Socialization of Providencia stuartii Enables Resistance to Environmental Insults
Abstract
1. Introduction
2. Methods
2.1. Strains
2.2. Production of Porins from P. stuartii in E. coli
2.3. Porin Extraction and Solubilization
2.4. Porin Purification
2.5. Liposome Preparation
2.6. Dynamic Light Scattering (DLS)
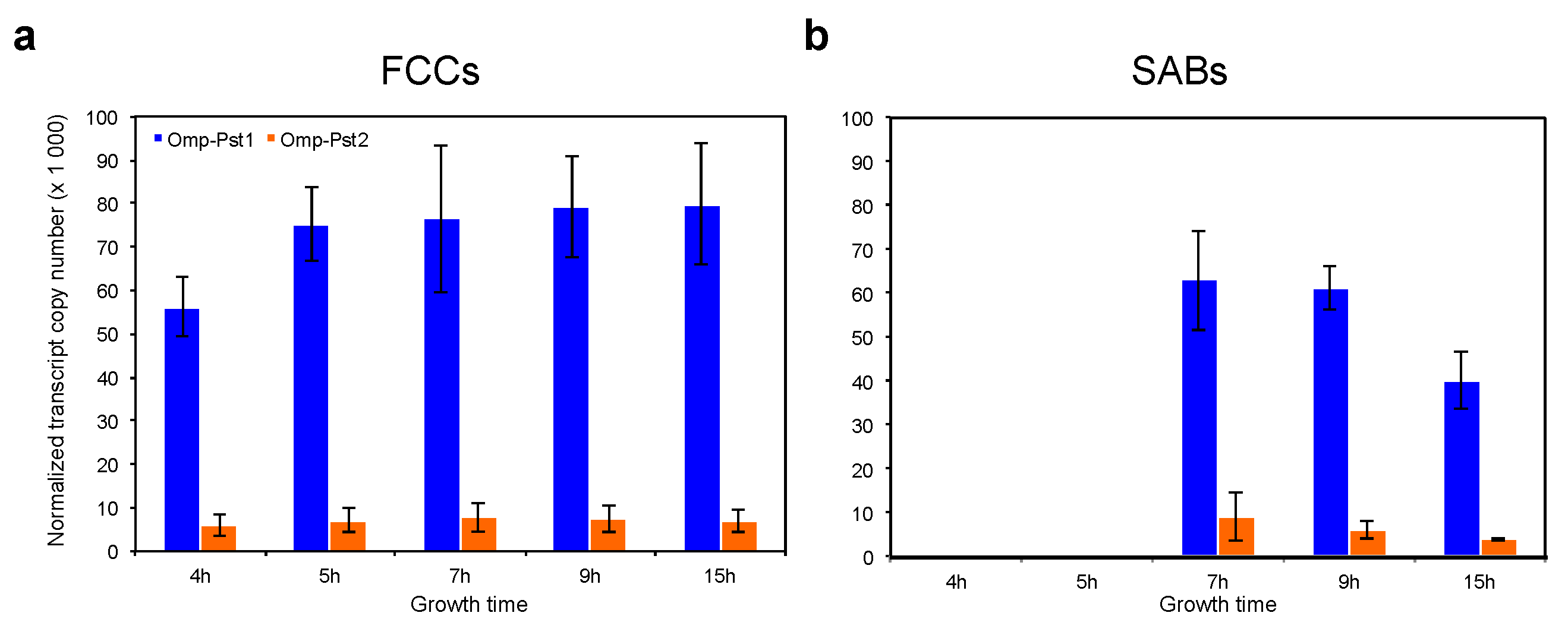
2.7. Gene Expression Study
2.8. Evaluation of Porin Abundance in the OM of P. stuartti Cells
2.9. Bacterial Growth Studies
2.10. Imaging
2.11. Statistical Analysis
3. Results
3.1. P. stuartii Cells Are Highly Resistant to Urea
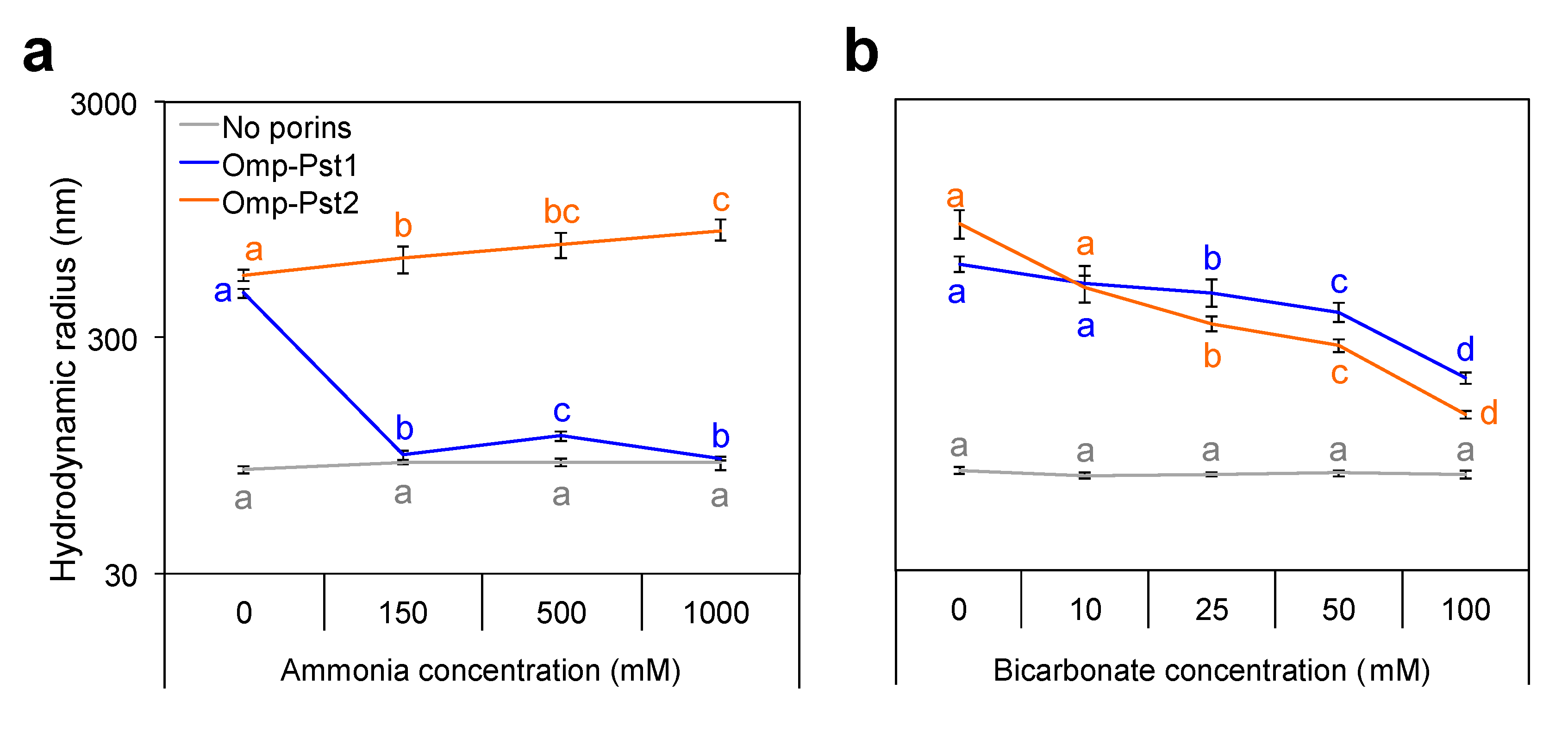
3.2. Resistance of P. stuartii to High Concentration of Ammonia Involves Omp-Pst2
3.3. P. stuartii Is Highly Sensitive to Bicarbonate but Unaffected by Creatinine
3.4. Variation of pH Hardly Affects P. stuartii Socialization and Fitness
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Costerton, J.W. Bacterial biofilms: A common cause of persistent infections. Science 1999, 284, 1318–1322. [Google Scholar] [CrossRef]
- Donlan, R.M.; Costerton, J.W. Biofilms: Survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 2002, 15, 167–193. [Google Scholar] [CrossRef]
- Das, T.; Sehar, S.; Manefield, M. The roles of extracellular DNA in the structural integrity of extracellular polymeric substance and bacterial biofilm development: The roles of eDNA in the bacterial biofilm development. Environ. Microbiol. Rep. 2013, 5, 778–786. [Google Scholar] [CrossRef]
- Okshevsky, M.; Regina, V.R.; Meyer, R.L. Extracellular DNA as a target for biofilm control. Curr. Opin. Biotechnol. 2015, 33, 73–80. [Google Scholar] [CrossRef] [PubMed]
- de la Fuente-Núñez, C.; Reffuveille, F.; Fernández, L.; Hancock, R.E. Bacterial biofilm development as a multicellular adaptation: Antibiotic resistance and new therapeutic strategies. Curr. Opin. Microbiol. 2013, 16, 580–589. [Google Scholar] [CrossRef]
- Thomas, D.N.; Dieckmann, G.S. Antarctic Sea ice—A habitat for extremophiles. Science 2002, 295, 641–644. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Miao, L.; Li, X.-C.; Xiao, X.; Qian, P.-Y. Antibacterial and antilarval activity of deep-sea bacteria from sediments of the West Pacific Ocean. Biofouling 2007, 23, 131–137. [Google Scholar] [CrossRef] [PubMed]
- Stickler, D.J. Bacterial biofilms in patients with indwelling urinary catheters. Nat. Clin. Pract. Urol. 2008, 5, 598–608. [Google Scholar] [CrossRef]
- Hall-Stoodley, L.; Stoodley, P. Evolving concepts in biofilm infections. Cell. Microbiol. 2009, 11, 1034–1043. [Google Scholar] [CrossRef]
- Lebeaux, D.; Ghigo, J.-M. Infections associées aux biofilms: Quelles perspectives thérapeutiques issues de la recherche fondamentale? Méd. Sci. 2012, 28, 727–739. [Google Scholar] [CrossRef]
- Wie, S.-H. Clinical significance of Providencia bacteremia or bacteriuria. Korean J. Intern. Med. 2015, 30, 167. [Google Scholar] [CrossRef]
- McHale, P.J.; Walker, F.; Scully, B.; English, L.; Keane, C.T. Providencia stuartii infections: A review of 117 cases over an eight year period. J. Hosp. Infect. 1981, 2, 155–165. [Google Scholar] [CrossRef]
- Gould, C.V.; Umscheid, C.A.; Agarwal, R.K.; Kuntz, G.; Pegues, D.A.; Healthcare Infection Control Practices Advisory Committee (HICPAC). Guideline for Prevention of Catheter-Associated Urinary Tract Infections 2009. Infect. Control Hosp. Epidemiol. 2010, 31, 319–326. [Google Scholar] [CrossRef]
- Chapman, C.; Gibson, G.R.; Rowland, I. Effects of single- and multi-strain probiotics on biofilm formation and in vitro adhesion to bladder cells by urinary tract pathogens. Anaerobe 2014, 27, 71–76. [Google Scholar] [CrossRef] [PubMed]
- De Vecchi, E.; Sitia, S.; Romanò, C.L.; Ricci, C.; Mattina, R.; Drago, L. Aetiology and antibiotic resistance patterns of urinary tract infections in the elderly: A 6-month study. J. Med. Microbiol. 2013, 62, 859–863. [Google Scholar] [CrossRef]
- Jacoby, G.A. AmpC beta-lactamases. Clin. Microbiol. Rev. 2009, 22, 161–182. [Google Scholar] [CrossRef] [PubMed]
- Nicolle, L.E. Resistant pathogens in urinary tract infections. J. Am. Geriatr. Soc. 2002, 50, S230–S235. [Google Scholar] [CrossRef]
- Warren, J.W. Providencia stuartii: A common cause of antibiotic-resistant bacteriuria in patients with long-term indwelling catheters. Rev. Infect. Dis. 1986, 8, 61–67. [Google Scholar] [CrossRef]
- Tumbarello, M. ESBL-producing multidrug-resistant Providencia stuartii infections in a university hospital. J. Antimicrob. Chemother. 2004, 53, 277–282. [Google Scholar] [CrossRef]
- Aubert, D.; Naas, T.; Lartigue, M.-F.; Nordmann, P. Novel genetic structure associated with an extended-spectrum beta-lactamase blaVEB gene in a Providencia stuartii clinical isolate from Algeria. Antimicrob. Agents Chemother. 2005, 49, 3590–3592. [Google Scholar] [CrossRef]
- Vlamakis, H.; Chai, Y.; Beauregard, P.; Losick, R.; Kolter, R. Sticking together: Building a biofilm the Bacillus subtilis way. Nat. Rev. Microbiol. 2013, 11, 157–168. [Google Scholar] [CrossRef] [PubMed]
- El Khatib, M.; Tran, Q.T.; Nasrallah, C.; Lopes, J.; Bolla, J.M.; Vivaudou, M.; Pagès, J.M.; Colletier, J.P. Providencia stuartii form biofilms and floating communities of cells that display high resistance to environmental insults. PLoS ONE 2017, 12, e0174213. [Google Scholar] [CrossRef] [PubMed]
- El-Khatib, M.; Nasrallah, C.; Lopes, J.; Tran, Q.-T.; Tetreau, G.; Basbous, H.; Fenel, D.; Gallet, B.; Lethier, M.; Bolla, J.-M.; et al. Porin self-association enables cell-to-cell contact in Providencia stuartii floating communities. Proc. Natl. Acad. Sci. USA 2018, 115, E2220–E2228. [Google Scholar] [CrossRef] [PubMed]
- Stevens, J.C.; Jones, N.B.; Fay, D.F. Apparatus for uniaxial testing of soft human tissues in vitro. Med. Biol. Eng. Comput. 1978, 16, 226–228. [Google Scholar] [CrossRef] [PubMed]
- Pagès, J.-M.; James, C.E.; Winterhalter, M. The porin and the permeating antibiotic: A selective diffusion barrier in Gram-negative bacteria. Nat. Rev. Microbiol. 2008, 6, 893–903. [Google Scholar] [CrossRef]
- Zeth, K.; Thein, M. Porins in prokaryotes and eukaryotes: Common themes and variations. Biochem. J. 2010, 431, 13–22. [Google Scholar] [CrossRef]
- Delcour, A.H. Outer membrane permeability and antibiotic resistance. Biochim. Biophys. Acta—Proteins Proteom. 2009, 1794, 808–816. [Google Scholar] [CrossRef]
- Pagès, J.-M. Porines bactériennes et sensibilité aux antibiotiques. Méd. Sci. 2004, 20, 346–351. [Google Scholar] [CrossRef][Green Version]
- Nikaido, H. Prevention of drug access to bacterial targets: Permeability barriers and active efflux. Science 1994, 264, 382–388. [Google Scholar] [CrossRef]
- Ficarra, F.A.; Grandellis, C.; Galván, E.M.; Ielpi, L.; Feil, R.; Lunn, J.E.; Gottig, N.; Ottado, J. Xanthomonas citri ssp. citri requires the outer membrane porin OprB for maximal virulence and biofilm formation: Xanthomonas citri requires OprB for virulence. Mol. Plant Pathol. 2017, 18, 720–733. [Google Scholar] [CrossRef]
- Cattelan, N.; Villalba, M.I.; Parisi, G.; Arnal, L.; Serra, D.O.; Aguilar, M.; Yantorno, O. Outer membrane protein OmpQ of Bordetella bronchiseptica is required for mature biofilm formation. Microbiology 2016, 162, 351–363. [Google Scholar] [CrossRef] [PubMed]
- Tran, Q.-T.; Mahendran, K.R.; Hajjar, E.; Ceccarelli, M.; Davin-Regli, A.; Winterhalter, M.; Weingart, H.; Pagès, J.-M. Implication of Porins in β-Lactam Resistance of Providencia stuartii. J. Biol. Chem. 2010, 285, 32273–32281. [Google Scholar] [CrossRef] [PubMed]
- Putnam, D.F. Composition and Concentrative Properties of Urine. NASA NTRS, NASA-CR-1802; NTRS 19710023044; NASA: Washington, DC, USA, 1971. [Google Scholar]
- Dallmann, R.; Viola, A.U.; Tarokh, L.; Cajochen, C.; Brown, S.A. The human circadian metabolome. Proc. Natl. Acad. Sci. USA 2012, 109, 2625–2629. [Google Scholar] [CrossRef] [PubMed]
- Brooks, T.; Keevil, C.W. A simple artificial urine for the growth of urinary pathogens. Lett. Appl. Microbiol. 1997, 24, 203–206. [Google Scholar] [CrossRef] [PubMed]
- Prilipov, A.; Phale, P.S.; Gelder, P.; Rosenbusch, J.P.; Koebnik, R. Coupling site-directed mutagenesis with high-level expression: Large scale production of mutant porins from E. coli. FEMS Microbiol. Lett. 1998, 163, 65–72. [Google Scholar] [CrossRef]
- Dijkstra, J.; Ryan, J.L.; Szoka, F.C. A procedure for the efficient incorporation of wild-type lipopolysaccharide into liposomes for use in immunological studies. J. Immunol. Methods 1988, 114, 197–205. [Google Scholar] [CrossRef]
- Zgurskaya, H.I.; Nikaido, H. Bypassing the periplasm: Reconstitution of the AcrAB multidrug efflux pump of Escherichia coli. Proc. Natl. Acad. Sci. USA 1999, 96, 7190–7195. [Google Scholar] [CrossRef]
- Kruip, J.; Karapetyan, N.V.; Terekhova, I.V.; Rögner, M. In vitro oligomerization of a membrane protein complex: Liposome-based reconstitution of trimeric photosystem I from isolated monomers. J. Biol. Chem. 1999, 274, 18181–18188. [Google Scholar] [CrossRef]
- Johnson, G.; Nour, A.A.; Nolan, T.; Huggett, J.; Bustin, S. Minimum information necessary for quantitative real-time PCR experiments. Methods Mol. Biol. 2014, 1160, 5–17. [Google Scholar]
- de Jong, I.G.; Beilharz, K.; Kuipers, O.P.; Veening, J.-W. Live cell imaging of bacillus subtilis and streptococcus pneumoniae using automated time-lapse microscopy. J. Vis. Exp. 2011, 53, e3145. [Google Scholar] [CrossRef]
- Seabold, S.; Perktold, J. Statsmodels: Econometric and Statistical Modeling with Python. In Proceedings of the 9th Python in Science Conference, Austin, TX, USA, 28 June–3 July 2010; Volume 57, p. 61. [Google Scholar]
- Raman, A.; Sreenevasan, G.A. Urinary osmolarity and excretion of sodium, calcium and magnesium in patients with renal calculi. Br. J. Urol. 1972, 44, 537–540. [Google Scholar] [CrossRef] [PubMed]
- Isaacson, L.C. Urinary composition in calcific nephrolithiasis. Investig. Urol. 1969, 6, 356–363. [Google Scholar]
- Leid, J.G.; Willson, C.J.; Shirtliff, M.E.; Hassett, D.J.; Parsek, M.R.; Jeffers, A.K. The exopolysaccharide alginate protects Pseudomonas aeruginosa biofilm bacteria from IFN-gamma-mediated macrophage killing. J. Immunol. 2005, 175, 7512–7518. [Google Scholar] [CrossRef] [PubMed]
- Khoury, A.E.; Lam, K.; Ellis, B.; Costerton, J.W. Prevention and control of bacterial infections associated with medical devices. ASAIO J. Am. Soc. Artif. Intern. Organs 1992, 38, M174–M178. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lopes, J.; Tetreau, G.; Pounot, K.; El Khatib, M.; Colletier, J.-P. Socialization of Providencia stuartii Enables Resistance to Environmental Insults. Microorganisms 2022, 10, 901. https://doi.org/10.3390/microorganisms10050901
Lopes J, Tetreau G, Pounot K, El Khatib M, Colletier J-P. Socialization of Providencia stuartii Enables Resistance to Environmental Insults. Microorganisms. 2022; 10(5):901. https://doi.org/10.3390/microorganisms10050901
Chicago/Turabian StyleLopes, Julie, Guillaume Tetreau, Kevin Pounot, Mariam El Khatib, and Jacques-Philippe Colletier. 2022. "Socialization of Providencia stuartii Enables Resistance to Environmental Insults" Microorganisms 10, no. 5: 901. https://doi.org/10.3390/microorganisms10050901
APA StyleLopes, J., Tetreau, G., Pounot, K., El Khatib, M., & Colletier, J.-P. (2022). Socialization of Providencia stuartii Enables Resistance to Environmental Insults. Microorganisms, 10(5), 901. https://doi.org/10.3390/microorganisms10050901






