatlis and R. aquatilis-Relared Strains. J. Chemoterapy 2000, 12, 30–39. [Google Scholar] [CrossRef]
- Penrose, D.M.; Glick, B.R. Methods for Isolating and Characterizing ACC Deaminase-Containing Plant Growth-Promoting Rhizobacteria. Physiol. Plant. 2003, 118, 10–15. [Google Scholar] [CrossRef] [Green Version]
- Dworkin, M.; Foster, J.W. Experiments with Some Microorganisms Which Utilize Ethane And Hydrogen. J. Bacteriol. 1958, 75, 592–603. [Google Scholar] [CrossRef] [Green Version]
- Rashid, S.; Charles, T.C.; Glick, B.R. Isolation and Characterization of New Plant Growth-Promoting Bacterial Endophytes. Appl. Soil Ecol. 2012, 61, 217–224. [Google Scholar] [CrossRef]
- Bradford, M.M. A Rapid and Sensitive Method for the Quantification of Micrograms Quantities of Proteins Utilizing the Principle of Protein-Dye Binding. Ann. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Bona, E.; Massa, N.; Novello, G.; Boatti, L.; Cesaro, P.; Todeschini, V.; Magnelli, V.; Manfredi, M.; Marengo, E.; Mignone, F.; et al. Metaproteomic Characterization of Vitis vinifera Rhizosphere. FEMS Microbiol. Ecol. 2019, 95, fiy204. [Google Scholar] [CrossRef]
- Bona, E.; Scarafoni, A.; Marsano, F.; Boatti, L.; Copetta, A.; Massa, N.; Gamalero, E.; D’Agostino, G.; Cesaro, P.; Cavaletto, M.; et al. Arbuscular Mycorrhizal Symbiosis Affects the Grain Proteome of Zea mays: A Field Study. Sci. Rep. 2016, 6, 26439. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Albanese, P.; Manfredi, M.; Re, A.; Marengo, E.; Saracco, G.; Pagliano, C. Thylakoid Proteome Modulation in Pea Plants Grown at Different Irradiances: Quantitative Proteomic Profiling in a Non-Model Organism Aided by Transcriptomic Data Integration. Plant. J. 2018, 96, 786–800. [Google Scholar] [CrossRef]
- Dalla Pozza, E.; Manfredi, M.; Brandi, J.; Buzzi, A.; Conte, E.; Pacchiana, R.; Cecconi, D.; Marengo, E.; Donadelli, M. Trichostatin A Alters Cytoskeleton and Energy Metabolism of Pancreatic Adenocarcinoma Cells: An in Depth Proteomic Study. J. Cell. Biochem. 2018, 119, 2696–2707. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, A.H. Bacterial Solubilization of Mineral Phosphates: Historical Perspective and Future Prospects. Am. J. Alt. Ag. 1986, 1, 51–57. [Google Scholar] [CrossRef]
- Roeder, H.A.; Fuller, B.; Scoular, S. Septic Shock Caused by Rahnella aquatilis Bacteremia in an Immunocompetent Adult. Am. J. Case Rep. 2021, 22, e930888-1. [Google Scholar] [CrossRef]
- Lee, W.J.; Song, Y.; Park, S.Y.; Kim, M.J. Bacteremia Due to Rahnella aquatilis in a Patient with a Chemoport. Infect. Chemother 2019, 51, 191. [Google Scholar] [CrossRef]
- Carrero, P.; Garrote, J.A.; Pacheco, S.; García, A.I.; Gil, R.; Carbajosa, S.G. Report of Six Cases of Human Infection by Serratia plymuthica. J. Clin. Microbiol. 1995, 33, 275–276. [Google Scholar] [CrossRef] [Green Version]
- Bakhshandeh, E.; Rahimian, H.; Pirdashti, H.; Nematzadeh, G.A. Phosphate Solubilization Potential and Modeling of Stress Tolerance of Rhizobacteria from Rice Paddy Soil in Northern Iran. World J. Microbiol. Biotechnol. 2014, 30, 2437–2447. [Google Scholar] [CrossRef] [PubMed]
- Tash, K. Rahnella aquatilis Bacteremia from a Suspected Urinary Source. J. Clin. Microbiol. 2005, 43, 2526–2528. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruimy, R.; Meziane-Cherif, D.; Momcilovic, S.; Arlet, G.; Andremont, A.; Courvalin, P. RAHN-2, a Chromosomal Extendedspectrum Class A b-Lactamase from Rahnella aquatilis. J Antimicrob. Chemother 2010, 65, 1619–1623. [Google Scholar] [CrossRef] [PubMed]
- Gin, K.Y.-H.; Goh, S.G. Modeling the Effect of Light and Salinity on Viable but Non-Culturable (VBNC) Enterococcus. Water Res. 2013, 47, 3315–3328. [Google Scholar] [CrossRef]
- Pumirat, P.; Saetun, P.; Sinchaikul, S.; Chen, S.-T.; Korbsrisate, S.; Thongboonkerd, V. Altered Secretome of Burkholderia pseudomallei Induced by Salt Stress. Biochim. Et Biophys. Acta (BBA)—Proteins Proteom. 2009, 1794, 898–904. [Google Scholar] [CrossRef]
- Rubiano-Labrador, C.; Bland, C.; Miotello, G.; Armengaud, J.; Baena, S. Salt Stress Induced Changes in the Exoproteome of the Halotolerant Bacterium Tistlia consotensis Deciphered by Proteogenomics. PLoS ONE 2015, 10, e0135065. [Google Scholar] [CrossRef] [Green Version]
- Díaz-Cárdenas, C.; Patel, B.K.C.; Baena, S. Tistlia consotensis Gen. Nov., Sp. Nov., an Aerobic, Chemoheterotrophic, Free-Living, Nitrogen-Fixing Alphaproteobacterium, Isolated from a Colombian Saline Spring. Int. J. Syst. Evol. Microbiol. 2010, 60, 1437–1443. [Google Scholar] [CrossRef] [Green Version]
- Rice, A.J.; Park, A.; Pinkett, H.W. Diversity in ABC Transporters: Type I, II and III Importers. Crit. Rev. Biochem. Mol. Biol. 2014, 49, 426–437. [Google Scholar] [CrossRef] [Green Version]
- Teichmann, L.; Kümmel, H.; Warmbold, B.; Bremer, E. OpuF, a New Bacillus Compatible Solute ABC Transporter with a Substrate-Binding Protein Fused to the Transmem.mbrane Domain. Appl. Env. Microbiol. 2018, 84. [Google Scholar] [CrossRef] [Green Version]
- Lennon, C.W.; Thamsen, M.; Friman, E.T.; Cacciaglia, A.; Sachsenhauser, V.; Sorgenfrei, F.A.; Wasik, M.A.; Bardwell, J.C.A. Folding Optimization In Vivo Uncovers New Chaperones. J. Mol. Biol. 2015, 427, 2983–2994. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W.; Sun, K.; Cheng, S.; Sun, L. Characterization of DegQ Vh, a Serine Protease and a Protective Immunogen from a Pathogenic Vibrio Harveyi Strain. Appl Env. Microbiol 2008, 74, 6254–6262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ünal, C.M.; Steinert, M. Microbial Peptidyl-Prolyl Cis/Trans. Isomerases (PPIases): Virulence Factors and Potential Alternative Drug Targets. Microbiol Mol. Biol Rev. 2014, 78, 544–571. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bontemps-Gallo, S.; Bohin, J.-P.; Lacroix, J.-M. Osmoregulated Periplasmic Glucans. EcoSal. Plus 2017, 7, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Typas, A.; Banzhaf, M.; van den Berg van Saparoea, B.; Verheul, J.; Biboy, J.; Nichols, R.J.; Zietek, M.; Beilharz, K.; Kannenberg, K.; von Rechenberg, M.; et al. Regulation of Peptidoglycan Synthesis by Outer-Membrane Proteins. Cell 2010, 143, 1097–1109. [Google Scholar] [CrossRef] [Green Version]
- He, Y.; Xu, T.; Fossheim, L.E.; Zhang, X.-H. FliC, a Flagellin Protein, Is Essential for the Growth and Virulence of Fish Pathogen Edwardsiella tarda. PLoS ONE 2012, 7, e45070. [Google Scholar] [CrossRef] [Green Version]
- Eismann, S.; Endres, R.G. Protein Connectivity in Chemotaxis Receptor Complexes. PLoS Comput. Biol. 2015, 11, e1004650. [Google Scholar] [CrossRef] [Green Version]
- Berg, A.K.; Srivastava, D.K. Delineation of Alternative Conformational States in Escherichia Coli Peptide Deformylase via Thermodynamic Studies for the Binding of Actinonin. Biochemistry 2009, 48, 1584–1594. [Google Scholar] [CrossRef] [Green Version]
- Ohnishi, K.; Ohto, Y.; Aizawa, S.; Macnab, R.M.; Iino, T. FlgD Is a Scaffolding Protein Needed for Flagellar Hook Assembly in Salmonella typhimurium. J. Bacteriol. 1994, 176, 2272–2281. [Google Scholar] [CrossRef] [Green Version]
- Muramoto, K.; Makishima, S.; Aizawa, S.-I.; Macnab, R.M. Effect of Hook Subunit Concentration on Assembly and Control of Length of the Flagellar Hook of Salmonella. J. Bacteriol 1999, 181, 5808–5813. [Google Scholar] [CrossRef] [Green Version]
- Vijayakumar, V.; Vanhove, A.S.; Pickering, B.S.; Liao, J.; Tierney, B.T.; Asara, J.M.; Bronson, R.; Watnick, P.I. Removal of a Membrane Anchor Reveals the Opposing Regulatory Functions of Vibrio cholerae Glucose-Specific Enzyme IIA in Biofilms and the Mammalian Intestine. mBio 2018, 9, e00858-18. [Google Scholar] [CrossRef] [Green Version]
- Den Besten, H.M.W.; Mols, M.; Moezelaar, R.; Zwietering, M.H.; Abee, T. Phenotypic and Transcriptomic Analyses of Mildly and Severely Salt-Stressed Bacillus cereus ATCC 14579 Cells. Appl. Env. Microbiol. 2009, 75, 4111–4119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Remonsellez, F.; Castro-Severyn, J.; Pardo-Esté, C.; Aguilar, P.; Fortt, J.; Salinas, C.; Barahona, S.; León, J.; Fuentes, B.; Areche, C.; et al. Characterization and Salt Response in Recurrent Halotolerant Exiguobacterium Sp. SH31 Isolated From Sediments of Salar de Huasco, Chilean Altiplano. Front. Microbiol. 2018, 9, 2228. [Google Scholar] [CrossRef] [PubMed]
- Grauschopf, U.; Winther, J.R.; Korber, P.; Zander, T.; Dallinger, P.; Bardwell, J.C.A. Why Is DsbA Such an Oxidizing Disulfide Catalyst? Cell 1995, 83, 947–955. [Google Scholar] [CrossRef] [Green Version]
- Suss, O.; Reichmann, D. Protein Plasticity Underlines Activation and Function of ATP-Independent Chaperones. Front. Mol. Biosci. 2015, 2, 43. [Google Scholar] [CrossRef] [Green Version]
- Havekes, L.M.; Hoekstra, W.P. Characterization of an Escherichia coli K-12 F-Con-Mutant. J. Bacteriol. 1976, 126, 593–600. [Google Scholar] [CrossRef] [Green Version]
- Lugtenberg, B.; Peterz, R.; Bernheimer, H.; Berendsen, W. Influence of Cultural Conditions and Mutations on the Composition of the Outer Membrane Proteins of Escherichia coli. Mol. Gen. Genet. 1976, 147, 251–262. [Google Scholar] [CrossRef]
- Nikaido, H.; Vaara, M. Molecular Basis of Bacterial Outer Membrane Permeability. Microbiol. Rev. 1985, 49, 32. [Google Scholar] [CrossRef]
- Sato, M.; Machida, K.; Arikado, E.; Saito, H.; Kakegawa, T.; Kobayashi, H. Expression of Outer Membrane Proteins in Escherichia coli Growing at Acid PH. Appl. Env. Microbiol. 2000, 66, 943–947. [Google Scholar] [CrossRef] [Green Version]
- Jung, J.U.; Gutierrez, C.; Martin, F.; Ardourel, M.; Villarejo, M. Transcription of OsmB, a Gene Encoding an Escherichia coli Lipoprotein, Is Regulated by Dual Signals. Osmotic Stress and Stationary Phase. J. Biol. Chem. 1990, 265, 10574–10581. [Google Scholar] [CrossRef]
- Gothel, S.F.; Marahiel, M.A. Peptidyl-Prolyl Cis-Trans Isomerases, a Superfamily of Ubiquitous Folding Catalysts. Cell. Mol. Life Sci. (CMLS) 1999, 55, 423–436. [Google Scholar] [CrossRef]
- Ikeda, T.; Oosawa, K.; Hotani, H. Self-Assembly of the Filament Capping Protein, FliD, of Bacterial Flagella into an Annular Structure. J. Mol. Biol. 1996, 259, 679–686. [Google Scholar] [CrossRef] [PubMed]
- Waters, R.C.; O’Toole, P.W.; Ryan, K.A. The FliK Protein and Flagellar Hook-Length Control. Protein Sci. 2007, 16, 769–780. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Strain | Strain Label | Host Plant | Sampling Site | Colony Description (YMA) | Gram Staining | DCP * | CAS @ | IAA § |
|---|---|---|---|---|---|---|---|---|
| FV_Fr4B | Ra1 | V. fava | Genova | Small colony, round shape, convex elevation, entire margin, mucoid and pink | Negative | 2.26 | 0.00 | 3 |
| FV_Fr4C | V. fava | Genova | Small colony, round shape, convex elevation, entire margin, mucoid and pink | Negative | 0.00 | 0.00 | neg | |
| FV_Fr5B | Ra2 | V. fava | Genova | Small colony, round shape, convex elevation, entire margin, mucoid with pink core | Negative | 2.89 | 0.00 | 1 |
| FV_Fr6A | V. fava | Genova | Round shape, entire margin, mucoid and orange | Negative | 0.00 | 0.00 | neg | |
| FV_Fr6B | V. fava | Genova | Small colony, round shape, convex elevation, entire margin, mucoid and pink | Negative | 0.00 | 0.00 | 0 | |
| FV_EB1A_1 | Sp1 | V. fava | Calliano | Small colony, round shape, convex elevation, entire margin, mucoid with pink core | Negative | 1.60 | 0.00 | 2 |
| FV_EB2A_1 | V. fava | Calliano | Large colony, entire margin, mucoid and pink | Negative | 2.82 | 0.00 | neg | |
| FV_EB2C_1 | V. fava | Calliano | Small colony, entire margin, mucoid and dark pink | Negative | 0.00 | 0.00 | neg | |
| FV_EB1B_1 | Sp2 | V. fava | Calliano | Small colony, round shape, convex elevation, entire margin, mucoid with dark orange core | Negative | 1.78 | 0.00 | 1 |
| FV_EB1C_1 | V. fava | Calliano | Small colony, round shape with a dark orange core | Negative | 1.75 | 0.00 | 0 | |
| PS_EB1A_1 | P. sativum | Calliano | Large colony, entire margin, mucoid and pink | Negative | 3.32 | 1.19 | neg | |
| PS_EB1A_2 | P. sativum | Calliano | Large colony, entire margin, mucoid and pink | Negative | 2.00 | 0.00 | 2 | |
| PS_EB1C_1 | P. sativum | Calliano | Mucoid, with a dark red core | Negative | 1.58 | 0.00 | neg | |
| FV_EB2B_1 | V. fava | Calliano | Mucoid, with a light pink core | Negative | 2.97 | 0.00 | neg | |
| FV_VT2B_1 | Ra3 | V. fava | Frassineto | Small colony, round shape, convex elevation, entire margin, mucoid with pink core | Negative | 2.93 | 0.00 | 2 |
| FV_VT1C_1 | Sp3 | V. fava | Frassineto | Large colony, entire margin, mucoid and pink | Negative | 2.34 | 0.00 | 0 |
| FV_VT1B_1 | V. fava | Frassineto | Mucoid with a red core | Negative | 2.90 | 0.00 | neg | |
| PS_EN1B_1 | P. sativum | Druento | Large colony, entire margin, mucoid and pink | Negative | 2.08 | 0.00 | 1 | |
| PS_EN1B_2 | P. sativum | Druento | Large colony, entire margin, mucoid and pink | Negative | 1.80 | 0.00 | neg | |
| PS_EN1A_1 | P. sativum | Druento | Round shape, smooth, with a dark red core | Negative | 2.06 | 0.00 | 1 | |
| PS_EN1C_1 | Ra4 | P. sativum | Druento | Large colony, entire margin, mucoid with a dark red core | Negative | 2.73 | 0.00 | 3 |
| PS_EN2B_1 | P. sativum | Druento | Large colony, entire margin, mucoid and pink with a red core | Negative | 2.94 | 1.02 | neg |
| Strain | Strain Label | BLAST Identification | Seq. Cov (%) a | Id (%) b | Genbank Accesion Number | 4 °C | 28 °C | 37 °C | 42 °C | Salt MIC (%) |
|---|---|---|---|---|---|---|---|---|---|---|
| FV_Fr4B | Ra1 | R. aquatilis | 99 | 99.3 | MK157042 | + | + | ± § | - | 6.4 |
| FV_Fr5B | Ra2 | R. aquatilis | 98 | 99.1 | MK156743 | + | + | ± | - | 6.4 |
| FV_VT2B1 | Ra3 | R. aquatilis | 98 | 99.5 | MK157017 | + | + | ± | - | 6.4 |
| PS_EN1C1 | Ra4 | R. aquatilis | 100 | 99.8 | OK614097 | + | + | ± | - | 5.8 |
| FV EB 1A1 | Sp1 | S. plymuthica | 98 | 99.5 | MK156722 | + | + | ± | - | 5.8 |
| FV EB 1B1 | Sp2 | S. plymuthica | 98 | 99.6 | MK157024 | + | + | ± | - | 6.2 |
| FV VT 1C1 | Sp3 | S. plymuthica | 98 | 99.1 | MK157016 | + | + | ± | - | 5.8 |
| Antibiotic | Ra1 | I 1 | I 2 | I 3 | Ra2 | I 1 | I 2 | I 3 | Ra3 | I 1 | I 2 | I 3 | Ra4 | I 1 | I 2 | I 3 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Amoxicillin | 6.0 ± 0.1 | R | 6.0 ± 0.1 | R | 6.0 ± 0.1 | R | 6.0 ± 0.1 | R | ||||||||
| Cefepime | 6.0 ± 0.1 | R | 6.0 ± 0.1 | R | 8.0 ± 0.1 | R | 6.0 ± 0.1 | R | ||||||||
| Cefotaxime | 18.0 ± 0.6 | I | 18.0 ± 0.7 | I | 17.0 ± 0.6 | I | 24.0 ± 0.8 | S | ||||||||
| Ceftazidime | 22.0 ± 0.3 | S | 22.0 ± 0.4 | S | 25.0 ± 0.3 | S | 22.0 ± 0.5 | S | ||||||||
| Chloramphenicol | 26.7 ± 1.8 | S | S | 26.7 ± 1.3 | S | S | 24.3 ± 0.3 | S | S | 24.0 ± 1.0 | S | S | ||||
| Ciprofloxacin | 34.0 ± 0.5 | S | 40.0 ± 0.7 | S | 28.0 ± 0.4 | S | 35.0 ± 0.6 | S | ||||||||
| Erythromycin | 8.3 ± 1.8 | R | R | 6.3 ± 0.3 | R | R | 8.0 ± 0.6 | R | R | 6.7 ± 0.3 | R | R | ||||
| Kanamycin | 16.3 ± 0.9 | S | S | 18.3 ± 0.3 | S | S | 19.0 ± 0.6 | S | S | 16.3 ± 0.3 | S | S | ||||
| Meropenem | 34.0 ± 0.7 | S | 18.0 ± 0.6 | I | 28.0 ± 0.7 | S | 29.0 ± 0.6 | S | ||||||||
| Neomycin | 10.3 ± 0.9 | S | S | 11.0 ± 0.6 | S | S | 11.7 ± 0.3 | S | S | 11.3 ± 0.3 | S | S | ||||
| Piperacillin/Tazobactam | 23.0 ± 0.5 | S | 22.0 ± 0.6 | S | 27.0 ± 0.6 | S | 24.0 ± 0.7 | S | ||||||||
| Rifampicin | 20.0 ± 0.0 | S | 19.0 ± 0.5 | S | 21.3 ± 0.7 | S | 22.3 ± 0.9 | S | ||||||||
| Streptomycin | 14.0 ± 1.0 | S | S | 16.7 ± 0.7 | S | S | 15.3 ± 0.3 | S | S | 15.0 ± 0.0 | S | S | ||||
| Trimetoprim/Sulfametoxazol | 24.0 ± 0.7 | 32.0 ± 0.6 | 28.0 ± 0.6 | 32.0 ± 0.5 |
| Antibiotic | Sp1 | I 1 | I 2 | Sp2 | I 1 | I 2 | Sp3 | I 1 | I 2 |
|---|---|---|---|---|---|---|---|---|---|
| Amoxicillin | 6.0 ± 0.1 | R | 6.0 ± 0.1 | R | 6.0 ± 0.1 | R | |||
| Cefepime | 7.0 ± 0.1 | R | 6.0 ± 0.1 | R | 6.0 ± 0.1 | R | |||
| Cefotaxime | 25.0 ± 0.7 | S | 24.0 ± 0.6 | S | 23.0 ± 0.7 | S | |||
| Ceftazidime | 22.0 ± 0.3 | S | 23.0 ± 0.3 | S | 19.0 ± 0.5 | I | |||
| Chloramphenicol | 17.0 ± 1.7 | S | 17.3 ± 1.8 | S | 24.0 ± 1.0 | S | |||
| Ciprofloxacin | 45.0 ± 0.5 | S | 44.0 ± 0.4 | S | 36.0 ± 0.6 | S | |||
| Erythromycin | 13.0 ± 1.5 | S | 6.0 ± 0.2 | R | 7.0 ± 0.3 | R | |||
| Kanamycin | 15.3 ± 0.7 | S | 17.3 ± 1.3 | S | 17.7 ± 0.3 | S | |||
| Meropenem | 24.0 ± 0.5 | S | 25.0 ± 0.6 | S | 25.0 ± 0.6 | S | |||
| Neomycin | 12.3 ± 1.5 | S | 9.7 ± 0.9 | S | 11.3 ± 0.3 | S | |||
| Piperacillin/Tazobactam | 22.0 ± 0.5 | S | 23.0 ± 0.6 | S | 21.0 ± 0.6 | S | |||
| Rifampicin | 12.7 ± 1.3 | S | 14.0 ± 1.2 | S | 22.0 ± 1.2 | S | |||
| Streptomycin | 15.0 ± 1.7 | S | 12.7 ± 1.5 | S | 15.3 ± 0.3 | S | |||
| Trimetoprim/Sulfametoxazol | 27.0 ± 0.8 | 32.0 ± 0.6 | 34.0 ± 0.7 |
| Antibiotic | Ra1 | I 1 | Ra2 | I 1 | Ra3 | I 1 | Ra4 | I 1 | Sp1 | I 2 | Sp2 | I 2 | Sp3 | I 2 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Amikacin | <=8 | S | <=8 | S | <=8 | S | <=8 | S | <=8 | S | <=8 | S | <=8 | S |
| Amoxicillin/Clavulanic Acid | <=4 | S | <=4 | S | <=4 | S | <=4 | S | <=4 | S | <=4 | S | 4 | S |
| Cefepime | <=1 | S | <=1 | S | <=0.5 | S | <=1 | S | <=1 | S | 2 | I | <=1 | S |
| Cefotaxime | <=0.5 | S | <=0.5 | S | <=0.5 | S | <=0.5 | S | ------- | ------- | ------ | |||
| Ceftazidime | <=0.5 | S | <=0.5 | S | <=0.2 | S | <=0.5 | S | <=0.5 | S | <=0.5 | S | <=0.5 | S |
| Ciprofloxacin | <=0.2 | S | <=0.2 | S | <=0.1 | S | <=0.2 | S | <=0.2 | S | <=0.2 | S | <=0.2 | S |
| Ertapenem | <=0.1 | S | <=0.1 | S | <=0.1 | S | <=0.1 | S | <=0.1 | S | <=0.1 | S | <=0.1 | S |
| Gentamicin | <=2 | S | <=2 | S | <=2 | S | <=2 | S | <=2 | S | <=2 | S | <=2 | S |
| Meropenem | <=0.1 | S | <=0.1 | S | <=0.1 | S | <=0.1 | S | <=0.1 | S | <=0.1 | S | ||
| Piperacillin/Tazobactam | <=4 | S | <=4 | S | <=4 | S | <=4 | S | <=4 | S | <=4 | S | <=4 | S |
| Trimetoprim/Sulfametoxazol | <=2 | S | <=2 | S | <=2 | S | <=2 | S | <=2 | S | <=2 | S | <=2 | S |
| Ampicillin | <=2 | S | 4 | S | >8 | R | 8 | S | 4 | S | 4 | S | 4 | S |
| Aztreonam | <=1 | S | <=1 | S | <=1 | S | <=1 | S | 2 | I | <=1 | S | ------- | |
| Colistin | <=2 | S | <=2 | S | <=2 | S | <=2 | S | >4 | R | >4 | R | <=2 | S |
| Levofloxacin | <=0.5 | S | <=0.5 | S | <=0.5 | S | <=0.5 | S | <=0.5 | S | <=0.5 | S | <=0.5 | S |
| Piperacillin | <=4 | S | <=4 | S | <=4 | S | <=4 | S | <=4 | S | <=4 | S | <=4 | S |
| Ticarcillin | <=8 | S | <=8 | S | <=8 | S | <=8 | S | <=8 | S | <=8 | S | <=8 | S |
| Tigecycline | <=0.5 | S | <=0.5 | S | <=0.5 | S | <=0.5 | S | <=0.5 | S | <=0.5 | S | <=0.5 | S |
| Tobramycin | <=2 | S | <=2 | S | <=2 | S | <=2 | S | <=2 | S | <=2 | S | <=2 | S |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Novello, G.; Gamalero, E.; Massa, N.; Cesaro, P.; Lingua, G.; Todeschini, V.; Caramaschi, A.; Favero, F.; Corà, D.; Manfredi, M.; et al. Proteome and Physiological Characterization of Halotolerant Nodule Endophytes: The Case of Rahnella aquatilis and Serratia plymuthica. Microorganisms 2022, 10, 890. https://doi.org/10.3390/microorganisms10050890
Novello G, Gamalero E, Massa N, Cesaro P, Lingua G, Todeschini V, Caramaschi A, Favero F, Corà D, Manfredi M, et al. Proteome and Physiological Characterization of Halotolerant Nodule Endophytes: The Case of Rahnella aquatilis and Serratia plymuthica. Microorganisms. 2022; 10(5):890. https://doi.org/10.3390/microorganisms10050890
Chicago/Turabian StyleNovello, Giorgia, Elisa Gamalero, Nadia Massa, Patrizia Cesaro, Guido Lingua, Valeria Todeschini, Alice Caramaschi, Francesco Favero, Davide Corà, Marcello Manfredi, and et al. 2022. "Proteome and Physiological Characterization of Halotolerant Nodule Endophytes: The Case of Rahnella aquatilis and Serratia plymuthica" Microorganisms 10, no. 5: 890. https://doi.org/10.3390/microorganisms10050890
APA StyleNovello, G., Gamalero, E., Massa, N., Cesaro, P., Lingua, G., Todeschini, V., Caramaschi, A., Favero, F., Corà, D., Manfredi, M., Marengo, E., Pelagi, M., Pangaro, L., Caffiero, G., Milano, F., & Bona, E. (2022). Proteome and Physiological Characterization of Halotolerant Nodule Endophytes: The Case of Rahnella aquatilis and Serratia plymuthica. Microorganisms, 10(5), 890. https://doi.org/10.3390/microorganisms10050890













