Targeting the Gut Microbiota and Host Immunity with a Bacilli-Species Probiotic during Antibiotic Exposure in Mice
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents
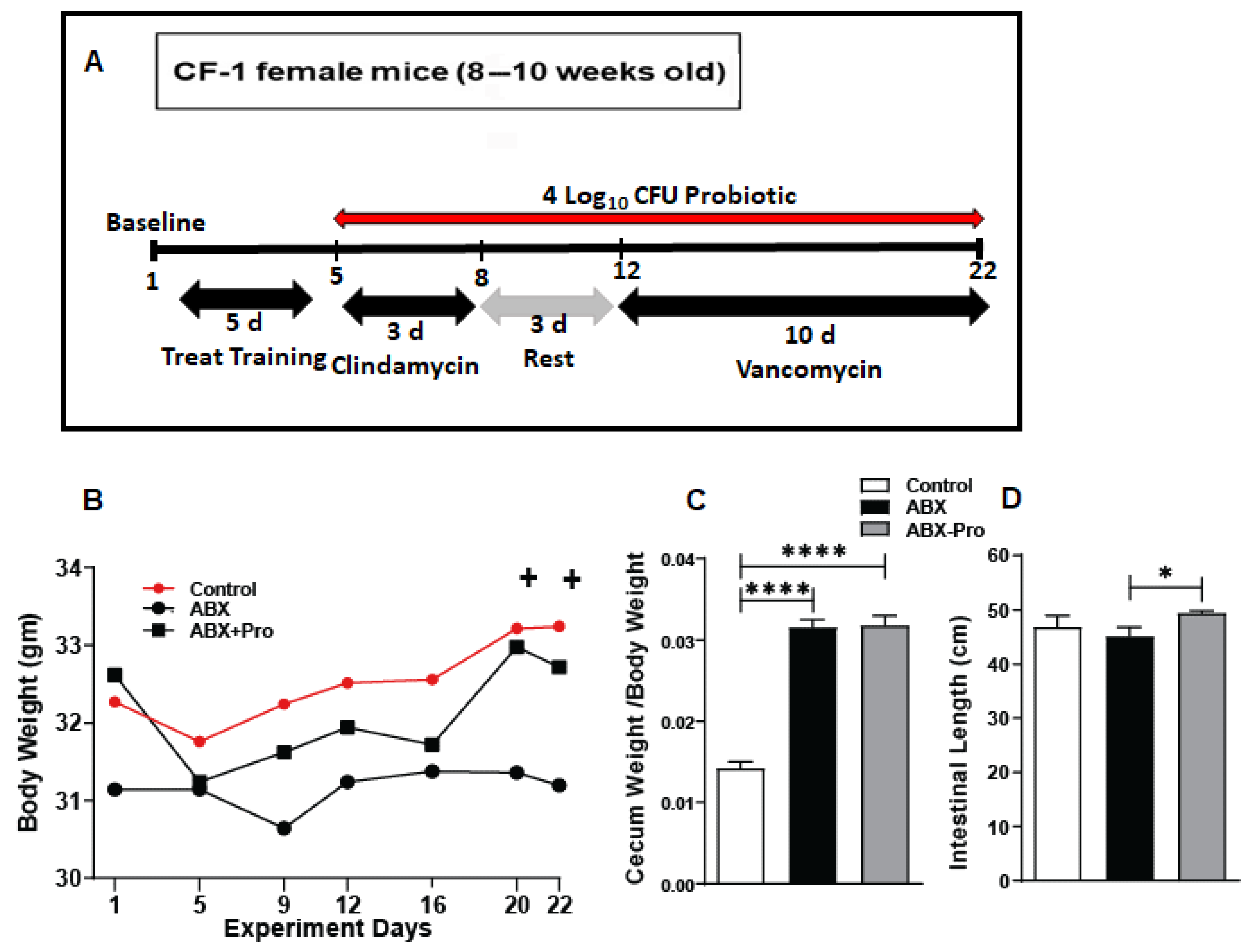
2.2. Mouse Model
2.3. Selective Bacterial Plating
2.4. Fecal qRT-PCR
2.5. Tissue qRT-PCR
2.6. Enzyme-Linked Immunoassay (ELISA)
2.7. Immunohistochemistry (IHC)
2.8. Statistics
3. Results
3.1. Bodyweight and Intestinal Assessment
3.2. Gut Microbiota Analysis
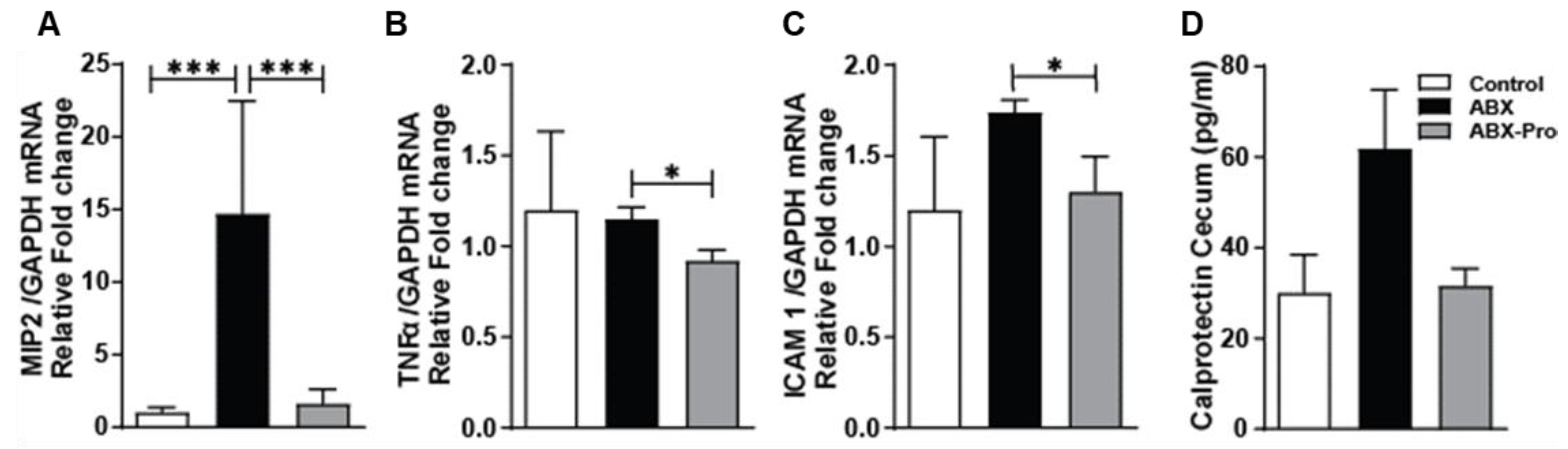
3.3. Gut Microbe and Host Immune Responses
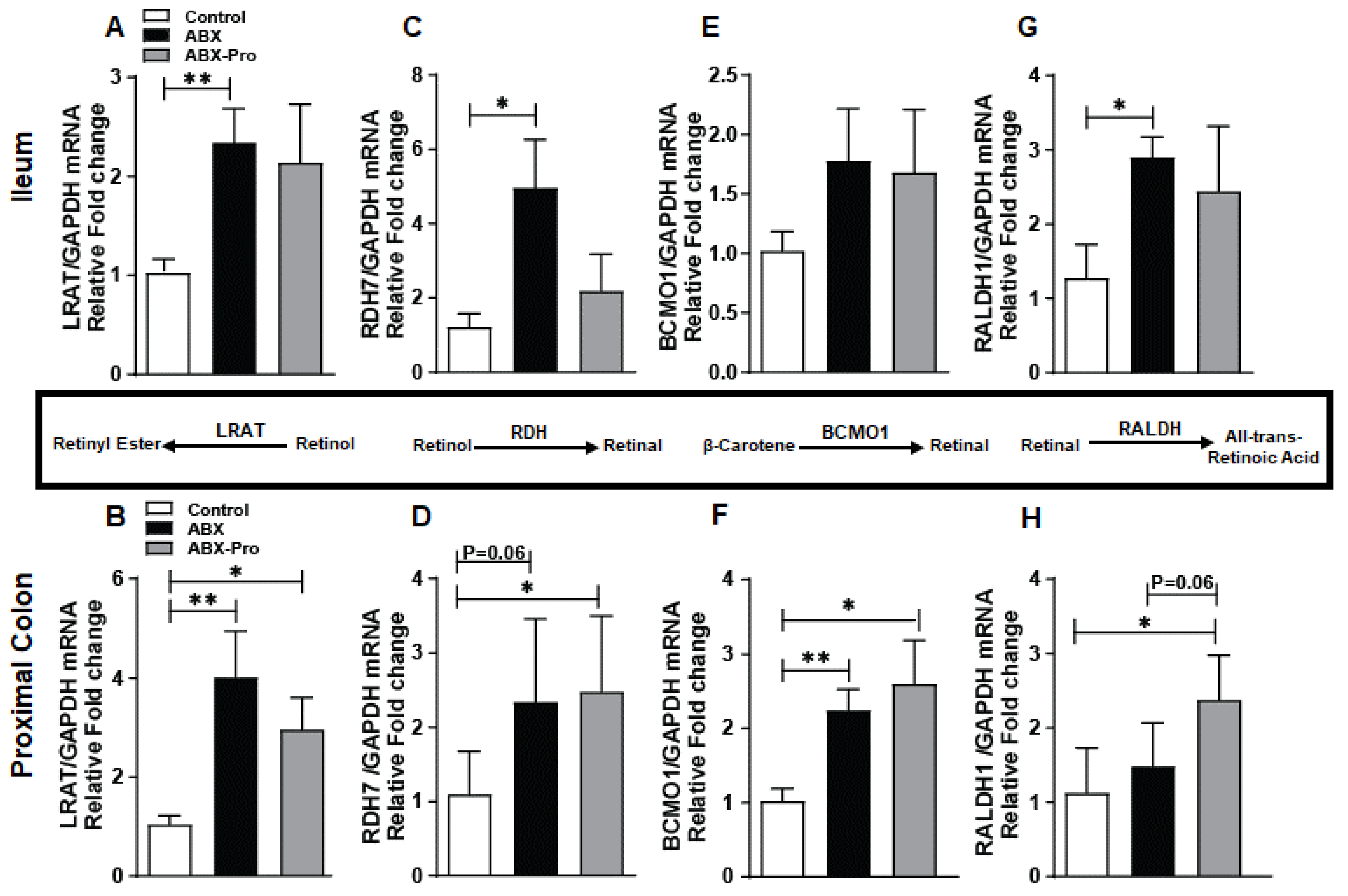
3.4. Vitamin A Metabolism
3.5. Serum Amyloid
3.6. Inflammatory Markers
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ayres, J.S. Cooperative Microbial Tolerance Behaviors in Host-Microbiota Mutualism. Cell 2016, 165, 1323–1331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Messer, J.S.; Chang, E.B. Microbial Physiology of the Digestive Tract and Its Role in Inflammatory Bowel Diseases. In Physiology of the Gastrointestinal Tract; Hamid, M.S., Ed.; Elsevier: Amsterdam, The Netherlands, 2018; pp. 795–810. [Google Scholar]
- Mantis, N.J.; Rol, N.; Corthésy, B. Secretory IgA’s Complex Roles in Immunity and Mucosal Homeostasis in the Gut. Mucosal Immunol. 2011, 4, 603–611. [Google Scholar] [CrossRef] [PubMed]
- Grizotte-Lake, M.; Zhong, G.; Duncan, K.; Kirkwood, J.; Iyer, N.; Smolenski, I.; Isoherranen, N.; Vaishnava, S. Commensals Suppress Intestinal Epithelial Cell Retinoic Acid Synthesis to Regulate Interleukin-22 Activity and Prevent Microbial Dysbiosis. Immunity 2018, 49, 1103–1115.e6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, D.; Liwinski, T.; Elinav, E. Interaction between Microbiota and Immunity in Health and Disease. Cell Res. 2020, 30, 492–506. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, J.; Guarner, F.; Fernandez, L.B.; Maruy, A.; Sdepanian, V.L.; Cohen, H. Antibiotics as Major Disruptors of Gut Microbiota. Front. Cell. Infect. Microbiol. 2020, 10, 572912. [Google Scholar] [CrossRef]
- Robak, O.H.; Heimesaat, M.M.; Kruglov, A.A.; Prepens, S.; Ninnemann, J.; Gutbier, B.; Reppe, K.; Hochrein, H.; Suter, M.; Kirschning, C.J.; et al. Antibiotic Treatment-Induced Secondary IgA Deficiency Enhances Susceptibility to Pseudomonas Aeruginosa Pneumonia. J. Clin. Investig. 2018, 128, 3535–3545. [Google Scholar] [CrossRef]
- Gattu, S.; Bang, Y.J.; Pendse, M.; Dende, C.; Chara, A.L.; Harris, T.A.; Wang, Y.; Ruhn, K.A.; Kuang, Z.; Sockanathan, S.; et al. Epithelial Retinoic Acid Receptor β Regulates Serum Amyloid A Expression and Vitamin A-Dependent Intestinal Immunity. Proc. Natl. Acad. Sci. USA 2019, 166, 10911–10916. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.Y.; Hall, J.A.; Kroehling, L.; Wu, L.; Najar, T.; Nguyen, H.H.; Lin, W.Y.; Yeung, S.T.; Silva, H.M.; Li, D.; et al. Serum Amyloid A Proteins Induce Pathogenic Th17 Cells and Promote Inflammatory Disease. Cell 2020, 180, 79–91.e16. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention Measuring Outpatient Antibiotic Prescribing. Available online: https://www.cdc.gov/antibiotic-use/data/outpatient-prescribing/index.html (accessed on 1 May 2022).
- Chiu, C.W.; Tsai, P.J.; Lee, C.C.; Ko, W.C.; Hung, Y.P. Inhibition of Spores to Prevent the Recurrence of Clostridioides difficile Infection—A Possibility or an Improbability? J. Microbiol. Immunol. Infect. 2021, 54, 1011–1017. [Google Scholar] [CrossRef]
- Blaabjerg, S.; Artzi, D.M.; Aabenhus, R. Probiotics for the Prevention of Antibiotic-Associated Diarrhea in Outpatients—A Systematic Review and Meta-Analysis. Antibiotics 2017, 6, 21. [Google Scholar] [CrossRef] [Green Version]
- Cao, J.; Yu, Z.; Liu, W.; Zhao, J.; Zhang, H.; Zhai, Q.; Chen, W. Probiotic Characteristics of Bacillus Coagulans and Associated Implications for Human Health and Diseases. J. Funct. Foods 2020, 64, 103643. [Google Scholar] [CrossRef]
- Casula, G.; Cutting, S.M. Bacillus Probiotics: Spore Germination in the Gastrointestinal Tract. Appl. Environ. Microbiol. 2002, 68, 2344–2352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kapourchali, F.R.; Glueck, B.; Han, Y.; Shapiro, D.; Fulmer, C.G.; Cresci, G.A.M. A Spore-Forming Probiotic Supplement Improves the Intestinal Immune Response and Protects the Intestinal Health During Recurrent Clostridioides difficile Colonization in Mice. J. Parenter. Enter. Nutr. 2020, 44, 1428–1438. [Google Scholar] [CrossRef] [PubMed]
- Sy, C.; Dangles, O.; Borel, P.; Caris-Veyrat, C. Stability of Bacterial Carotenoids in the Presence of Iron in a Model of the Gastric Compartment—Comparison with Dietary Reference Carotenoids. Arch. Biochem. Biophys. 2015, 572, 89–100. [Google Scholar] [CrossRef] [PubMed]
- Donskey, C.J.; Hanrahan, J.A.; Hutton, R.A.; Rice, L.B. Effect of Parenteral Antibiotic Administration on Persistence of Vancomycin-Resistant Enterococcus Faecium in the Mouse Gastrointestinal Tract. J. Infect. Dis. 1999, 180, 384–390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roychowdhury, S.; Glueck, B.; Han, Y.; Mohammad, M.A.; Cresci, G.A.M. A Designer Synbiotic Attenuates Chronic-Binge Ethanol-Induced Gut-Liver Injury in Mice. Nutrients 2019, 11, 97. [Google Scholar] [CrossRef] [Green Version]
- Cresci, G.; Nagy, L.E.; Ganapathy, V. Lactobacillus GG and Tributyrin Supplementation Reduce Antibiotic-Induced Intestinal Injury. J. Parenter. Enter. Nutr. 2013, 37, 763–774. [Google Scholar] [CrossRef] [Green Version]
- Pabst, O.; Slack, E. IgA and the Intestinal Microbiota: The Importance of Being Specific. Mucosal Immunol. 2020, 13, 12–21. [Google Scholar] [CrossRef] [Green Version]
- D’Ambrosio, D.N.; Clugston, R.D.; Blaner, W.S. Vitamin A Metabolism: An Update. Nutrients 2011, 3, 63–103. [Google Scholar] [CrossRef] [Green Version]
- Eroglu, A.; Harrison, E.H. Carotenoid Metabolism in Mammals, Including Man: Formation, Occurrence, and Function of Apocarotenoids. J. Lipid Res. 2013, 54, 1719–1730. [Google Scholar] [CrossRef] [Green Version]
- Iyer, N.; Vaishnava, S. Vitamin A at the Interface of Host– Commensal–Pathogen Interactions. PLoS Pathog. 2019, 15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walsham, N.E.; Sherwood, R.A. Fecal Calprotectin in Inflammatory Bowel Disease. Clin. Exp. Gastroenterol. 2016, 9, 21–29. [Google Scholar] [PubMed] [Green Version]
- Smieja, M. Current Indications for the Use of Clindamycin: A Critical Review. Can. J. Inf. Dis. 1998, 9, 22–28. [Google Scholar] [CrossRef] [Green Version]
- Stojanov, S.; Berlec, A.; Štrukelj, B. The Influence of Probiotics on the Firmicutes/Bacteroidetes Ratio in the Treatment of Obesity and Inflammatory Bowel Disease. Microorganisms 2020, 8, 1715. [Google Scholar] [CrossRef]
- Vallianou, N.; Dalamaga, M.; Stratigou, T.; Karampela, I.; Tsigalou, C.; Tsigalou xtsigalou, C. Do Antibiotics Cause Obesity through Long-Term Alterations in the Gut Microbiome? A Review of Current Evidence. Curr. Obes. Rep. 2021, 10, 244–262. [Google Scholar] [CrossRef]
- Mirpuri, J.; Raetz, M.; Sturge, C.R.; Wilhelm, C.L.; Benson, A.; Savani, R.C.; Hooper, L.V.; Yarovinsky, F. Proteobacteria-Specific IgA Regulates Maturation of the Intestinal Microbiota. Gut Microbes 2013, 5, 28–39. [Google Scholar] [CrossRef] [Green Version]
- Lyu, Y.; Wu, L.; Wang, F.; Shen, X.; Lin, D. Carotenoid Supplementation and Retinoic Acid in Immunoglobulin A Regulation of the Gut Microbiota Dysbiosis. Exp. Biol. Med. 2018, 243, 613–620. [Google Scholar] [CrossRef] [Green Version]
- Bang, Y.J.; Hu, Z.; Li, Y.; Gattu, S.; Ruhn, K.A.; Raj, P.; Herz, J.; Hooper, L.V. Serum Amyloid a Delivers Retinol to Intestinal Myeloid Cells to Promote Adaptive Immunity. Science 2021, 373, eabf9232. [Google Scholar] [CrossRef]
- Karaduta, O.; Dvanajscak, Z.; Zybailov, B. Metaproteomics—An Advantageous Option in Studies of Host–Microbiota Interaction. Microorganisms 2021, 9, 980. [Google Scholar] [CrossRef]
- Valle-Gough, R.E.; Samaniego-Gámez, B.Y.; Apodaca-Hernández, J.E.; Chiappa-Carrara, F.X.; Rodríguez-Dorantes, M.; Arena-Ortiz, M.L. RNA-Seq Analysis on the Microbiota Associated with the White Shrimp (Litopenaeus Vannamei) in Different Stages of Development. Appl. Sci. 2022, 12, 2483. [Google Scholar] [CrossRef]





| Target | Type | Sequence |
|---|---|---|
| Universal | Total | ACTCCTACGGGAGGCAGCAG |
| ATTACCGCGGCTGCTGG | ||
| Actinobacteria | Phyla | CGCGGCCTATCAGCTTGTTG |
| ATTACCGCGGCTGCTGG | ||
| Bacteroidetes | Phyla | GGCGACCGGCGCACGGG |
| GRCCTTCCTCTCAGAACCC | ||
| Firmicutes | Phyla | GGAGYATGTGGTTTAATTCGAAGCA |
| AGCTGACGACAACCATGCAC | ||
| Alphaproteobacteria | Class | ACTCCTACGGGAGGCAGCAG |
| TCTACGRATTTCACCYCTAC | ||
| Betaproteobacteria | Class | CCGCACAGTTGGCGAGATGA |
| CGACAGTTATGACGCCCTCC | ||
| Gammaproteobacteria | Class | GAGTTTGATCATGGCTCA |
| GTATTACCGCGGCTGCTG | ||
| Bifidobacterium | Genus | GGGTGGTAATGCCGGATG |
| TAAGCCATGGACTTTCACACC | ||
| Escherichia coli | Species | GTGTGATATCTACCCGCTTCGC |
| AGAACGCTTTGTGGTTAATCAGGA |
| Target | Primer | Sequence |
|---|---|---|
| Β-Carotene 15, 15′ Monooxygenase 1 | BCMO1 F | GAGCAAGTACAACCATTGGT |
| BCMO1 R | AACTCAGACACCACGATTC | |
| Glyceraldehyde-3-Phosphate Dehydrogenase | GAPDH F | AGGTCGGTGTGAACGGATTTG |
| GAPDH R | TGTAGACCATGTAGTTGAGGTCA | |
| Intercellular Adhesion Molecule 1 | ICAM1 F | GTGATGCTCAGGTATCCATCCA |
| ICAM1 R | CACAGTTCTCAAAGCACAGCG | |
| Macrophage Inflammatory Protein | MIP2 F | GCGCCCAGACAGAAGTCATAG |
| MIP2 R | AGCCTTGCCTTTGTTCAGTAT C | |
| Lecithin Retinol Acyltransferase | LRAT F | GCAGTTGGGACTGACTCCAT |
| LRAT R | GCAGTTGGGACTGACTCCAT | |
| Retinoic Acid Receptor-alpha | RARα F | AGAGCAGCAGTTCCGAAGAG |
| RARα R | AGTGGTAGCCGGATGATTTG | |
| Retinoic Acid Receptor-beta | RARβ F | AATGCCACCTCTCATTCAGG |
| RARβ R | GTCTGCAACAGCTGGAAATG | |
| Retinoic Acid Receptor-gamma | RARƴ F | CCACCAAATGCATCATCAAG |
| RARƴ R | ATCCGCAGCATTAGGATGTC | |
| Retinoic Aldehyde Dehydrogenase 1 | RALDH1 F | ATACTTGTCGGATTTAGGAGGCT |
| RALDH1 R | GGGCCTATCTTCCAAATGAACA | |
| Retinol Dehydrogenase 7 | RDH7 F | CCTGGCGGGTTCAGGACTA |
| RDH7 R | CGAGGATGTCTGGTCCCAC | |
| Serum Amyloid A 1 | SAA1 F | ATCACCAGATCTGCCCAGGA |
| SAA1 R | CCTTGGAAAGCCTCGTGAAC | |
| Serum Amyloid A 2 | SAA2 F | ACCAGATCTGCCCAGGAGAC |
| SAA2 R | GCATGGAAGTATTTGTCTCCATCT | |
| Serum Amyloid A 3 | SAA3 F | GACATGTGGCGAGCCTACTC |
| SAA3 R | TTGGCAAACTGGTCAGCTCT |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shapiro, D.; Kapourchali, F.R.; Santilli, A.; Han, Y.; Cresci, G.A.M. Targeting the Gut Microbiota and Host Immunity with a Bacilli-Species Probiotic during Antibiotic Exposure in Mice. Microorganisms 2022, 10, 1178. https://doi.org/10.3390/microorganisms10061178
Shapiro D, Kapourchali FR, Santilli A, Han Y, Cresci GAM. Targeting the Gut Microbiota and Host Immunity with a Bacilli-Species Probiotic during Antibiotic Exposure in Mice. Microorganisms. 2022; 10(6):1178. https://doi.org/10.3390/microorganisms10061178
Chicago/Turabian StyleShapiro, David, Fatemeh Ramezani Kapourchali, Anthony Santilli, Yingchun Han, and Gail A. M. Cresci. 2022. "Targeting the Gut Microbiota and Host Immunity with a Bacilli-Species Probiotic during Antibiotic Exposure in Mice" Microorganisms 10, no. 6: 1178. https://doi.org/10.3390/microorganisms10061178






