Some Clues about Enzymes from Psychrophilic Microorganisms
Abstract
:1. Introduction
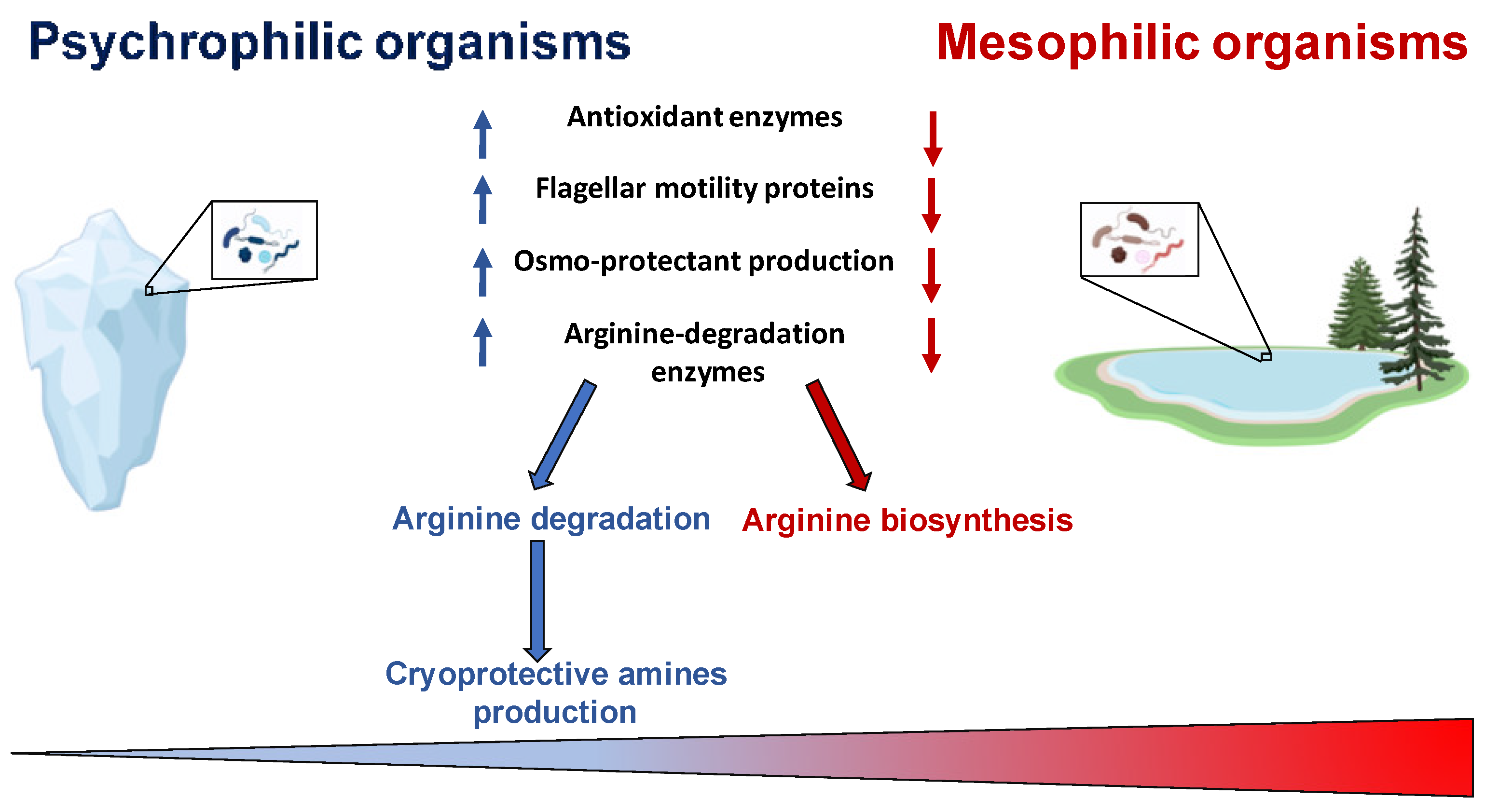
2. Metabolism Adaptation
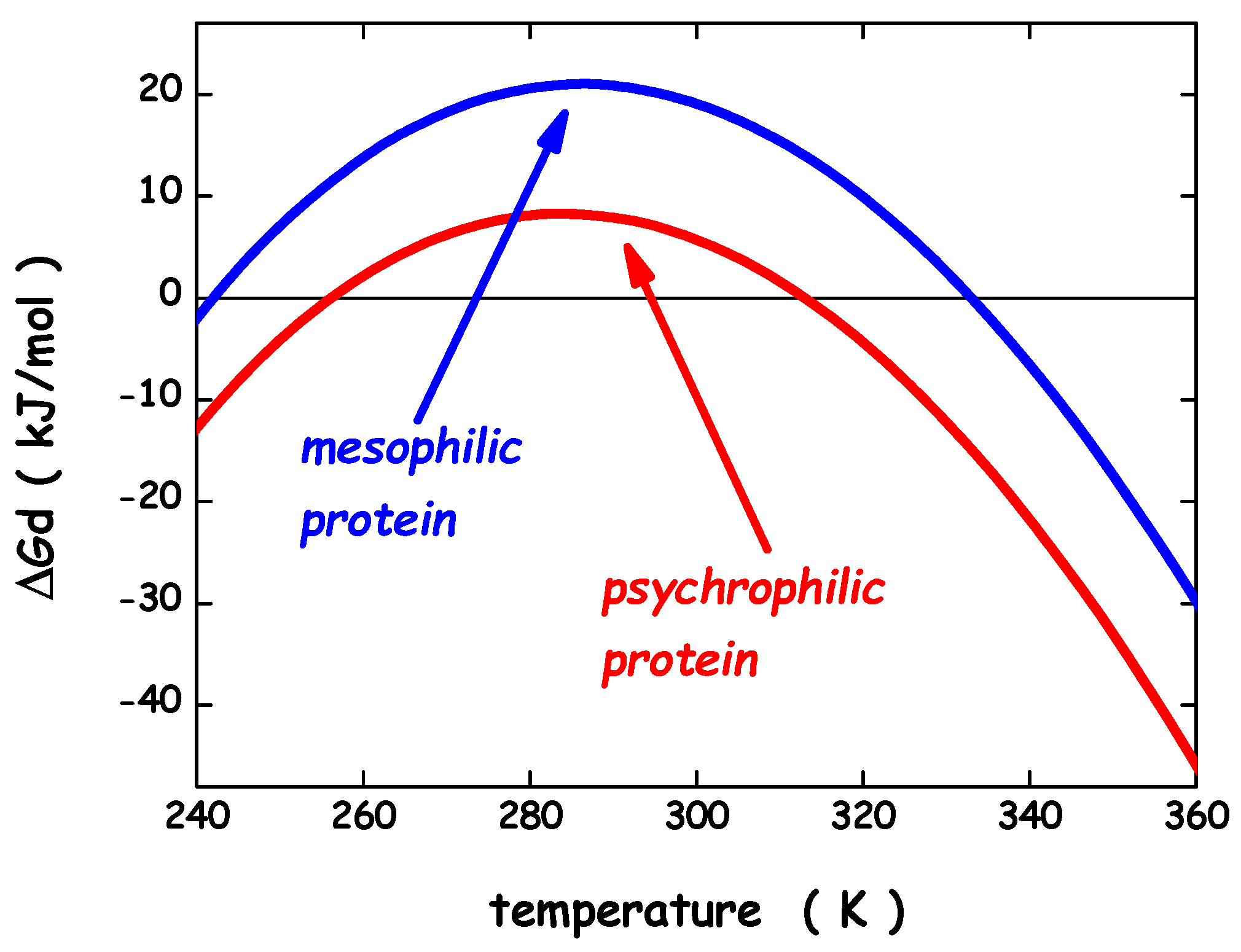
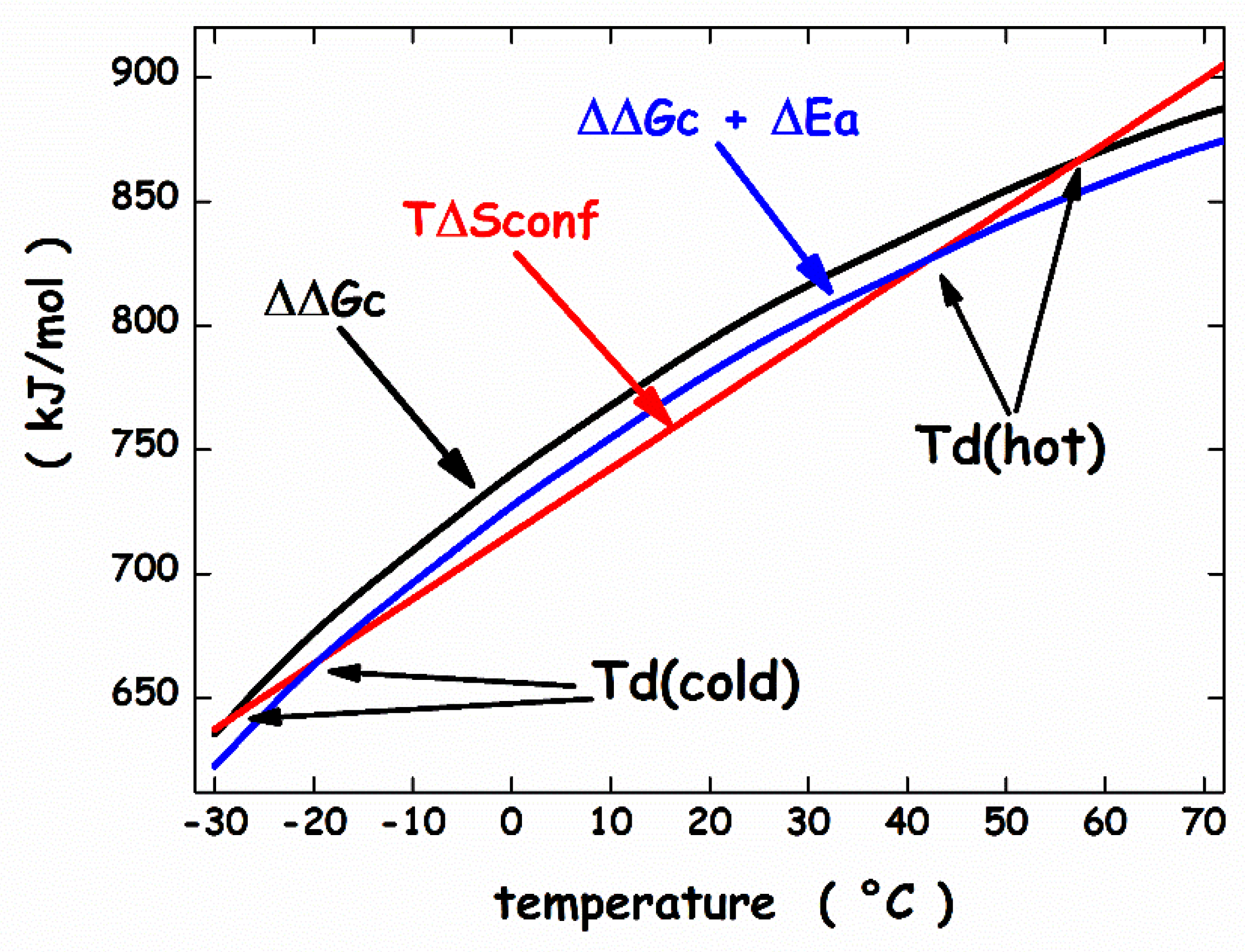
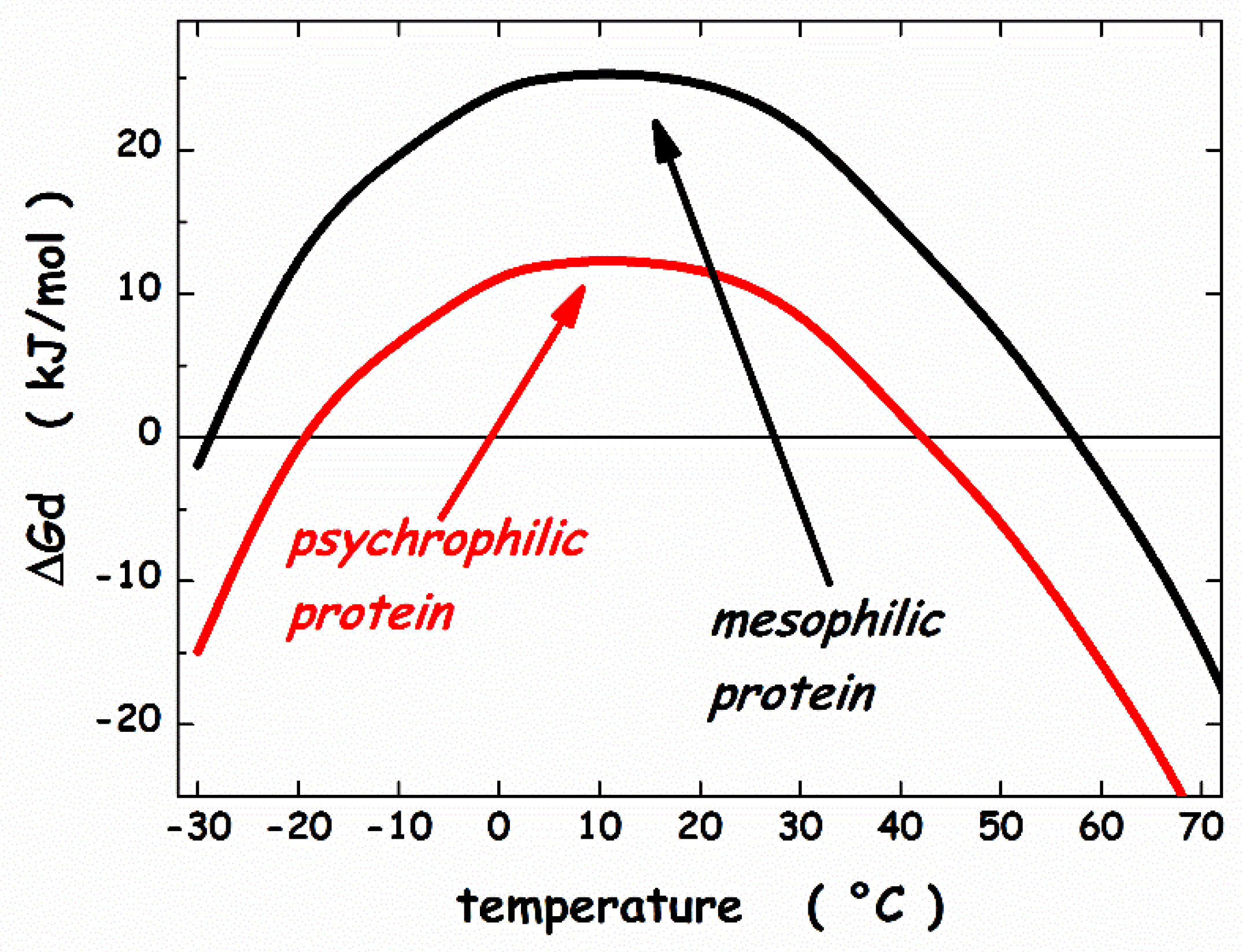
3. Thermodynamic Features
4. Enzymatic Adaptation
4.1. α-Amylase
4.2. Citrate Synthase
4.3. β-Galactosidase
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Coker, J.A. Recent advances in understanding extremophiles. F1000Res 2019, 8, 1917. [Google Scholar] [CrossRef] [PubMed]
- Casanueva, A.; Tuffin, M.; Cary, C.; Cowan, D.A. Molecular adaptations to psychrophily: The impact of ‘omic’ technologies. Trends Microbiol. 2010, 18, 374–381. [Google Scholar] [CrossRef]
- Margesin, R.; Miteva, V. Diversity and ecology of psychrophilic microorganisms. Res. Microbiol. 2011, 162, 346–361. [Google Scholar] [CrossRef] [PubMed]
- Murray, A.E.; Kenig, F.; Fritsen, C.H.; McKay, C.P.; Cawley, K.M.; Edwards, R.; Kuhn, E.; McKnight, D.M.; Ostrom, N.E.; Peng, V.; et al. Microbial life at −13 degrees C in the brine of an ice-sealed Antarctic lake. Proc. Natl. Acad. Sci. USA 2012, 109, 20626–20631. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clarke, A.; Morris, G.J.; Fonseca, F.; Murray, B.J.; Acton, E.; Price, H.C. A Low Temperature Limit for Life on Earth. PLoS ONE 2013, 8, e66207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodrigues, D.F.; Tiedje, J.M. Coping with our cold planet. Appl. Environ. Microbiol. 2008, 74, 1677–1686. [Google Scholar] [CrossRef] [Green Version]
- D’Amico, S.; Collins, T.; Marx, J.C.; Feller, G.; Gerday, C. Psychrophilic microorganisms: Challenges for life. EMBO Rep. 2006, 7, 385–389. [Google Scholar] [CrossRef]
- Rizvi, A.; Ahmed, B.; Khan, M.S.; Umar, S.; Lee, J. Psychrophilic Bacterial Phosphate-Biofertilizers: A Novel Extremophile for Sustainable Crop Production under Cold Environment. Microorganisms 2021, 9, 2451. [Google Scholar] [CrossRef] [PubMed]
- Piette, F.; D’Amico, S.; Mazzucchelli, G.; Danchin, A.; Leprince, P.; Feller, G. Life in the cold: A proteomic study of cold-repressed proteins in the antarctic bacterium pseudoalteromonas haloplanktis TAC125. Appl. Environ. Microbiol. 2011, 77, 3881–3883. [Google Scholar] [CrossRef] [Green Version]
- Duarte, A.W.F.; Dos Santos, J.A.; Vianna, M.V.; Vieira, J.M.F.; Mallagutti, V.H.; Inforsato, F.J.; Wentzel, L.C.P.; Lario, L.D.; Rodrigues, A.; Pagnocca, F.C.; et al. Cold-adapted enzymes produced by fungi from terrestrial and marine Antarctic environments. Crit. Rev. Biotechnol. 2018, 38, 600–619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morita, R.Y. Psychrophilic bacteria. Bacteriol. Rev. 1975, 39, 144–167. [Google Scholar] [CrossRef]
- De Maayer, P.; Anderson, D.; Cary, C.; Cowan, D.A. Some like it cold: Understanding the survival strategies of psychrophiles. EMBO Rep. 2014, 15, 508–517. [Google Scholar] [CrossRef] [PubMed]
- Tendulkar, S.; Hattiholi, A.; Chavadar, M.; Dodamani, S. Psychrophiles: A journey of hope. J. BioSci. 2021, 46, 64. [Google Scholar] [CrossRef] [PubMed]
- Jaenicke, R. Stability and folding of ultrastable proteins: Eye lens crystallins and enzymes from thermophiles. Faseb J. 1996, 10, 84–92. [Google Scholar] [CrossRef]
- Khan, S.; Farooq, U.; Kurnikova, M. Exploring Protein Stability by Comparative Molecular Dynamics Simulations of Homologous Hyperthermophilic, Mesophilic, and Psychrophilic Proteins. J. Chem. Inf. Model. 2016, 56, 2129–2139. [Google Scholar] [CrossRef]
- Peak, M.J.; Robb, F.T.; Peak, J.G. Extreme resistance to thermally induced DNA backbone breaks in the hyperthermophilic archaeon Pyrococcus furiosus. J. Bacteriol. 1995, 177, 6316–6318. [Google Scholar] [CrossRef] [Green Version]
- Del Vecchio, P.; Graziano, G.; Granata, V.; Barone, G.; Mandrich, L.; Manco, G.; Rossi, M. Temperature- and denaturant-induced unfolding of two thermophilic esterases. Biochemistry 2002, 41, 1364–1371. [Google Scholar] [CrossRef] [PubMed]
- Granata, V.; Graziano, G.; Ruggiero, A.; Raimo, G.; Masullo, M.; Arcari, P.; Vitagliano, L.; Zagari, A. Stability against temperature of Sulfolobus solfataricus elongation factor 1 alpha, a multi-domain protein. Bioch. Biophys. Acta 2008, 1784, 573–581. [Google Scholar] [CrossRef]
- Bialkowska, A.; Majewska, E.; Olczak, A.; Twarda-Clapa, A. Ice Binding Proteins: Diverse Biological Roles and Applications in Different Types of Industry. Biomolecules 2020, 10, 274. [Google Scholar] [CrossRef] [Green Version]
- Munoz, P.A.; Marquez, S.L.; Gonzalez-Nilo, F.D.; Marquez-Miranda, V.; Blamey, J.M. Structure and application of antifreeze proteins from Antarctic bacteria. Microb. Cell Fact. 2017, 16, 138. [Google Scholar] [CrossRef] [Green Version]
- Eskandari, A.; Leow, T.C.; Rahman, M.B.A.; Oslan, S.N. Antifreeze Proteins and Their Practical Utilization in Industry, Medicine, and Agriculture. Biomolecules 2020, 10, 1649. [Google Scholar] [CrossRef] [PubMed]
- Russell, N.J. Toward a molecular understanding of cold activity of enzymes from psychrophiles. Extremophiles 2000, 4, 83–90. [Google Scholar] [CrossRef]
- Arcus, V.L.; Mulholland, A.J. Temperature, Dynamics, and Enzyme-Catalyzed Reaction Rates. Annu. Rev. Biophys. 2020, 49, 163–180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feller, G.; Gerday, C. Psychrophilic enzymes: Molecular basis of cold adaptation. Cell. Mol. Life Sci. 1997, 53, 830–841. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- D’Amico, S.; Claverie, P.; Collins, T.; Georlette, D.; Gratia, E.; Hoyoux, A.; Meuwis, M.A.; Feller, G.; Gerday, C. Molecular basis of cold adaptation. Philos. Trans. R Soc. Lond. B Biol. Sci. 2002, 357, 917–925. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Struvay, C.; Feller, G. Optimization to low temperature activity in psychrophilic enzymes. Int. J. Mol. Sci. 2012, 13, 11643–11665. [Google Scholar] [CrossRef] [Green Version]
- Gianese, G.; Bossa, F.; Pascarella, S. Comparative structural analysis of psychrophilic and meso- and thermophilic enzymes. Proteins 2002, 47, 236–249. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui, K.S.; Cavicchioli, R. Cold-adapted enzymes. Annu. Rev. Biochem. 2006, 75, 403–433. [Google Scholar] [CrossRef] [Green Version]
- Jahandideh, S.; Abdolmaleki, P.; Jahandideh, M.; Barzegari Asadabadi, E. Sequence and structural parameters enhancing adaptation of proteins to low temperatures. J. Theor. Biol. 2007, 246, 159–166. [Google Scholar] [CrossRef]
- De Vendittis, E.; Castellano, I.; Cotugno, R.; Ruocco, M.R.; Raimo, G.; Masullo, M. Adaptation of model proteins from cold to hot environments involves continuous and small adjustments of average parameters related to amino acid composition. J. Theor. Biol. 2008, 250, 156–171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paredes, D.I.; Watters, K.; Pitman, D.J.; Bystroff, C.; Dordick, J.S. Comparative void-volume analysis of psychrophilic and mesophilic enzymes: Structural bioinformatics of psychrophilic enzymes reveals sources of core flexibility. BMC Struct. Biol. 2011, 11, 42. [Google Scholar] [CrossRef] [Green Version]
- Siddiqui, K.S. Defying the activity-stability trade-off in enzymes: Taking advantage of entropy to enhance activity and thermostability. Crit. Rev. Biotechnol. 2017, 37, 309–322. [Google Scholar] [CrossRef] [PubMed]
- Parvizpour, S.; Hussin, N.; Shamsir, M.S.; Razmara, J. Psychrophilic enzymes: Structural adaptation, pharmaceutical and industrial applications. Appl. Microbiol. Biotechnol. 2021, 105, 899–907. [Google Scholar] [CrossRef] [PubMed]
- Shokrollahzade, S.; Sharifi, F.; Vaseghi, A.; Faridounnia, M.; Jahandideh, S. Protein cold adaptation: Role of physico-chemical parameters in adaptation of proteins to low temperatures. J. Theor. Biol. 2015, 383, 130–137. [Google Scholar] [CrossRef] [PubMed]
- Metpally, R.P.; Reddy, B.V. Comparative proteome analysis of psychrophilic versus mesophilic bacterial species: Insights into the molecular basis of cold adaptation of proteins. BMC Genom. 2009, 10, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, A.; Mukhia, S.; Kumar, R. Industrial applications of cold-adapted enzymes: Challenges, innovations and future perspective. 3 Biotech. 2021, 11, 426. [Google Scholar] [CrossRef] [PubMed]
- Mhetras, N.; Mapare, V.; Gokhale, D. Cold Active Lipases: Biocatalytic Tools for Greener Technology. Appl. Biochem. Biotechnol. 2021, 193, 2245–2266. [Google Scholar] [CrossRef]
- Park, H.J.; Lee, C.W.; Kim, D.; Do, H.; Han, S.J.; Kim, J.E.; Koo, B.H.; Lee, J.H.; Yim, J.H. Crystal structure of a cold-active protease (Pro21717) from the psychrophilic bacterium, Pseudoalteromonas arctica PAMC 21717, at 1.4 A resolution: Structural adaptations to cold and functional analysis of a laundry detergent enzyme. PLoS ONE 2018, 13, e0191740. [Google Scholar] [CrossRef]
- Yang, G.; Mozzicafreddo, M.; Ballarini, P.; Pucciarelli, S.; Miceli, C. An In-Silico Comparative Study of Lipases from the Antarctic Psychrophilic Ciliate Euplotes focardii and the Mesophilic Congeneric Species Euplotes crassus: Insight into Molecular Cold-Adaptation. Mar. Drugs 2021, 19, 67. [Google Scholar] [CrossRef]
- Al-Ghanayem, A.A.; Joseph, B. Current prospective in using cold-active enzymes as eco-friendly detergent additive. Appl. Microbiol. Biotechnol. 2020, 104, 2871–2882. [Google Scholar] [CrossRef]
- Xue, Y.; Braslavsky, I.; Quake, S.R. Temperature effect on polymerase fidelity. J. Biol. Chem. 2021, 297, 101270. [Google Scholar] [CrossRef] [PubMed]
- Feller, G. Psychrophilic enzymes: From folding to function and biotechnology. Scientifica 2013, 2013, 512840. [Google Scholar] [CrossRef] [PubMed]
- Dhaulaniya, A.S.; Balan, B.; Kumar, M.; Agrawal, P.K.; Singh, D.K. Cold survival strategies for bacteria, recent advancement and potential industrial applications. Arch. Microbiol. 2019, 201, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Furhan, J. Adaptation, production, and biotechnological potential of cold-adapted proteases from psychrophiles and psychrotrophs: Recent overview. J. Genet. Eng. Biotechnol. 2020, 18, 36. [Google Scholar] [CrossRef] [PubMed]
- Mesbah, N.M. Industrial Biotechnology Based on Enzymes From Extreme Environments. Front. Bioeng. Biotechnol. 2022, 10, 870083. [Google Scholar] [CrossRef]
- Corkrey, R.; Macdonald, C.; McMeekin, T. The Biokinetic Spectrum for Temperature and optimal Darwinian fitness. J. Theor. Biol. 2019, 462, 171–183. [Google Scholar] [CrossRef] [PubMed]
- Corkrey, R.; McMeekin, T.A.; Bowman, J.P.; Ratkowsky, D.A.; Olley, J.; Ross, T. The Biokinetic Spectrum for Temperature. PLoS ONE 2016, 11, e0153343. [Google Scholar] [CrossRef]
- Corkrey, R.; McMeekin, T.A.; Bowman, J.P.; Olley, J.; Ratkowsky, D.; Ross, T. The maximum growth rate of life on Earth. Int. J. Astrobiol. 2018, 17, 17–33. [Google Scholar] [CrossRef]
- Tribelli, P.M.; Lopez, N.I. Reporting Key Features in Cold-Adapted Bacteria. Life 2018, 8, 8. [Google Scholar] [CrossRef] [Green Version]
- Phadtare, S. Recent developments in bacterial cold-shock response. Curr. Issues Mol. Biol. 2004, 6, 125–136. [Google Scholar]
- Collins, T.; Margesin, R. Psychrophilic lifestyles: Mechanisms of adaptation and biotechnological tools. Appl. Microbiol. Biotechnol. 2019, 103, 2857–2871. [Google Scholar] [CrossRef] [PubMed]
- Brininger, C.; Spradlin, S.; Cobani, L.; Evilia, C. The more adaptive to change, the more likely you are to survive: Protein adaptation in extremophiles. Semin. Cell Dev. Biol. 2018, 84, 158–169. [Google Scholar] [CrossRef] [PubMed]
- Piszkin, L.; Bowman, J. Extremophile enzyme optimization for low temperature and high salinity are fundamentally incompatible. Extremophiles 2021, 26, 5. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Lucas, J.; Acebron, I.; Wu, R.Y.; Alfaro, Y.; Acosta, J.; Kaminski, P.A.; Arroyo, M.; Joachimiak, A.; Nocek, B.P.; De la Mata, I.; et al. Biochemical and structural studies of two tetrameric nucleoside 2′-deoxyribosyltransferases from psychrophilic and mesophilic bacteria: Insights into cold-adaptation. Int. J. Biol. Macromol. 2021, 192, 138–150. [Google Scholar] [CrossRef]
- Mock, T.; Thomas, D.N. Recent advances in sea-ice microbiology. Environ. Microbiol. 2005, 7, 605–619. [Google Scholar] [CrossRef]
- Beckering, C.L.; Steil, L.; Weber, M.H.; Volker, U.; Marahiel, M.A. Genomewide transcriptional analysis of the cold shock response in Bacillus subtilis. J. Bacteriol. 2002, 184, 6395–6402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castellano, I.; Di Maro, A.; Ruocco, M.R.; Chambery, A.; Parente, A.; Di Martino, M.T.; Parlato, G.; Masullo, M.; De Vendittis, E. Psychrophilic superoxide dismutase from Pseudoalteromonas haloplanktis: Biochemical characterization and identification of a highly reactive cysteine residue. Biochimie 2006, 88, 1377–1389. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Yu, H.; Li, L.; Hu, S.; Dong, X. The genome and transcriptome of a newly described psychrophilic archaeon, Methanolobus psychrophilus R15, reveal its cold adaptive characteristics. Environ. Microbiol. Rep. 2012, 4, 633–641. [Google Scholar] [CrossRef]
- Gao, H.; Yang, Z.K.; Wu, L.; Thompson, D.K.; Zhou, J. Global transcriptome analysis of the cold shock response of Shewanella oneidensis MR-1 and mutational analysis of its classical cold shock proteins. J. Bacteriol. 2006, 188, 4560–4569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Campanaro, S.; Williams, T.J.; Burg, D.W.; De Francisci, D.; Treu, L.; Lauro, F.M.; Cavicchioli, R. Temperature-dependent global gene expression in the Antarctic archaeon Methanococcoides burtonii. Environ. Microbiol. 2011, 13, 2018–2038. [Google Scholar] [CrossRef]
- Kumar, S.; Suyal, D.C.; Yadav, A.; Shouche, Y.; Goel, R. Psychrophilic Pseudomonas helmanticensis proteome under simulated cold stress. Cell Stress Chaperones 2020, 25, 1025–1032. [Google Scholar] [CrossRef]
- Hou, Y.; Qiao, C.; Wang, Y.; Wang, Y.; Ren, X.; Wei, Q.; Wang, Q. Cold-Adapted Glutathione S-Transferases from Antarctic Psychrophilic Bacterium Halomonas sp. ANT108: Heterologous Expression, Characterization, and Oxidative Resistance. Mar. Drugs 2019, 17, 147. [Google Scholar] [CrossRef] [Green Version]
- Kandror, O.; DeLeon, A.; Goldberg, A.L. Trehalose synthesis is induced upon exposure of Escherichia coli to cold and is essential for viability at low temperatures. Proc. Natl. Acad. Sci. USA 2002, 99, 9727–9732. [Google Scholar] [CrossRef] [Green Version]
- Kaushik, J.K.; Bhat, R. Why is trehalose an exceptional protein stabilizer? An analysis of the thermal stability of proteins in the presence of the compatible osmolyte trehalose. J. Biol. Chem. 2003, 278, 26458–26465. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Limsuwun, K.; Jones, P.G. Spermidine acetyltransferase is required to prevent spermidine toxicity at low temperatures in Escherichia coli. J. Bacteriol 2000, 182, 5373–5380. [Google Scholar] [CrossRef] [Green Version]
- Koh, H.Y.; Park, H.; Lee, J.H.; Han, S.J.; Sohn, Y.C.; Lee, S.G. Proteomic and transcriptomic investigations on cold-responsive properties of the psychrophilic Antarctic bacterium Psychrobacter sp. PAMC 21119 at subzero temperatures. Environ. Microbiol. 2017, 19, 628–644. [Google Scholar] [CrossRef] [PubMed]
- Strubbe-Rivera, J.O.; Schrad, J.R.; Pavlov, E.V.; Conway, J.F.; Parent, K.N.; Bazil, J.N. The mitochondrial permeability transition phenomenon elucidated by cryo-EM reveals the genuine impact of calcium overload on mitochondrial structure and function. Sci. Rep. 2021, 11, 1037. [Google Scholar] [CrossRef]
- Anchordoguy, T.J.; Rudolph, A.S.; Carpenter, J.F.; Crowe, J.H. Modes of interaction of cryoprotectants with membrane phospholipids during freezing. Cryobiology 1987, 24, 324–331. [Google Scholar] [CrossRef]
- Privalov, P.L. Stability of proteins: Small globular proteins. Adv. Protein Chem. 1979, 33, 167–241. [Google Scholar] [CrossRef]
- Makhatadze, G.I.; Privalov, P.L. Energetics of protein structure. Adv. Protein Chem. 1995, 47, 307–425. [Google Scholar] [CrossRef]
- Spassov, V.Z.; Karshikoff, A.D.; Ladenstein, R. The optimization of protein-solvent interactions: Thermostability and the role of hydrophobic and electrostatic interactions. Protein Sci. 1995, 4, 1516–1527. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Privalov, P.L. Thermodynamic problems of protein structure. Annu. Rev. Biophys. Biophys. Chem. 1989, 18, 47–69. [Google Scholar] [CrossRef] [PubMed]
- Becktel, W.J.; Schellman, J.A. Protein stability curves. Biopolymers 1987, 26, 1859–1877. [Google Scholar] [CrossRef] [PubMed]
- Privalov, P.L.; Griko Yu, V.; Venyaminov, S.; Kutyshenko, V.P. Cold denaturation of myoglobin. J. Mol. Biol. 1986, 190, 487–498. [Google Scholar] [CrossRef]
- Pastore, A.; Martin, S.R.; Politou, A.; Kondapalli, K.C.; Stemmler, T.; Temussi, P.A. Unbiased cold denaturation: Low- and high-temperature unfolding of yeast frataxin under physiological conditions. J. Am. Chem. Soc. 2007, 129, 5374–5375. [Google Scholar] [CrossRef] [Green Version]
- Robertson, A.D.; Murphy, K.P. Protein Structure and the Energetics of Protein Stability. Chem. Rev. 1997, 97, 1251–1268. [Google Scholar] [CrossRef] [PubMed]
- Sawle, L.; Ghosh, K. How do thermophilic proteins and proteomes withstand high temperature? Biophys. J. 2011, 101, 217–227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rees, D.C.; Robertson, A.D. Some thermodynamic implications for the thermostability of proteins. Protein Sci. 2001, 10, 1187–1194. [Google Scholar] [CrossRef] [Green Version]
- D’Amico, S.; Marx, J.C.; Gerday, C.; Feller, G. Activity-stability relationships in extremophilic enzymes. J. Biol. Chem. 2003, 278, 7891–7896. [Google Scholar] [CrossRef] [Green Version]
- Graziano, G. On the molecular origin of cold denaturation of globular proteins. Phys. Chem. Chem. Phys. 2010, 12, 14245–14252. [Google Scholar] [CrossRef] [PubMed]
- Graziano, G. On the mechanism of cold denaturation. Phys. Chem. Chem. Phys. 2014, 16, 21755–21767. [Google Scholar] [CrossRef] [PubMed]
- Pica, A.; Graziano, G. Shedding light on the extra thermal stability of thermophilic proteins. Biopolymers 2016, 105, 856–863. [Google Scholar] [CrossRef]
- Graziano, G. Why small proteins tend to have high denaturation temperatures. Phys. Chem. Chem. Phys. 2020, 22, 16258–16266. [Google Scholar] [CrossRef] [PubMed]
- Graziano, G. Contrasting the hydration thermodynamics of methane and methanol. Phys. Chem. Chem. Phys. 2019, 21, 21418–21430. [Google Scholar] [CrossRef]
- Graziano, G. A purely geometric derivation of the scaled particle theory formula for the work of cavity creation in a liquid. Chem. Phys. Lett. 2007, 440, 221–223. [Google Scholar] [CrossRef]
- Lee, B.; Richards, F.M. The interpretation of protein structures: Estimation of static accessibility. J. Mol. Biol. 1971, 55, 379–400. [Google Scholar] [CrossRef]
- Graziano, G. On the cavity size distribution in water and n-hexane. Biophys. Chem. 2003, 104, 393–405. [Google Scholar] [CrossRef]
- Royer, C.A. Revisiting volume changes in pressure-induced protein unfolding. Biochim. Biophys. Acta 2002, 1595, 201–209. [Google Scholar] [CrossRef]
- Chalikian, T.V. Volumetric properties of proteins. Annu. Rev. Biophys. Biomol. Struct. 2003, 32, 207–235. [Google Scholar] [CrossRef] [PubMed]
- Graziano, G. The Gibbs energy cost of cavity creation depends on geometry. J. Mol. Liq. 2015, 211, 1047–1051. [Google Scholar] [CrossRef]
- Graziano, G. Shedding light on the hydrophobicity puzzle. Pure Appl. Chem. 2016, 88, 177–188. [Google Scholar] [CrossRef]
- Rose, G.D. Protein folding—Seeing is deceiving. Protein Sci. Publ. Protein Soc. 2021, 30, 1606–1616. [Google Scholar] [CrossRef] [PubMed]
- Kell, G.S. Density, thermal expansivity, and compressibility of liquid water from 0.deg. to 150.deg. Correlations and tables for atmospheric pressure and saturation reviewed and expressed on 1968 temperature scale. J. Chem. Eng. Data 1975, 20, 97–105. [Google Scholar] [CrossRef]
- Tadokoro, T.; Matsushita, K.; Abe, Y.; Rohman, M.S.; Koga, Y.; Takano, K.; Kanaya, S. Remarkable stabilization of a psychrotrophic RNase HI by a combination of thermostabilizing mutations identified by the suppressor mutation method. Biochemistry 2008, 47, 8040–8047. [Google Scholar] [CrossRef] [PubMed]
- D’Amico, S.; Gerday, C.; Feller, G. Structural determinants of cold adaptation and stability in a large protein. J. Biol. Chem. 2001, 276, 25791–25796. [Google Scholar] [CrossRef] [Green Version]
- Ben-Naim, A. Hydrophobic interaction and structural changes in the solvent. Biopolym. Orig. Res. Biomol. 1975, 14, 1337–1355. [Google Scholar] [CrossRef]
- Yu, H.A.; Karplus, M. A thermodynamic analysis of solvation. J. Chem. Phys. 1988, 89, 2366–2379. [Google Scholar] [CrossRef]
- Lee, B. Enthalpy-entropy compensation in the thermodynamics of hydrophobicity. Biophys. Chem. 1994, 51, 271–278. [Google Scholar] [CrossRef]
- Dunitz, J.D. Win some, lose some: Enthalpy-entropy compensation in weak intermolecular interactions. Chem. Biol. 1995, 2, 709–712. [Google Scholar] [CrossRef] [Green Version]
- Graziano, G. Case study of enthalpy-entropy noncompensation. J. Chem. Phys. 2004, 120, 4467–4471. [Google Scholar] [CrossRef] [PubMed]
- Liquori, A.M.; Sadun, C. Close packing of amino acid residues in globular proteins: Specific volume and site binding of water molecules. Int. J. Biol. Macromol. 1981, 3, 56–59. [Google Scholar] [CrossRef]
- Cotter, M.A.; Martire, D.E. Statistical Mechanics of Rodlike Particles. II. A Scaled Particle Investigation of the Aligned→Isotropic Transition in a Fluid of Rigid Spherocylinders. J. Chem. Phys. 1970, 52, 1909–1919. [Google Scholar] [CrossRef]
- Head-Gordon, T.; Hura, G. Water structure from scattering experiments and simulation. Chem. Rev. 2002, 102, 2651–2670. [Google Scholar] [CrossRef] [Green Version]
- Baxa, M.C.; Haddadian, E.J.; Jumper, J.M.; Freed, K.F.; Sosnick, T.R. Loss of conformational entropy in protein folding calculated using realistic ensembles and its implications for NMR-based calculations. Proc. Natl. Acad. Sci. USA 2014, 111, 15396–15401. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tomasi, J.; Persico, M. Molecular interactions in solution: An overview of methods based on continuous distributions of the solvent. Chem. Rev. 1994, 94, 2027–2094. [Google Scholar] [CrossRef]
- Hummer, G.; Garde, S.; Garcia, A.; Paulaitis, M.E.; Pratt, L.R. Hydrophobic effects on a molecular scale. J. Phys. Chem. B 1998, 102, 10469–10482. [Google Scholar] [CrossRef] [Green Version]
- Graves, J.E.; Somero, G.N. Electrophoretic and Functional Enzymic Evolution in Four Species of Eastern Pacific Barracudas from Different Thermal Environments. Evolution 1982, 36, 97–106. [Google Scholar] [CrossRef]
- Somero, G.N. Proteins and temperature. Annu. Rev. Physiol. 1995, 57, 43–68. [Google Scholar] [CrossRef] [PubMed]
- Jaenicke, R.; Zavodszky, P. Proteins under extreme physical conditions. FEBS Lett. 1990, 268, 344–349. [Google Scholar] [CrossRef] [Green Version]
- Svingor, A.; Kardos, J.; Hajdu, I.; Nemeth, A.; Zavodszky, P. A better enzyme to cope with cold. Comparative flexibility studies on psychrotrophic, mesophilic, and thermophilic IPMDHs. J. Biol. Chem. 2001, 276, 28121–28125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Na, H.; Song, G.; ben-Avraham, D. Universality of vibrational spectra of globular proteins. Phys. Biol. 2016, 13, 016008. [Google Scholar] [CrossRef] [Green Version]
- Callender, R.; Dyer, R.B. The dynamical nature of enzymatic catalysis. Acc. Chem. Res. 2015, 48, 407–413. [Google Scholar] [CrossRef] [PubMed]
- Chalopin, Y. The physical origin of rate promoting vibrations in enzymes revealed by structural rigidity. Sci. Rep. 2020, 10, 17465. [Google Scholar] [CrossRef] [PubMed]
- Saavedra, H.G.; Wrabl, J.O.; Anderson, J.A.; Li, J.; Hilser, V.J. Dynamic allostery can drive cold adaptation in enzymes. Nature 2018, 558, 324–328. [Google Scholar] [CrossRef]
- Dong, Y.W.; Liao, M.L.; Meng, X.L.; Somero, G.N. Structural flexibility and protein adaptation to temperature: Molecular dynamics analysis of malate dehydrogenases of marine molluscs. Proc. Natl. Acad Sci. USA 2018, 115, 1274–1279. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feller, G.; Payan, F.; Theys, F.; Qian, M.; Haser, R.; Gerday, C. Stability and structural analysis of alpha-amylase from the antarctic psychrophile Alteromonas haloplanctis A23. Eur. J. Biochem. 1994, 222, 441–447. [Google Scholar] [CrossRef] [PubMed]
- Aghajari, N.; Feller, G.; Gerday, C.; Haser, R. Structures of the psychrophilic Alteromonas haloplanctis alpha-amylase give insights into cold adaptation at a molecular level. Structure 1998, 6, 1503–1516. [Google Scholar] [CrossRef] [Green Version]
- Feller, G.; d’Amico, D.; Gerday, C. Thermodynamic stability of a cold-active alpha-amylase from the Antarctic bacterium Alteromonas haloplanctis. Biochemistry 1999, 38, 4613–4619. [Google Scholar] [CrossRef]
- Cipolla, A.; Delbrassine, F.; Da Lage, J.L.; Feller, G. Temperature adaptations in psychrophilic, mesophilic and thermophilic chloride-dependent alpha-amylases. Biochimie 2012, 94, 1943–1950. [Google Scholar] [CrossRef]
- D’Amico, S.; Gerday, C.; Feller, G. Temperature adaptation of proteins: Engineering mesophilic-like activity and stability in a cold-adapted alpha-amylase. J. Mol. Biol. 2003, 332, 981–988. [Google Scholar] [CrossRef] [PubMed]
- Cipolla, A.; D’Amico, S.; Barumandzadeh, R.; Matagne, A.; Feller, G. Stepwise adaptations to low temperature as revealed by multiple mutants of psychrophilic alpha-amylase from Antarctic Bacterium. J. Biol. Chem. 2011, 286, 38348–38355. [Google Scholar] [CrossRef] [Green Version]
- Li, Q.; Yan, Y.; Liu, X.; Zhang, Z.; Tian, J.; Wu, N. Enhancing thermostability of a psychrophilic alpha-amylase by the structural energy optimization in the trajectories of molecular dynamics simulations. Int. J. Biol. Macromol. 2020, 142, 624–633. [Google Scholar] [CrossRef] [PubMed]
- Russell, R.J.; Hough, D.W.; Danson, M.J.; Taylor, G.L. The crystal structure of citrate synthase from the thermophilic archaeon, Thermoplasma acidophilum. Structure 1994, 2, 1157–1167. [Google Scholar] [CrossRef] [Green Version]
- Gerike, U.; Danson, M.J.; Hough, D.W. Cold-active citrate synthase: Mutagenesis of active-site residues. Protein Eng. 2001, 14, 655–661. [Google Scholar] [CrossRef] [PubMed]
- Russell, R.J.; Ferguson, J.M.; Hough, D.W.; Danson, M.J.; Taylor, G.L. The crystal structure of citrate synthase from the hyperthermophilic archaeon pyrococcus furiosus at 1.9 A resolution. Biochemistry 1997, 36, 9983–9994. [Google Scholar] [CrossRef] [PubMed]
- Russell, R.J.; Gerike, U.; Danson, M.J.; Hough, D.W.; Taylor, G.L. Structural adaptations of the cold-active citrate synthase from an Antarctic bacterium. Structure 1998, 6, 351–361. [Google Scholar] [CrossRef]
- Bell, G.S.; Russell, R.J.; Connaris, H.; Hough, D.W.; Danson, M.J.; Taylor, G.L. Stepwise adaptations of citrate synthase to survival at life’s extremes. From psychrophile to hyperthermophile. Eur. J. Biochem. 2002, 269, 6250–6260. [Google Scholar] [CrossRef] [PubMed]
- Juers, D.H.; Matthews, B.W.; Huber, R.E. LacZ beta-galactosidase: Structure and function of an enzyme of historical and molecular biological importance. Protein Sci. 2012, 21, 1792–1807. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Triveni Piasad Shukla, L.E.W. Beta-galactosidase technology: A solution to the lactose problem. CRC Crit. Rev. Food Technol. 1975, 5, 325–356. [Google Scholar] [CrossRef]
- Li, S.; Zhu, X.; Xing, M. A New beta-Galactosidase from the Antarctic Bacterium Alteromonas sp. ANT48 and Its Potential in Formation of Prebiotic Galacto-Oligosaccharides. Mar. Drugs 2019, 17, 599. [Google Scholar] [CrossRef] [Green Version]
- Mangiagalli, M.; Lotti, M. Cold-Active beta-Galactosidases: Insight into Cold Adaption Mechanisms and Biotechnological Exploitation. Mar. Drugs 2021, 19, 43. [Google Scholar] [CrossRef] [PubMed]
- Bruno, S.; Coppola, D.; di Prisco, G.; Giordano, D.; Verde, C. Enzymes from Marine Polar Regions and Their Biotechnological Applications. Mar. Drugs 2019, 17, 544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mangiagalli, M.; Brocca, S.; Orlando, M.; Lotti, M. The “cold revolution”. Present and future applications of cold-active enzymes and ice-binding proteins. New Biotechnol. 2020, 55, 5–11. [Google Scholar] [CrossRef] [PubMed]
- Hoyoux, A.; Jennes, I.; Dubois, P.; Genicot, S.; Dubail, F.; Francois, J.M.; Baise, E.; Feller, G.; Gerday, C. Cold-adapted beta-galactosidase from the Antarctic psychrophile Pseudoalteromonas haloplanktis. Appl. Environ. Microbiol. 2001, 67, 1529–1535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jacobson, R.H.; Zhang, X.J.; DuBose, R.F.; Matthews, B.W. Three-dimensional structure of beta-galactosidase from E. coli. Nature 1994, 369, 761–766. [Google Scholar] [CrossRef] [PubMed]
- Mangiagalli, M.; Lapi, M.; Maione, S.; Orlando, M.; Brocca, S.; Pesce, A.; Barbiroli, A.; Camilloni, C.; Pucciarelli, S.; Lotti, M.; et al. The co-existence of cold activity and thermal stability in an Antarctic GH42 beta-galactosidase relies on its hexameric quaternary arrangement. FEBS J. 2021, 288, 546–565. [Google Scholar] [CrossRef]
- Rutkiewicz, M.; Bujacz, A.; Wanarska, M.; Wierzbicka-Wos, A.; Cieslinski, H. Active Site Architecture and Reaction Mechanism Determination of Cold Adapted beta-d-galactosidase from Arthrobacter sp. 32cB. Int. J. Mol. Sci. 2019, 20, 4301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rutkiewicz, M.; Wanarska, M.; Bujacz, A. Mapping the Transglycosylation Relevant Sites of Cold-Adapted beta-d-Galactosidase from Arthrobacter sp. 32cB. Int. J. Mol. Sci. 2020, 21, 5354. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rapuano, R.; Graziano, G. Some Clues about Enzymes from Psychrophilic Microorganisms. Microorganisms 2022, 10, 1161. https://doi.org/10.3390/microorganisms10061161
Rapuano R, Graziano G. Some Clues about Enzymes from Psychrophilic Microorganisms. Microorganisms. 2022; 10(6):1161. https://doi.org/10.3390/microorganisms10061161
Chicago/Turabian StyleRapuano, Roberta, and Giuseppe Graziano. 2022. "Some Clues about Enzymes from Psychrophilic Microorganisms" Microorganisms 10, no. 6: 1161. https://doi.org/10.3390/microorganisms10061161
APA StyleRapuano, R., & Graziano, G. (2022). Some Clues about Enzymes from Psychrophilic Microorganisms. Microorganisms, 10(6), 1161. https://doi.org/10.3390/microorganisms10061161






