Effect of Gut Microbiota Biotransformation on Dietary Tannins and Human Health Implications
Abstract
:1. Introduction
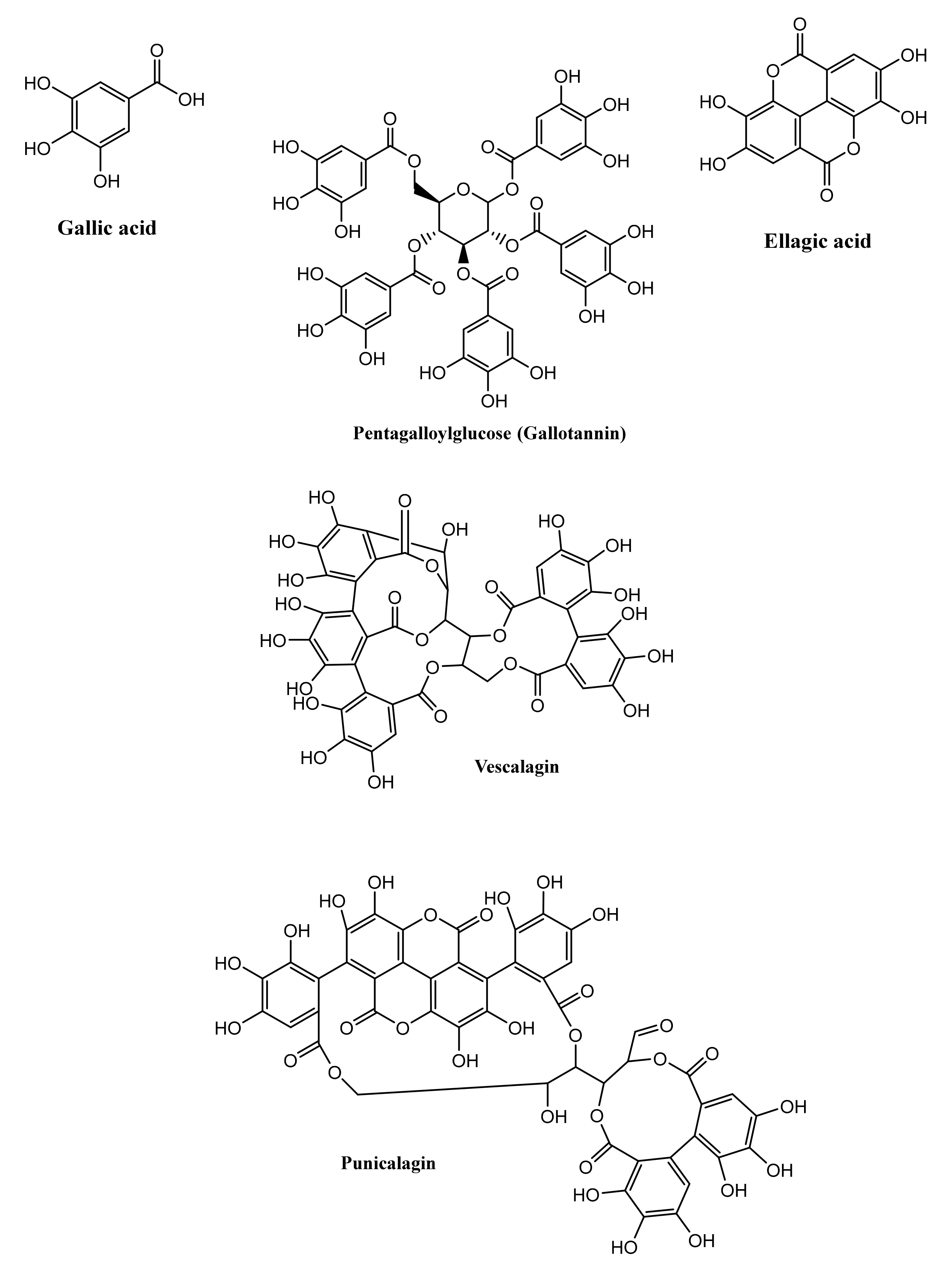
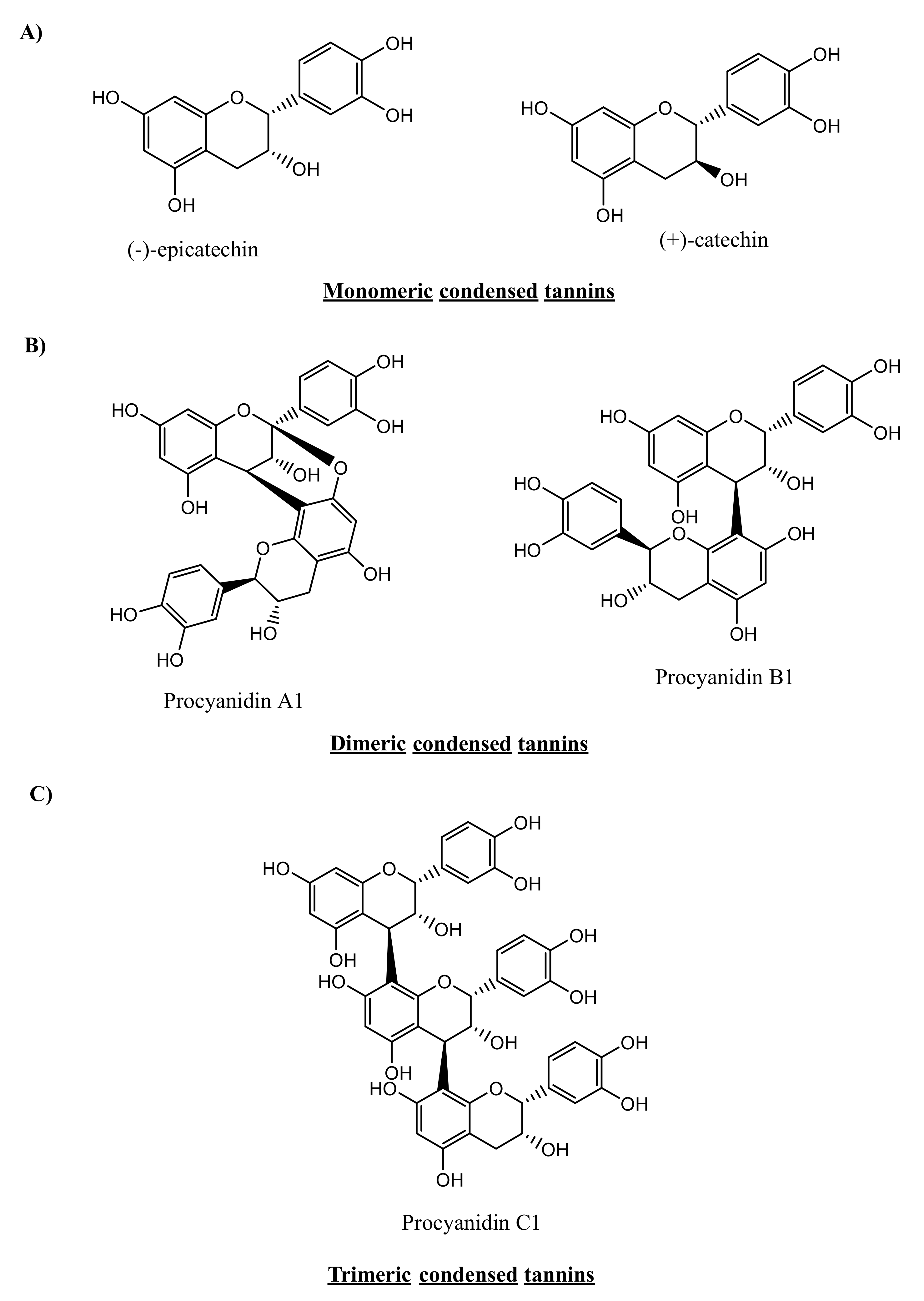
2. Chemistry and Diversity of Tannins
2.1. Hydrolysable Tannins
2.2. Condensed Tannins
3. Metabolism of Tannins
3.1. Non-Microbiota-Mediated Metabolism of Tannins
3.1.1. Hydrolysable Tannins
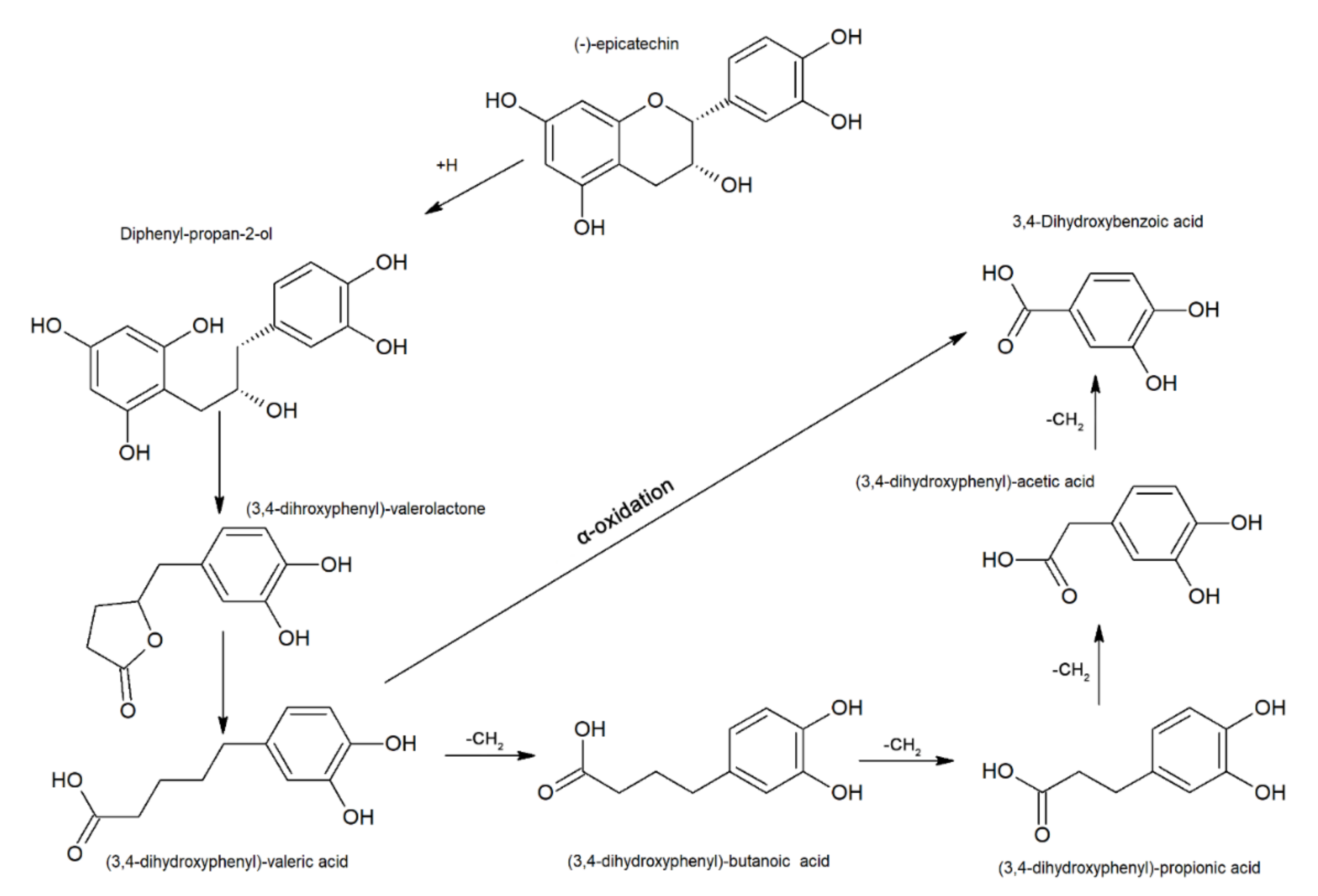
3.1.2. Condensed Tannins
3.2. Microbiota-Mediated Metabolism of Tannins
3.2.1. Composition of the Gut Microbiota
3.2.2. Metabolic Potential of the Gut Microbiota
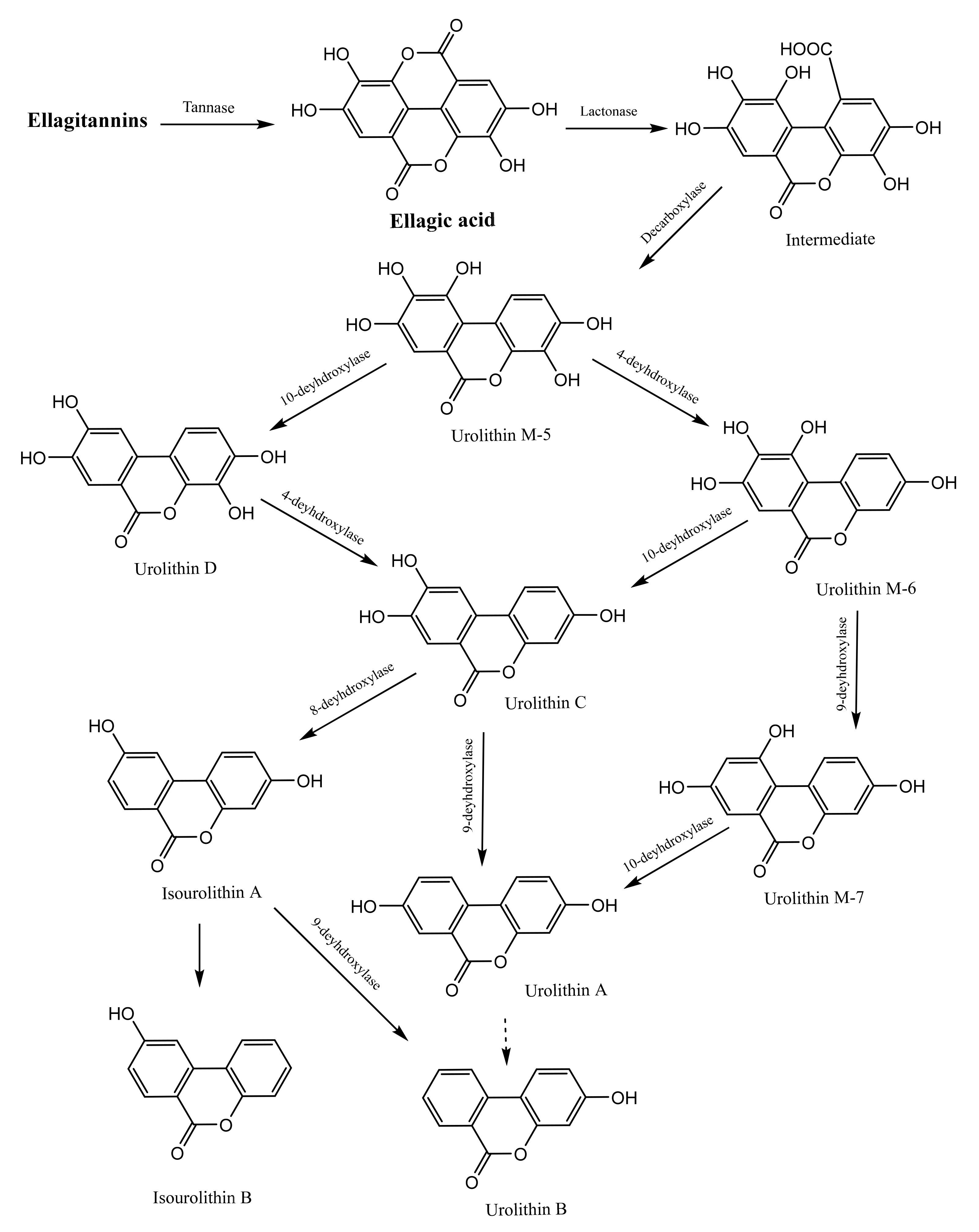
3.2.3. Metabolic Effect of the Gut Microbiota on Tannins
Gut Microbiota-Mediated Metabolism of Hydrolysable Tannins
Gut Microbiota-mediated Metabolism of Condensed Tannins
| Section 1: Metabolite Identification | ||||||
| Category | Code | Name | Category | Code | Name | |
| Valeric acid derivatives | 1 | (−)-5-(3′,4′,5′-Trihydroxyphenyl)-γ-valerolactone | 36 | 3-Hydroxybenzoic acid sulfate | ||
| 2 | (−)-5-(3′,4′,5′-Trihydroxyphenyl)-γ-valerolactone glucuronide | 37 | 4-Hydroxybenzoic acid sulfate | |||
| 3 | (−)-5-(3′,4′,5′-Trihydroxyphenyl)-γ-valerolactone sulfate | Procyanidin monomers metabolites/conjugates | 38 | (+)-Catechin | ||
| 4 | (−)-5-(3′,4′Dihydroxyphenyl)-γ-valerolactone | 39 | (−)-Epicatechin | |||
| 5 | (−)-5-(3′,4′Dihydroxyphenyl)-γ-valerolactone glucuronide | 40 | (–)-Epicatechin glucuronide | |||
| 6 | (−)-5-(3′,4′Dihydroxyphenyl)-γ-valerolactone methyl glucuronide | 41 | (-)-Epicatechin sulfate | |||
| 7 | (−)-5-(3′,4′Dihydroxyphenyl)-γ-valerolactone sulfate | 42 | (−)-Epicatechin-5/7-O -sulfate | |||
| 8 | 5-(3′-Hydroxy phenyl)-γ-valerolactone | 43 | 5/7-O -Sulfate-(−)-epicatechin-glucuronide | |||
| 9 | 5-Hydroxyphenyl-γ-valerolactone-O-glucuronide | 44 | 3′-O -Methyl-epicatechin | |||
| 10 | 5-(3′-Hydroxyphenyl)-γ-valerolactone-4′-O –sulfate | 45 | 4′-O -Methyl-epicatechin | |||
| 11 | 5-(3′,4′-Dihydroxy phenyl)valeric acid | 46 | 3′-O -Methyl-(−)-epicatechin-5/7-O -sulfate | |||
| 12 | 5-(3′,4′-Dihydroxy phenyl)valeric acid-O -sulfate | 47 | 4′-O -methyl-(–)-epicatechin-3′-O -beta-glucuronide | |||
| 13 | 5-(3′-Hydroxy phenyl)valeric acid | 48 | (–)-Epigallocatechin glucuronide | |||
| 14 | 4-Hydroxy-5-(3′,4′-dihydroxyphenyl)valeric acid | 49 | Methylated epigallocatechin glucuronide | |||
| 15 | 4-Hydroxy-5-(3′,4′-dihydroxyphenyl)valeric acid-O -sulfate | 50 | Methylated epigallocatechin sulfate | |||
| 16 | 3-O -Methyl-4-hydroxy-5-(3′,4′-dihydroxyphenyl)valeric acid-O –sulfate | Procyanidin dimers and other polyphenol metabolites/conjugates | 51 | Procyanidin dimers metabolites * | ||
| Propionic acid metabolites/conjugates | 17 | 3,4-Dihydroxyphenyl propan-2-ol | 52 | Procyanidin dimers metabolites ** | ||
| 18 | 3,4-Dihydroxyphenyl propan-2-ol. Dihydrate | 53 | Vanillic acid | |||
| 19 | 3,4-Dihydroxyphenyl propan-2-ol-O-glucuronide | 54 | Homovanillic acid | |||
| 20 | 1-(3′,4′-Dihydroxyphenyl)-3-(2′′,4′′,6′′- trihydroxyphenyl)propan-2-ol | 55 | Homovanillyl alcohol | |||
| 21 | 3-(3′,4′-Dihydroxy phenyl)propionic acid | 56 | Gallic acid | |||
| 22 | 3-(3,4-Dihytdroxyphenyl)propionic acid sulfate | 57 | 3-O -Methyl gallic acid | |||
| 23 | 3-(3′-Hydroxy phenyl)propionic acid | 58 | M-Coumaric sulfate | |||
| 24 | 3-(3′-Hydroxy phenyl)propionic acid sulfate | 59 | p-Coumaric sulfate | |||
| 25 | 3-(4-Hytdroxyphenyl)propionic acid | 60 | Ferulic acid sulfate | |||
| 26 | 3-(4-Hydroxyphenyl)propionic acid sulfate | 61 | 3-O -Protocatechuic acid sulfate | |||
| 27 | 3-Phenylpropionic acid | Hydrolysable tannins metabolites/conjugates | 62 | Urolithin A | ||
| Acetic and benzoic acid metabolites/conjugates | 28 | 2-(3,4-Dihydroxyphenyl)acetic acid | 63 | Hydroxyl urolithin A | ||
| 29 | 2-(3,4-Dihydroxyphenyl)acetic acid sulfate | 64 | Urolithin A glucuronide | |||
| 30 | 2-(3′-Hydroxyphenyl)acetic acid | 65 | Urolithin B | |||
| 31 | 2-(3′-Hydroxyphenyl)acetic acid sulfate | 66 | Urolithin B glucuronide | |||
| 32 | 2-(4′-Hydroxyphenyl)acetic acid | 67 | Urolithin C | |||
| 33 | 2-Phenylacetic acid | 68 | Urolithin D | |||
| 34 | Benzoic acid | 69 | Ellagic acid | |||
| 35 | 3-Hydroxybenzoic acid | 70 | Dimethylellagic acid glucuronide | |||
| Section 2: Material, detection and reference | ||||||
| Source | Metabolite code | Detected in | Detection mode | Reference | ||
| Filipendula ulmaria, Geranium pratense, Geranium robertianum, Geum urbanum root and rhizome, Lythrum salicaria, Potentilla anserina, Potentilla erecta rhizome, Quercus robur, Rubus idaeus leaf, Rubus fruticosus L. and pure ellagitannin vescalagin | 62-65-67 *** | Fermentation with human microbiota | LC-MS | [7] | ||
| Punicalagin | 62-65 | Fermentation with human microbiota | LC-MS | [19] | ||
| Red raspberries (Rubus idaeus L.) | 62-65 | Urine (human) | LC-MS | [75] | ||
| Cocoa powder | 4-28-30 | Urine (human) | LC-MS | [80] | ||
| Proanthocyanidin dimers | 4-8-13-20-21-23-28-30-32 | Fermentation with human microbiota | [81] | |||
| Green tea | 2-3-40-48-49-50 | Urine (human) | LC-MS | [85] | ||
| (-)-epicatechin | 4 | Urine (rats) | LC-ECD | [86] | ||
| EC, PC B1 and Polymeric PC fraction of cocoa | 4-14-32-44-45-51 | Urine/plasma (human) | LC-MS, GC-MS | [102] | ||
| Epicatechin, catechin, procyanidin B2 | 4-8-15-23-27-28-30-32-33-34 | Fermentation with human microbiota | GC-MS | [103] | ||
| Grape seed proanthocyanidin extract | 4-21-23-26-27-28-30-32-33-34-35-38-39-53-54-55-56-57 | Plasma (rats) | LC-MS | [87] | ||
| Procyanidin B2, Epicatechin | 4-8-11-14-15-20-21-23-30-33-39 | Fermentation with human microbiota | [104] | |||
| Apple polyphenol extract | 5-6-12-14-21 | Urine/plasma (human) | [105] | |||
| Procyanidin B2 | 7-15-16-17-18-19-22-24-29-31-36-37-41-42-43-46-51-58-59-60-61 | Urine (rats) | LC-DAD-MS | [88] | ||
| Partially purified apple procyanidin (PPCP) | 9-11-15-47 | Plasma (rats) | LC-MS, NMR | [106] | ||
| Carnberry juice | 10 | Plasma (human) | LC-MS | [107] | ||
| Pomegranate and walnuts | 62-65 | Urine/plasma/feces (human) | LC-MS | [108] | ||
| Strawberries, red raspberries, walnuts and oak-aged red wine. | 62-65 | Urine (human) | LC-MS | [12] | ||
| Pomegranate juice | 62-65-69-70 | Urine/plasma (human) | LC-MS | [8] | ||
| 62-63-64-65-70 | Plasma (human) | LC-MS | [77] | |||
| Strawberry | 62-64-65-66 | Urine (human) | LC-MS | [11] | ||
| Walnuts (Juglans regia L.), hazelnuts (Corylus avellana L.), and almonds (Prunus dulcis Mill.) | 62-65-67-68 | Urine (human) | LC-MS | [76] | ||
3.2.4. Gut Metabotypes and Tannin Metabolism
4. Biological Effects of Gut-Biotransformed Metabolites of Tannins
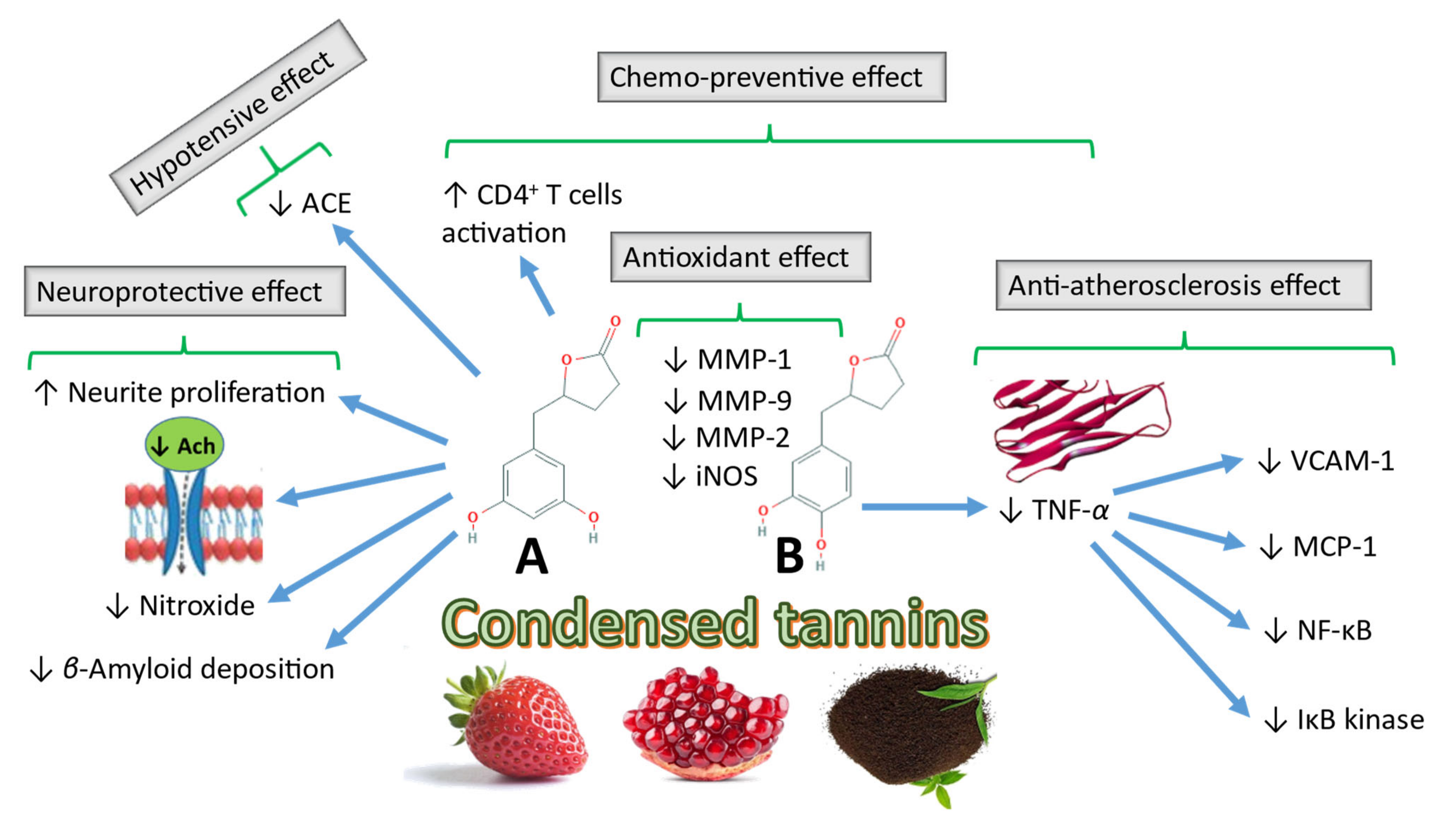
4.1. Biological Effects of Gut-Biotransformed Metabolites of CTs
4.1.1. Chemopreventive Activities of CT Metabolites
4.1.2. Other Biological Activities of CT Gut-biotransformed Metabolites
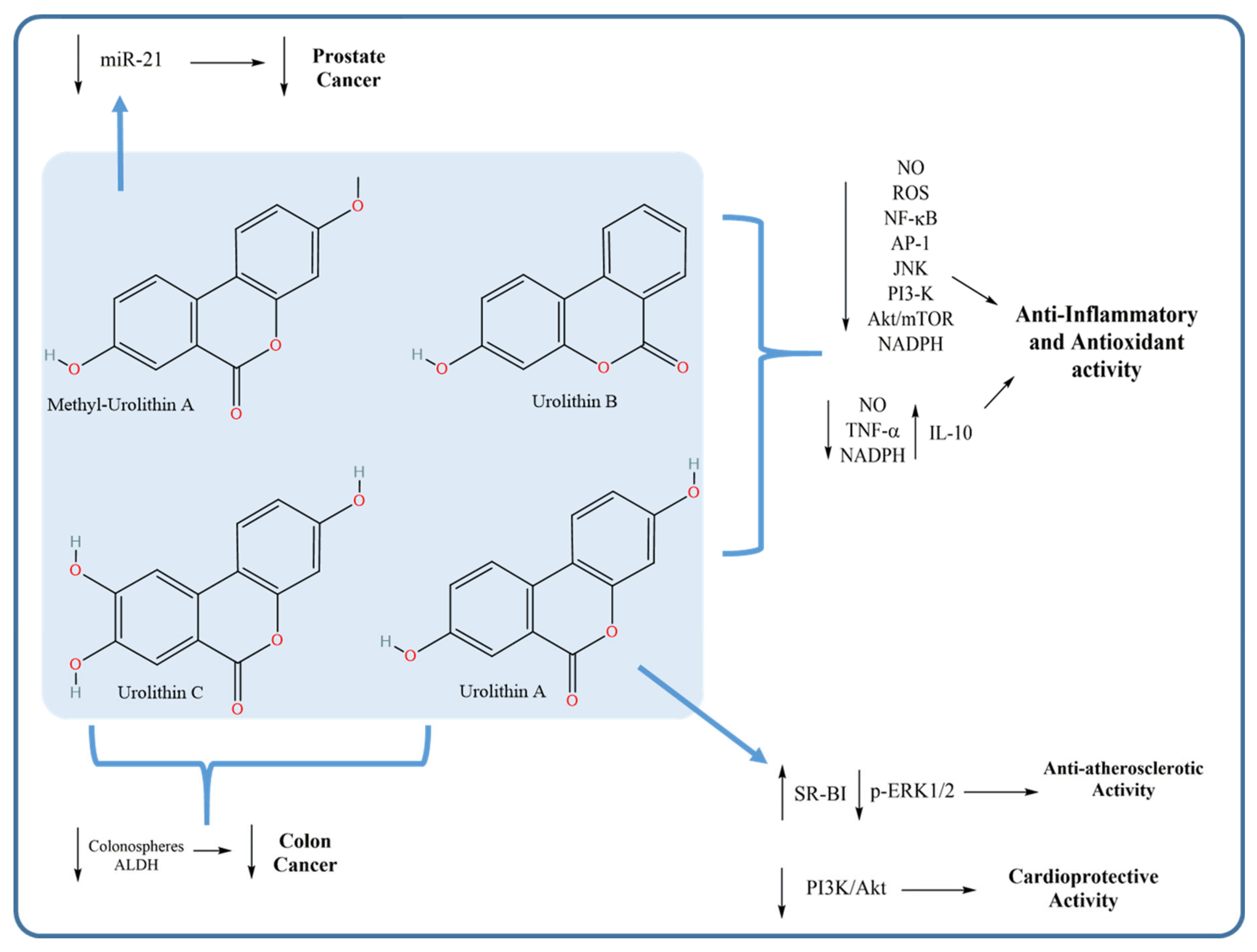
4.2. Biological Effects of Gut-Biotransformed HT Metabolites
4.2.1. Anti-Inflammatory and Antioxidant Activities of Urolithins
4.2.2. Anticancer Activity of Urolithins
4.2.3. Anti-Atherosclerotic and Cardioprotective Activities of Urolithins
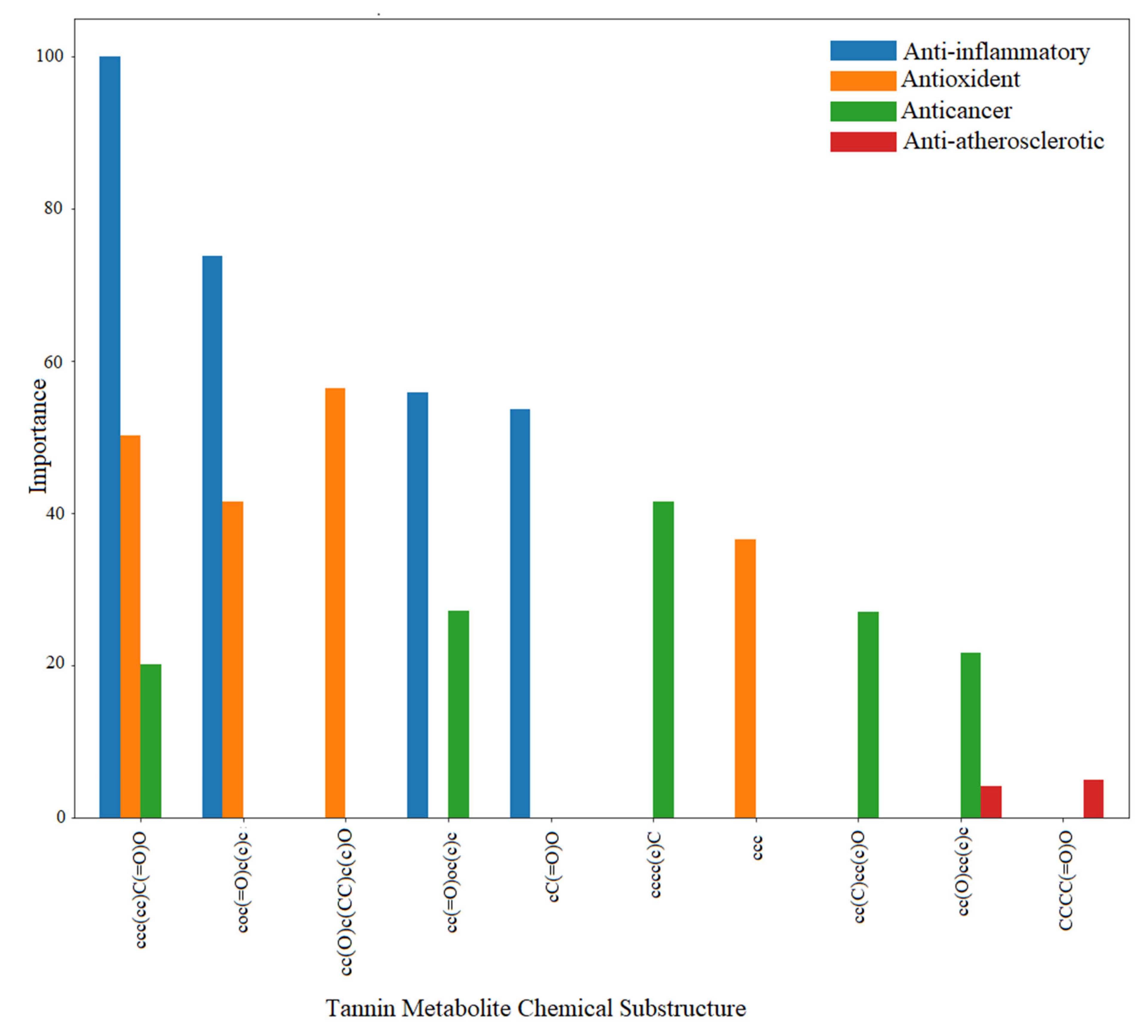
4.3. Mining the Correlation between the Chemical Space of Gut-Biotransformed Tannin Metabolites and Biological Effects
5. Conclusions and Future Directions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Falcão, L.; Araújo, M.E.M. Vegetable tannins used in the manufacture of historic leathers. Molecules 2018, 23, 1081. [Google Scholar] [CrossRef] [Green Version]
- Smeriglio, A.; Barreca, D.; Bellocco, E.; Trombetta, D. Proanthocyanidins and hydrolysable tannins: Occurrence, dietary intake and pharmacological effects. Br. J. Pharm. 2017, 174, 1244–1262. [Google Scholar] [CrossRef] [Green Version]
- Sathya, R.; Kanaga, N.; Sankar, P.; Jeeva, S. Antioxidant properties of phlorotannins from brown seaweed Cystoseira trinodis (Forsskål) C. Agardh. Arab. J. Chem. 2017, 10, S2608–S2614. [Google Scholar] [CrossRef] [Green Version]
- Cardona, F.; Andrés-Lacueva, C.; Tulipani, S.; Tinahones, F.J.; Queipo-Ortuño, M.I. Benefits of polyphenols on gut microbiota and implications in human health. J. Nutr. Biochem. 2013, 24, 1415–1422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vazquez-Fresno, R.; Llorach, R.; Perera, A.; Mandal, R.; Feliz, M.; Tinahones, F.J.; Wishart, D.S.; Andres-Lacueva, C. Clinical phenotype clustering in cardiovascular risk patients for the identification of responsive metabotypes after red wine polyphenol intake. J. Nutr. Biochem 2016, 28, 114–120. [Google Scholar] [CrossRef] [Green Version]
- Ozdal, T.; Sela, D.A.; Xiao, J.; Boyacioglu, D.; Chen, F.; Capanoglu, E. The reciprocal interactions between polyphenols and gut microbiota and effects on bioaccessibility. Nutrients 2016, 8, 78. [Google Scholar] [CrossRef] [PubMed]
- Piwowarski, J.P.; Granica, S.; Zwierzyńska, M.; Stefańska, J.; Schopohl, P.; Melzig, M.F.; Kiss, A.K. Role of human gut microbiota metabolism in the anti-inflammatory effect of traditionally used ellagitannin-rich plant materials. J. Ethnopharmacol. 2014, 155, 801–809. [Google Scholar] [CrossRef] [PubMed]
- Jiménez-Luna, J.; Grisoni, F.; Schneider, G. Drug discovery with explainable artificial intelligence. Nat. Mach. Intell. 2020, 2, 573–584. [Google Scholar] [CrossRef]
- Huang, Q.; Liu, X.; Zhao, G.; Hu, T.; Wang, Y. Potential and challenges of tannins as an alternative to in-feed antibiotics for farm animal production. Anim. Nutr. 2018, 4, 137–150. [Google Scholar] [CrossRef] [PubMed]
- Seeram, N.P.; Henning, S.M.; Zhang, Y.; Suchard, M.; Li, Z.; Heber, D. Pomegranate juice ellagitannin metabolites are present in human plasma and some persist in urine for up to 48 hours. J. Nutr. 2006, 136, 2481–2485. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Truchado, P.; Larrosa, M.; García-Conesa, M.T.; Cerdá, B.; Vidal-Guevara, M.L.; Tomás-Barberán, F.A.; Espín, J.C. Strawberry processing does not affect the production and urinary excretion of urolithins, ellagic acid metabolites, in humans. J. Agric. Food Chem. 2012, 60, 5749–5754. [Google Scholar] [CrossRef] [PubMed]
- Cerda, B.; Tomas-Barberan, F.A.; Espin, J.C. Metabolism of antioxidant and chemopreventive ellagitannins from strawberries, raspberries, walnuts, and oak-aged wine in humans: Identification of biomarkers and individual variability. J. Agric. Food Chem. 2005, 53, 227–235. [Google Scholar] [CrossRef] [PubMed]
- Okuda, T.; Ito, H. Tannins of constant structure in medicinal and food plants—Hydrolyzable tannins and polyphenols related to tannins. Molecules 2011, 16, 2191. [Google Scholar] [CrossRef]
- Moctezuma, C.; Hammerbacher, A.; Heil, M.; Gershenzon, J.; Mendez-Alonzo, R.; Oyama, K. Specific polyphenols and tannins are associated with defense against insect herbivores in the tropical oak Quercus oleoides. J. Chem. Ecol. 2014, 40, 458–467. [Google Scholar] [CrossRef] [PubMed]
- Stoupi, S.; Williamson, G.; Drynan, J.W.; Barron, D.; Clifford, M.N. Procyanidin B2 catabolism by human fecal microflora: Partial characterization of ‘dimeric’ intermediates. Arch. Biochem. Biophys. 2010, 501, 73–78. [Google Scholar] [CrossRef]
- Deprez, S.; Mila, I.; Huneau, J.F.; Tome, D.; Scalbert, A. Transport of proanthocyanidin dimer, trimer, and polymer across monolayers of human intestinal epithelial Caco-2 cells. Antioxid. Redox Signal. 2001, 3, 957–967. [Google Scholar] [CrossRef]
- Sieniawska, E.; Baj, T. Chapter 10—Tannins. In Pharmacognosy; Badal, S., Delgoda, R., Eds.; Academic Press: Boston, MA, USA, 2017; pp. 199–232. [Google Scholar] [CrossRef]
- Lee, H.J.; Choi, I.H.; Kim, D.H.; Amanullah, S.M.; Kim, S.C. Nutritional characterization of tannin rich chestnut (Castanea) and its meal for pig. J. Appl. Anim. Res. 2016, 44, 258–262. [Google Scholar] [CrossRef] [Green Version]
- Heber, D. Multitargeted therapy of cancer by ellagitannins. Cancer Lett. 2008, 269, 262–268. [Google Scholar] [CrossRef]
- Rue, E.A.; Rush, M.D.; van Breemen, R.B. Procyanidins: A comprehensive review encompassing structure elucidation via mass spectrometry. Phytochem. Rev. 2018, 17, 1–16. [Google Scholar] [CrossRef]
- McSweeney, C.S.; Palmer, B.; McNeill, D.M.; Krause, D.O. Microbial interactions with tannins: Nutritional consequences for ruminants. Anim. Feed Sci. Technol. 2001, 91, 83–93. [Google Scholar] [CrossRef]
- Ou, K.; Gu, L. Absorption and metabolism of proanthocyanidins. J. Funct. Foods 2014, 7, 43–53. [Google Scholar] [CrossRef]
- Monagas, M.; Urpi-Sarda, M.; Sanchez-Patan, F.; Llorach, R.; Garrido, I.; Gomez-Cordoves, C.; Andres-Lacueva, C.; Bartolome, B. Insights into the metabolism and microbial biotransformation of dietary flavan-3-ols and the bioactivity of their metabolites. Food Funct. 2010, 1, 233–253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chai, W.; Ou-Yang, C.; Ma, Z.; Song, S.; Huang, Q.; Wei, Q.; Peng, Y. Anti-α-glucosidase and antityrosinase activity of condensed tannins from the bark of Clausena lansium (Lour.) Skeels with antiproliferative and apoptotic properties in B16 mouse melanoma cells. Process. Biochem. 2019, 86, 205–214. [Google Scholar] [CrossRef]
- Jiang, Y.; Zhang, H.; Qi, X.; Wu, G. Structural characterization and antioxidant activity of condensed tannins fractionated from sorghum grain. J. Cereal Sci. 2020, 92, 102918. [Google Scholar] [CrossRef]
- Shi, J.; Wang, Y.; Wei, H.; Hu, J.; Gao, M.-T. Structure analysis of condensed tannin from rice straw and its inhibitory effect on Staphylococcus aureus. Ind. Crop. Prod. 2020, 145, 112130. [Google Scholar] [CrossRef]
- Facchi, S.P.; de Oliveira, A.C.; Bezerra, E.O.T.; Vlcek, J.; Hedayati, M.; Reynolds, M.M.; Kipper, M.J.; Martins, A.F. Polycationic condensed tannin/polysaccharide-based polyelectrolyte multilayers prevent microbial adhesion and proliferation. Eur. Polym. J. 2020, 130, 109677. [Google Scholar] [CrossRef]
- Abu Zarin, M.; Wan, H.Y.; Isha, A.; Armania, N. Antioxidant, antimicrobial and cytotoxic potential of condensed tannins from Leucaena leucocephala hybrid-Rendang. Food Sci. Hum. Wellness 2016, 5, 65–75. [Google Scholar] [CrossRef] [Green Version]
- Serrano, J.; Puupponen-Pimia, R.; Dauer, A.; Aura, A.M.; Saura-Calixto, F. Tannins: Current knowledge of food sources, intake, bioavailability and biological effects. Mol. Nutr. Food Res. 2009, 53 (Suppl. 2), S310–S329. [Google Scholar] [CrossRef] [Green Version]
- Konishi, Y.; Hitomi, Y.; Yoshioka, E. Intestinal absorption of p-coumaric and gallic acids in rats after oral administration. J. Agric. Food Chem. 2004, 52, 2527–2532. [Google Scholar] [CrossRef] [PubMed]
- Shahrzad, S.; Aoyagi, K.; Winter, A.; Koyama, A.; Bitsch, I. Pharmacokinetics of gallic acid and its relative bioavailability from tea in healthy humans. J. Nutr. 2001, 131, 1207–1210. [Google Scholar] [CrossRef]
- Konishi, Y.; Zhao, Z.; Shimizu, M. Phenolic acids are absorbed from the rat stomach with different absorption rates. J. Agric. Food Chem. 2006, 54, 7539–7543. [Google Scholar] [CrossRef] [PubMed]
- Seeram, N.P.; Lee, R.; Heber, D. Bioavailability of ellagic acid in human plasma after consumption of ellagitannins from pomegranate (Punica granatum L.) juice. Clin. Chim. Acta 2004, 348, 63–68. [Google Scholar] [CrossRef] [Green Version]
- Lei, F.; Xing, D.M.; Xiang, L.; Zhao, Y.N.; Wang, W.; Zhang, L.J.; Du, L.J. Pharmacokinetic study of ellagic acid in rat after oral administration of pomegranate leaf extract. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2003, 796, 189–194. [Google Scholar] [CrossRef]
- Whitley, A.C.; Stoner, G.D.; Darby, M.V.; Walle, T. Intestinal epithelial cell accumulation of the cancer preventive polyphenol ellagic acid--extensive binding to protein and DNA. Biochem. Pharmacol. 2003, 66, 907–915. [Google Scholar] [CrossRef]
- Bell, J.R.; Donovan, J.L.; Wong, R.; Waterhouse, A.L.; German, J.B.; Walzem, R.L.; Kasim-Karakas, S.E. (+)-Catechin in human plasma after ingestion of a single serving of reconstituted red wine. Am. J. Clin. Nutr. 2000, 71, 103–108. [Google Scholar] [CrossRef]
- Ottaviani, J.I.; Momma, T.Y.; Heiss, C.; Kwik-Uribe, C.; Schroeter, H.; Keen, C.L. The stereochemical configuration of flavanols influences the level and metabolism of flavanols in humans and their biological activity in vivo. Free Radic. Biol. Med. 2011, 50, 237–244. [Google Scholar] [CrossRef]
- Requena, T.; Monagas, M.; Pozo-Bayón, M.A.; Martín-Álvarez, P.J.; Bartolomé, B.; del Campo, R.; Ávila, M.; Martínez-Cuesta, M.C.; Peláez, C.; Moreno-Arribas, M.V. Perspectives of the potential implications of wine polyphenols on human oral and gut microbiota. Trends Food Sci. Technol. 2010, 21, 332–344. [Google Scholar] [CrossRef]
- Ottaviani, J.I.; Momma, T.Y.; Kuhnle, G.K.; Keen, C.L.; Schroeter, H. Structurally related (−)-epicatechin metabolites in humans: Assessment using de novo chemically synthesized authentic standards. Free Radic. Biol. Med. 2012, 52, 1403–1412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Romanov-Michailidis, F.; Viton, F.; Fumeaux, R.; Lévèques, A.; Actis-Goretta, L.; Rein, M.; Williamson, G.; Barron, D. Epicatechin B-ring conjugates: First enantioselective synthesis and evidence for their occurrence in human biological fluids. Org. Lett. 2012, 14, 3902–3905. [Google Scholar] [CrossRef]
- Actis-Goretta, L.; Lévèques, A.; Giuffrida, F.; Romanov-Michailidis, F.; Viton, F.; Barron, D.; Duenas-Paton, M.; Gonzalez-Manzano, S.; Santos-Buelga, C.; Williamson, G.; et al. Elucidation of (−)-epicatechin metabolites after ingestion of chocolate by healthy humans. Free Radic. Biol. Med. 2012, 53, 787–795. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Meng, X.; Yang, C.S. Enzymology of methylation of tea catechins and inhibition of catechol-O-methyltransferase by (−)-epigallocatechin gallate. Drug Metab. Dispos. 2003, 31, 572–579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borges, G.; Ottaviani, J.I.; van der Hooft, J.J.J.; Schroeter, H.; Crozier, A. Absorption, metabolism, distribution and excretion of (−)-epicatechin: A review of recent findings. Mol. Asp. Med. 2018, 61, 18–30. [Google Scholar] [CrossRef] [PubMed]
- Soares, S.; Brandão, E.; Mateus, N.; de Freitas, V. Interaction between red wine procyanidins and salivary proteins: Effect of stomach digestion on the resulting complexes. RSC Adv. 2015, 5, 12664–12670. [Google Scholar] [CrossRef]
- Clifford, M.N. Diet-derived phenols in plasma and tissues and their implications for health. Planta Med. 2004, 70, 1103–1114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guinane, C.M.; Cotter, P.D. Role of the gut microbiota in health and chronic gastrointestinal disease: Understanding a hidden metabolic organ. Ther. Adv. Gastroenterol. 2013, 6, 295–308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caballero, S.; Pamer, E.G. Microbiota-mediated inflammation and antimicrobial defense in the intestine. Annu. Rev. Immunol. 2015, 33, 227–256. [Google Scholar] [CrossRef] [Green Version]
- Vaiserman, A.M.; Koliada, A.K.; Marotta, F. Gut microbiota: A player in aging and a target for anti-aging intervention. Ageing Res. Rev. 2017, 35, 36–45. [Google Scholar] [CrossRef]
- Hakansson, A.; Molin, G. Gut microbiota and inflammation. Nutrients 2011, 3, 637–682. [Google Scholar] [CrossRef] [PubMed]
- Lynch, S.V.; Pedersen, O. The Human Intestinal Microbiome in Health and Disease. N. Engl. J. Med. 2016, 375, 2369–2379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jandhyala, S.M.; Talukdar, R.; Subramanyam, C.; Vuyyuru, H.; Sasikala, M.; Reddy, D.N. Role of the normal gut microbiota. World J. Gastroenterol. 2015, 21, 8787. [Google Scholar] [CrossRef]
- Duggan, C.; Gannon, J.; Walker, W.A. Protective nutrients and functional foods for the gastrointestinal tract. Am. J. Clin. Nutr. 2002, 75, 789–808. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, M.; Zhang, X.; Zhu, J.; Cheng, L.; Cao, J.; Wu, Z.; Weng, P.; Zheng, X. A metagenomics approach to the intestinal microbiome structure and function in high fat diet-induced obesity mice fed with oolong tea polyphenols. Food Funct. 2018, 9, 1079–1087. [Google Scholar] [CrossRef]
- Turnbaugh, P.J.; Quince, C.; Faith, J.J.; McHardy, A.C.; Yatsunenko, T.; Niazi, F.; Affourtit, J.; Egholm, M.; Henrissat, B.; Knight, R.; et al. Organismal, genetic, and transcriptional variation in the deeply sequenced gut microbiomes of identical twins. Proc. Natl. Acad. Sci. USA 2010, 107, 7503–7508. [Google Scholar] [CrossRef] [Green Version]
- Qin, J.; Li, R.; Raes, J.; Arumugam, M.; Burgdorf, K.S.; Manichanh, C.; Nielsen, T.; Pons, N.; Levenez, F.; Yamada, T.; et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010, 464, 59–65. [Google Scholar] [CrossRef] [Green Version]
- Zhu, B.; Wang, X.; Li, L. Human gut microbiome: The second genome of human body. Protein Cell 2010, 1, 718–725. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giuffrè, M.; Campigotto, M.; Campisciano, G.; Comar, M.; Crocè, L.S. A story of liver and gut microbes: How does the intestinal flora affect liver disease? A review of the literature. Am. J. Physiol. Gastrointest. Liver Physiol. 2020, 318, G889–G906. [Google Scholar] [CrossRef] [PubMed]
- Flint, H.J. The impact of nutrition on the human microbiome. Nutr. Rev. 2012, 70, S10–S13. [Google Scholar] [CrossRef]
- Rizkallah, M.R.; Saad, R.; Aziz, R.K. The Human Microbiome Project, personalized medicine and the birth of pharmacomicrobiomics. Curr. Pharm. Pers. Med. 2010, 8, 182–193. [Google Scholar] [CrossRef] [Green Version]
- Aziz, R.K.; Hegazy, S.M.; Yasser, R.; Rizkallah, M.R.; ElRakaiby, M.T. Drug pharmacomicrobiomics and toxicomicrobiomics: From scattered reports to systematic studies of drug–microbiome interactions. Expert Opin. Drug Metab. Toxicol. 2018, 14, 1043–1055. [Google Scholar] [CrossRef]
- Sharma, A.; Buschmann, M.M.; Gilbert, J.A. Pharmacomicrobiomics: The Holy Grail to Variability in Drug Response? Clin. Pharmacol. Ther. 2019, 106, 317–328. [Google Scholar] [CrossRef]
- Giuffrè, M.; Moretti, R.; Campisciano, G.; Silveira, A.; Monda, V.; Comar, M.; Di Bella, S.; Antonello, R.; Luzzati, R.; Croce, S. You Talking to Me? Says the Enteric Nervous System (ENS) to the Microbe. How Intestinal Microbes Interact with the ENS. J. Clin. Med. 2020, 9, 3705. [Google Scholar] [CrossRef]
- Abdelsalam, N.A.; Ramadan, A.T.; ElRakaiby, M.T.; Aziz, R.K. Toxicomicrobiomics: The human microbiome vs. pharmaceutical, dietary, and environmental xenobiotics. Front. Pharmacol. 2020, 11, 390. [Google Scholar] [CrossRef]
- Rechner, A.R.; Smith, M.A.; Kuhnle, G.; Gibson, G.R.; Debnam, E.S.; Srai, S.K.; Moore, K.P.; Rice-Evans, C.A. Colonic metabolism of dietary polyphenols: Influence of structure on microbial fermentation products. Free Radic. Biol. Med. 2004, 36, 212–225. [Google Scholar] [CrossRef] [PubMed]
- Hamauzu, Y. Non-extractable polyphenols in fruit: Distribution, changes, and potential health effects. In Non-Extractable Polyphenols and Carotenoids; Royal Society of Chemistry: London, UK, 2018; pp. 284–306. [Google Scholar] [CrossRef]
- Bouayed, J.; Hoffmann, L.; Bohn, T. Total phenolics, flavonoids, anthocyanins and antioxidant activity following simulated gastro-intestinal digestion and dialysis of apple varieties: Bioaccessibility and potential uptake. Food Chem. 2011, 128, 14–21. [Google Scholar] [CrossRef]
- Walle, T.; Browning, A.M.; Steed, L.L.; Reed, S.G.; Walle, U.K. Flavonoid glucosides are hydrolyzed and thus activated in the oral cavity in humans. J. Nutr. 2005, 135, 48–52. [Google Scholar] [CrossRef] [Green Version]
- Daniel, E.M.; Ratnayake, S.; Kinstle, T.; Stoner, G.D. The effects of pH and rat intestinal contents on the liberation of ellagic acid from purified and crude ellagitannins. J. Nat. Prod. 1991, 54, 946–952. [Google Scholar] [CrossRef] [PubMed]
- Espín, J.C.; Larrosa, M.; García-Conesa, M.T.; Tomás-Barberán, F. Biological significance of urolithins, the gut microbial ellagic acid-derived metabolites: The evidence so far. Evid. Based Complement. Altern. Med. 2013, 2013, 270418. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gaya, P.; Peirotén, Á.; Medina, M.; Álvarez, I.; Landete, J.M. Bifidobacterium pseudocatenulatum INIA P815: The first bacterium able to produce urolithins A and B from ellagic acid. J. Funct. Foods 2018, 45, 95–99. [Google Scholar] [CrossRef]
- Selma, M.V.; Beltran, D.; Garcia-Villalba, R.; Espin, J.C.; Tomas-Barberan, F.A. Description of urolithin production capacity from ellagic acid of two human intestinal Gordonibacter species. Food Funct. 2014, 5, 1779–1784. [Google Scholar] [CrossRef] [Green Version]
- Romo-Vaquero, M.; García-Villalba, R.; González-Sarrías, A.; Beltrán, D.; Tomás-Barberán, F.A.; Espín, J.C.; Selma, M.V. Interindividual variability in the human metabolism of ellagic acid: Contribution of Gordonibacter to urolithin production. J. Funct. Foods 2015, 17, 785–791. [Google Scholar] [CrossRef]
- Selma, M.V.; Tomas-Barberan, F.A.; Beltran, D.; Garcia-Villalba, R.; Espin, J.C. Gordonibacter urolithinfaciens sp. nov., a urolithin-producing bacterium isolated from the human gut. Int. J. Syst. Evol. Microbiol. 2014, 64, 2346–2352. [Google Scholar] [CrossRef]
- Martinez-Blanch, J.F.; Ramon, D.; Beltran, D.; Romo-Vaquero, M.; Garcia-Villalba, R.; Espin, J.C.; Tomas-Barberan, F.A.; Codoner, F.M.; Selma, M.V. Complete genome sequence of the new urolithin-producing bacterium Gordonibacter urolithinfaciens DSM 27213(T). Genome Announc. 2017, 5. [Google Scholar] [CrossRef] [Green Version]
- Ludwig, I.A.; Mena, P.; Calani, L.; Borges, G.; Pereira-Caro, G.; Bresciani, L.; Del Rio, D.; Lean, M.E.; Crozier, A. New insights into the bioavailability of red raspberry anthocyanins and ellagitannins. Free Radic. Biol. Med. 2015, 89, 758–769. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tulipani, S.; Urpi-Sarda, M.; Garcia-Villalba, R.; Rabassa, M.; Lopez-Uriarte, P.; Bullo, M.; Jauregui, O.; Tomas-Barberan, F.; Salas-Salvado, J.; Espin, J.C.; et al. Urolithins are the main urinary microbial-derived phenolic metabolites discriminating a moderate consumption of nuts in free-living subjects with diagnosed metabolic syndrome. J. Agric. Food Chem. 2012, 60, 8930–8940. [Google Scholar] [CrossRef] [PubMed]
- Mertens-Talcott, S.U.; Jilma-Stohlawetz, P.; Rios, J.; Hingorani, L.; Derendorf, H. Absorption, metabolism, and antioxidant effects of pomegranate (Punica granatum l.) polyphenols after ingestion of a standardized extract in healthy human volunteers. J. Agric. Food Chem. 2006, 54, 8956–8961. [Google Scholar] [CrossRef]
- Seeram, N.P.; Aronson, W.J.; Zhang, Y.; Henning, S.M.; Moro, A.; Lee, R.P.; Sartippour, M.; Harris, D.M.; Rettig, M.; Suchard, M.A.; et al. Pomegranate ellagitannin-derived metabolites inhibit prostate cancer growth and localize to the mouse prostate gland. J. Agric. Food Chem. 2007, 55, 7732–7737. [Google Scholar] [CrossRef] [PubMed]
- Spencer, J.P.E. Metabolism of Tea Flavonoids in the Gastrointestinal Tract. J. Nutr. 2003, 133, 3255S–3261S. [Google Scholar] [CrossRef] [Green Version]
- Urpi-Sarda, M.; Monagas, M.; Khan, N.; Llorach, R.; Lamuela-Raventos, R.M.; Jauregui, O.; Estruch, R.; Izquierdo-Pulido, M.; Andres-Lacueva, C. Targeted metabolic profiling of phenolics in urine and plasma after regular consumption of cocoa by liquid chromatography-tandem mass spectrometry. J. Chromatogr. A 2009, 1216, 7258–7267. [Google Scholar] [CrossRef]
- Appeldoorn, M.M.; Vincken, J.P.; Aura, A.M.; Hollman, P.C.; Gruppen, H. Procyanidin dimers are metabolized by human microbiota with 2-(3,4-dihydroxyphenyl)acetic acid and 5-(3,4-dihydroxyphenyl)-gamma-valerolactone as the major metabolites. J. Agric. Food Chem. 2009, 57, 1084–1092. [Google Scholar] [CrossRef]
- Takagaki, A.; Otani, S.; Nanjo, F. Antioxidative Activity of Microbial Metabolites of (−)-Epigallocatechin Gallate Produced in Rat Intestines. Biosci. Biotechnol. Biochem. 2011, 75, 582–585. [Google Scholar] [CrossRef]
- Gonthier, M.P.; Cheynier, V.; Donovan, J.L.; Manach, C.; Morand, C.; Mila, I.; Lapierre, C.; Remesy, C.; Scalbert, A. Microbial aromatic acid metabolites formed in the gut account for a major fraction of the polyphenols excreted in urine of rats fed red wine polyphenols. J. Nutr. 2003, 133, 461–467. [Google Scholar] [CrossRef] [Green Version]
- Rios, L.Y.; Gonthier, M.-P.; Rémésy, C.; Mila, I.; Lapierre, C.; Lazarus, S.A.; Williamson, G.; Scalbert, A. Chocolate intake increases urinary excretion of polyphenol-derived phenolic acids in healthy human subjects. Am. J. Clin. Nutr. 2003, 77, 912–918. [Google Scholar] [CrossRef] [Green Version]
- Sang, S.; Lee, M.-J.; Yang, I.; Buckley, B.; Yang, C.S. Human urinary metabolite profile of tea polyphenols analyzed by liquid chromatography/electrospray ionization tandem mass spectrometry with data-dependent acquisition. Rapid Commun. Mass Spectrom. 2008, 22, 1567–1578. [Google Scholar] [CrossRef]
- Unno, T.; Tamemoto, K.; Yayabe, F.; Kakuda, T. Urinary excretion of 5-(3′,4′-dihydroxyphenyl)-gamma-valerolactone, a ring-fission metabolite of (−)-epicatechin, in rats and its in vitro antioxidant activity. J. Agric. Food Chem. 2003, 51, 6893–6898. [Google Scholar] [CrossRef]
- Margalef, M.; Pons, Z.; Bravo, F.I.; Muguerza, B.; Arola-Arnal, A. Tissue distribution of rat flavanol metabolites at different doses. J. Nutr. Biochem. 2015, 26, 987–995. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Hu, Z.; Yin, Z.; Zhou, Y.; Liu, T.; Zhou, X.; Chang, D. Profiling and Distribution of Metabolites of Procyanidin B2 in Mice by UPLC-DAD-ESI-IT-TOF-MS(n) Technique. Front. Pharm. 2017, 8, 231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, H.; Garrett, T.J.; Tayyari, F.; Gu, L. Profiling the metabolome changes caused by cranberry procyanidins in plasma of female rats using (1) H NMR and UHPLC-Q-Orbitrap-HRMS global metabolomics approaches. Mol. Nutr. Food Res. 2015, 59, 2107–2118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borges, G.; van der Hooft, J.J.J.; Crozier, A. A comprehensive evaluation of the [2-14C](–)-epicatechin metabolome in rats. Free Radic. Biol. Med. 2016, 99, 128–138. [Google Scholar] [CrossRef] [PubMed]
- Saito, A. Challenges and complexity of functionality evaluation of flavan-3-ol derivatives. Biosci. Biotechnol. Biochem. 2017, 81, 1055–1060. [Google Scholar] [CrossRef] [Green Version]
- Mena, P.; Ludwig, I.A.; Tomatis, V.B.; Acharjee, A.; Calani, L.; Rosi, A.; Brighenti, F.; Ray, S.; Griffin, J.L.; Bluck, L.J.; et al. Inter-individual variability in the production of flavan-3-ol colonic metabolites: Preliminary elucidation of urinary metabotypes. Eur. J. Nutr. 2019, 58, 1529–1543. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Donovan, J.L.; Manach, C.; Faulks, R.M.; Kroon, P.A. Absorption and metabolism of dietary plant secondary metabolites. In Plant Secondary Metabolites: Occurrence, Structure and Role in the Human Diet; Blackwell Publishing Ltd.: Oxford, UK, 2006. [Google Scholar] [CrossRef]
- Zheng, C.-J.; Liu, R.; Xue, B.; Luo, J.; Gao, L.; Wang, Y.; Ou, S.; Li, S.; Peng, X. Impact and consequences of polyphenols and fructooligosaccharide interplay on gut microbiota in rats. Food Funct. 2017, 8, 1925–1932. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.S.; Hattori, M. Isolation and characterization of a human intestinal bacterium Eggerthella sp. CAT-1 capable of cleaving the C-ring of (+)-catechin and (-)-epicatechin, followed by p-dehydroxylation of the B-ring. Biol. Pharm. Bull. 2012, 35, 2252–2256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kutschera, M.; Engst, W.; Blaut, M.; Braune, A. Isolation of catechin-converting human intestinal bacteria. J. Appl. Microbiol. 2011, 111, 165–175. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Dhakan, D.B.; Maji, A.; Saxena, R.; PK, V.P.; Mahajan, S.; Pulikkan, J.; Kurian, J.; Gomez, A.M.; Scaria, J.; et al. Association of Flavonifractor plautii, a flavonoid-degrading bacterium, with the gut microbiome of colorectal cancer patients in India. mSystems 2019, 4. [Google Scholar] [CrossRef] [Green Version]
- Marín, L.; Miguélez, E.M.; Villar, C.J.; Lombó, F. Bioavailability of dietary polyphenols and gut microbiota metabolism: Antimicrobial properties. Biomed. Res. Int. 2015, 2015, 905215. [Google Scholar] [CrossRef] [Green Version]
- Braune, A.; Blaut, M. Bacterial species involved in the conversion of dietary flavonoids in the human gut. Gut Microbes 2016, 7, 216–234. [Google Scholar] [CrossRef] [Green Version]
- Braune, A.; Engst, W.; Blaut, M. Identification and functional expression of genes encoding flavonoid O-and C-glycosidases in intestinal bacteria. Environ. Microbiol. 2016, 18, 2117–2129. [Google Scholar] [CrossRef]
- Mena, P.; Bresciani, L.; Brindani, N.; Ludwig, I.A.; Pereira-Caro, G.; Angelino, D.; Llorach, R.; Calani, L.; Brighenti, F.; Clifford, M.N.; et al. Phenyl-γ-valerolactones and phenylvaleric acids, the main colonic metabolites of flavan-3-ols: Synthesis, analysis, bioavailability, and bioactivity. Nat. Prod. Rep. 2019. [Google Scholar] [CrossRef]
- Wiese, S.; Esatbeyoglu, T.; Winterhalter, P.; Kruse, H.-P.; Winkler, S.; Bub, A.; Kulling, S.E. Comparative biokinetics and metabolism of pure monomeric, dimeric, and polymeric flavan-3-ols: A randomized cross-over study in humans. Mol. Nutr. Food Res. 2015, 59, 610–621. [Google Scholar] [CrossRef]
- Ou, K.; Sarnoski, P.; Schneider, K.R.; Song, K.; Khoo, C.; Gu, L. Microbial catabolism of procyanidins by human gut microbiota. Mol. Nutr. Food Res. 2014, 58, 2196–2205. [Google Scholar] [CrossRef]
- Stoupi, S.; Williamson, G.; Drynan, J.W.; Barron, D.; Clifford, M.N. A comparison of the in vitro biotransformation of (-)-epicatechin and procyanidin B2 by human faecal microbiota. Mol. Nutr. Food Res. 2010, 54, 747–759. [Google Scholar] [CrossRef]
- Trost, K.; Ulaszewska, M.M.; Stanstrup, J.; Albanese, D.; De Filippo, C.; Tuohy, K.M.; Natella, F.; Scaccini, C.; Mattivi, F. Host: Microbiome co-metabolic processing of dietary polyphenols—An acute, single blinded, cross-over study with different doses of apple polyphenols in healthy subjects. Food Res. Int. 2018, 112, 108–128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; Gao, F.; Ji, B.; Wang, R.; Yang, J.; Liu, H.; Zhou, F. Anthocyanins-rich extract of wild Chinese blueberry protects glucolipotoxicity-induced INS832/13 beta-cell against dysfunction and death. J. Food Sci. Technol. 2015, 52, 3022–3029. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodriguez-Mateos, A.; Feliciano, R.P.; Boeres, A.; Weber, T.; Dos Santos, C.N.; Ventura, M.R.; Heiss, C. Cranberry (poly)phenol metabolites correlate with improvements in vascular function: A double-blind, randomized, controlled, dose-response, crossover study. Mol. Nutr. Food Res. 2016, 60, 2130–2140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Selma, M.V.; Gonzalez-Sarrias, A.; Salas-Salvado, J.; Andres-Lacueva, C.; Alasalvar, C.; Orem, A.; Tomas-Barberan, F.A.; Espin, J.C. The gut microbiota metabolism of pomegranate or walnut ellagitannins yields two urolithin-metabotypes that correlate with cardiometabolic risk biomarkers: Comparison between normoweight, overweight-obesity and metabolic syndrome. Clin. Nutr. 2018, 37, 897–905. [Google Scholar] [CrossRef] [PubMed]
- Bolca, S.; Wyns, C.; Possemiers, S.; Depypere, H.; De Keukeleire, D.; Bracke, M.; Verstraete, W.; Heyerick, A. Cosupplementation of isoflavones, prenylflavonoids, and lignans alters human exposure to phytoestrogen-derived 17 β-estradiol equivalents. J. Nutr. 2009, 139, 2293–2300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gross, G.; Jacobs, D.M.; Peters, S.; Possemiers, S.; van Duynhoven, J.; Vaughan, E.E.; Van de Wiele, T. In vitro bioconversion of polyphenols from black tea and red wine/grape juice by human intestinal microbiota displays strong interindividual variability. J. Agric. Food Chem. 2010, 58, 10236–10246. [Google Scholar] [CrossRef] [Green Version]
- Tzounis, X.; Rodriguez-Mateos, A.; Vulevic, J.; Gibson, G.R.; Kwik-Uribe, C.; Spencer, J.P. Prebiotic evaluation of cocoa-derived flavanols in healthy humans by using a randomized, controlled, double-blind, crossover intervention study. Am. J. Clin. Nutr. 2011, 93, 62–72. [Google Scholar] [CrossRef] [Green Version]
- James, L.P. Metabolomics: Integration of a new “omics” with clinical pharmacology. Clin. Pharm. Ther. 2013, 94, 547–551. [Google Scholar] [CrossRef]
- Kaddurah-Daouk, R.; Weinshilboum, R.M.; Pharmacometabolomics Research, N. Pharmacometabolomics: Implications for clinical pharmacology and systems pharmacology. Clin. Pharm. Ther. 2014, 95, 154–167. [Google Scholar] [CrossRef]
- Tomás-Barberán, F.A.; Selma, M.V.; Espín, J.C. Interactions of gut microbiota with dietary polyphenols and consequences to human health. Curr. Opin. Clin. Nutr. Metab. Care 2016, 19, 471–476. [Google Scholar] [CrossRef]
- Takagaki, A.; Nanjo, F. Bioconversion of (−)-epicatechin, (+)-epicatechin, (−)-catechin, and (+)-catechin by (−)-epigallocatechin-metabolizing bacteria. Biol. Pharm. Bull. 2015, 38, 789–794. [Google Scholar] [CrossRef] [Green Version]
- Tomas-Barberan, F.A.; Garcia-Villalba, R.; Gonzalez-Sarrias, A.; Selma, M.V.; Espin, J.C. Ellagic acid metabolism by human gut microbiota: Consistent observation of three urolithin phenotypes in intervention trials, independent of food source, age, and health status. J. Agric. Food Chem. 2014, 62, 6535–6538. [Google Scholar] [CrossRef] [PubMed]
- Tomas-Barberan, F.A.; Gonzalez-Sarrias, A.; Garcia-Villalba, R.; Nunez-Sanchez, M.A.; Selma, M.V.; Garcia-Conesa, M.T.; Espin, J.C. Urolithins, the rescue of “old” metabolites to understand a “new” concept: Metabotypes as a nexus among phenolic metabolism, microbiota dysbiosis, and host health status. Mol. Nutr. Food Res. 2017, 61. [Google Scholar] [CrossRef] [PubMed]
- Selma, M.V.; Romo-Vaquero, M.; García-Villalba, R.; González-Sarrías, A.; Tomás-Barberán, F.A.; Espín, J.C. The human gut microbial ecology associated with overweight and obesity determines ellagic acid metabolism. Food Funct. 2016, 7, 1769–1774. [Google Scholar] [CrossRef] [PubMed]
- Mora-Cubillos, X.; Tulipani, S.; Garcia-Aloy, M.; Bulló, M.; Tinahones, F.J.; Andres-Lacueva, C. Plasma metabolomic biomarkers of mixed nuts exposure inversely correlate with severity of metabolic syndrome. Mol. Nutr. Food Res. 2015, 59, 2480–2490. [Google Scholar] [CrossRef] [Green Version]
- Gonzalez-Sarrias, A.; Garcia-Villalba, R.; Romo-Vaquero, M.; Alasalvar, C.; Orem, A.; Zafrilla, P.; Tomas-Barberan, F.A.; Selma, M.V.; Espin, J.C. Clustering according to urolithin metabotype explains the interindividual variability in the improvement of cardiovascular risk biomarkers in overweight-obese individuals consuming pomegranate: A randomized clinical trial. Mol. Nutr. Food Res. 2017, 61. [Google Scholar] [CrossRef] [PubMed]
- Brenner, D.; Gescher, A. Cancer chemoprevention: Lessons learned and future directions. Br. J. Cancer 2005, 93, 735. [Google Scholar] [CrossRef] [Green Version]
- Neuwirthová, J.; Gál, B.; Smilek, P.; Urbánková, P. Coffee in Cancer Chemoprevention. Klin. Onkol. Cas. Ceske A Slov. Onkol. Spol. 2017, 30, 106–114. [Google Scholar] [CrossRef]
- Surh, Y.-j. M11-03: Natural agents for chemoprevention. J. Thorac. Oncol. 2007, 2, S184–S185. [Google Scholar] [CrossRef] [Green Version]
- Steward, W.P.; Brown, K. Cancer chemoprevention: A rapidly evolving field. Br. J. Cancer 2013, 109, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y. Cancer Chemopreventive Potential of Procyanidin. Toxicol. Res. 2017, 33, 273–282. [Google Scholar] [CrossRef] [PubMed]
- Lambert, J.D.; Hong, J.; Yang, G.-y.; Liao, J.; Yang, C.S. Inhibition of carcinogenesis by polyphenols: Evidence from laboratory investigations. Am. J. Clin. Nutr. 2005, 81, 284S–291S. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hara-Terawaki, A.; Takagaki, A.; Kobayashi, H.; Nanjo, F. Inhibitory activity of catechin metabolites produced by intestinal microbiota on proliferation of HeLa cells. Biol. Pharm. Bull. 2017, 40, 1331–1335. [Google Scholar] [CrossRef] [Green Version]
- Kim, Y.H.; Won, Y.-S.; Yang, X.; Kumazoe, M.; Yamashita, S.; Hara, A.; Takagaki, A.; Goto, K.; Nanjo, F.; Tachibana, H. Green tea catechin metabolites exert immunoregulatory effects on CD4+ T cell and natural killer cell activities. J. Agric. Food Chem. 2016, 64, 3591–3597. [Google Scholar] [CrossRef]
- Grimm, T.; Schäfer, A.; Högger, P. Antioxidant activity and inhibition of matrix metalloproteinases by metabolites of maritime pine bark extract (pycnogenol). Free Radic. Biol. Med. 2004, 36, 811–822. [Google Scholar] [CrossRef]
- Uhlenhut, K.; Högger, P. Facilitated cellular uptake and suppression of inducible nitric oxide synthase by a metabolite of maritime pine bark extract (Pycnogenol). Free Radic. Biol. Med. 2012, 53, 305–313. [Google Scholar] [CrossRef]
- Adnan, S.; Nelson, J.W.; Ajami, N.J.; Venna, V.R.; Petrosino, J.F.; Bryan Jr, R.M.; Durgan, D.J. Alterations in the gut microbiota can elicit hypertension in rats. Physiol. Genom. 2016, 49, 96–104. [Google Scholar] [CrossRef] [Green Version]
- Van Rymenant, E.; Grootaert, C.; Beerens, K.; Needs, P.; Kroon, P.; Kerimi, A.; Williamson, G.; García-Villalba, R.; González-Sarrías, A.; Tomas-Barberan, F. Vasorelaxant activity of twenty-one physiologically relevant (poly) phenolic metabolites on isolated mouse arteries. Food Funct. 2017, 8, 4331–4335. [Google Scholar] [CrossRef]
- Lee, C.C.; Kim, J.H.; Kim, J.S.; Oh, Y.S.; Han, S.M.; Park, J.H.Y.; Lee, K.W.; Lee, C.Y. 5-(3′,4′-Dihydroxyphenyl-γ-valerolactone), a major microbial metabolite of proanthocyanidin, attenuates THP-1 monocyte-endothelial adhesion. Int. J. Mol. Sci. 2017, 18, 1363. [Google Scholar] [CrossRef] [Green Version]
- Mele, L.; Carobbio, S.; Brindani, N.; Curti, C.; Rodriguez-Cuenca, S.; Bidault, G.; Mena, P.; Zanotti, I.; Vacca, M.; Vidal-Puig, A. Phenyl-γ-valerolactones, flavan-3-ol colonic metabolites, protect brown adipocytes from oxidative stress without affecting their differentiation or function. Mol. Nutr. Food Res. 2017, 61, 1700074. [Google Scholar] [CrossRef] [PubMed]
- Peron, G.; Sut, S.; Pellizzaro, A.; Brun, P.; Voinovich, D.; Castagliuolo, I.; Dall’Acqua, S. The antiadhesive activity of cranberry phytocomplex studied by metabolomics: Intestinal PAC-A metabolites but not intact PAC-A are identified as markers in active urines against uropathogenic Escherichia coli. Fitoterapia 2017, 122, 67–75. [Google Scholar] [CrossRef] [PubMed]
- Unno, K.; Pervin, M.; Nakagawa, A.; Iguchi, K.; Hara, A.; Takagaki, A.; Nanjo, F.; Minami, A.; Nakamura, Y. Blood-brain barrier permeability of green tea catechin metabolites and their neuritogenic activity in human neuroblastoma SH-SY5Y Cells. Mol. Nutr. Food Res. 2017, 61, 1700294. [Google Scholar] [CrossRef] [PubMed]
- Fernando, W.; Somaratne, G.; Goozee, K.G.; Williams, S.; Singh, H.; Martins, R.N. Diabetes and Alzheimer’s disease: Can tea phytochemicals play a role in prevention? J. Alzheimer Dis. 2017, 59, 481–501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, T.; Ma, H.; Liu, W.; Niesen, D.B.; Shah, N.; Crews, R.; Rose, K.N.; Vattem, D.A.; Seeram, N.P. Pomegranate’s neuroprotective effects against Alzheimer’s disease are mediated by urolithins, its ellagitannin-gut microbial derived metabolites. ACS Chem. Neurosci. 2016, 7, 26–33. [Google Scholar] [CrossRef]
- Nakajima, A.; Ueda, Y.; Matsuda, E.; Sameshima, H.; Ikenoue, T. Enhancement of in vivo antioxidant ability in the brain of rats fed tannin. Neurochem. Res. 2013, 38, 1360–1364. [Google Scholar] [CrossRef]
- Ide, K.; Matsuoka, N.; Yamada, H.; Furushima, D.; Kawakami, K. Effects of tea catechins on Alzheimer’s disease: Recent updates and perspectives. Molecules 2018, 23, 2357. [Google Scholar] [CrossRef] [Green Version]
- Boakye, Y.D.; Groyer, L.; Heiss, E.H. An increased autophagic flux contributes to the anti-inflammatory potential of urolithin A in macrophages. Biochim. Biophys. Acta Gen. Subj. 2018, 1862, 61–70. [Google Scholar] [CrossRef]
- Komatsu, W.; Kishi, H.; Yagasaki, K.; Ohhira, S. Urolithin A attenuates pro-inflammatory mediator production by suppressing PI3-K/Akt/NF-kappaB and JNK/AP-1 signaling pathways in lipopolysaccharide-stimulated RAW264 macrophages: Possible involvement of NADPH oxidase-derived reactive oxygen species. Eur. J. Pharmacol. 2018, 833, 411–424. [Google Scholar] [CrossRef]
- Lee, G.; Park, J.S.; Lee, E.J.; Ahn, J.H.; Kim, H.S. Anti-inflammatory and antioxidant mechanisms of urolithin B in activated microglia. Phytomedicine 2019, 55, 50–57. [Google Scholar] [CrossRef]
- Núñez-Sánchez, M.Á.; Karmokar, A.; González-Sarrías, A.; García-Villalba, R.; Tomás-Barberán, F.A.; García-Conesa, M.T.; Brown, K.; Espín, J.C. In vivo relevant mixed urolithins and ellagic acid inhibit phenotypic and molecular colon cancer stem cell features: A new potentiality for ellagitannin metabolites against cancer. Food Chem. Toxicol. 2016, 92, 8–16. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Wang, J.; Zheng, G.; Qiu, Z. Methylated urolithin A, the modified ellagitannin-derived metabolite, suppresses cell viability of DU145 human prostate cancer cells via targeting miR-21. Food Chem. Toxicol. 2016, 97, 375–384. [Google Scholar] [CrossRef] [PubMed]
- Cui, G.H.; Chen, W.Q.; Shen, Z.Y. Urolithin A shows anti-atherosclerotic activity via activation of class B scavenger receptor and activation of Nef2 signaling pathway. Pharm. Rep. 2018, 70, 519–524. [Google Scholar] [CrossRef] [PubMed]
- Tang, L.; Mo, Y.; Li, Y.; Zhong, Y.; He, S.; Zhang, Y.; Tang, Y.; Fu, S.; Wang, X.; Chen, A. Urolithin A alleviates myocardial ischemia/reperfusion injury via PI3K/Akt pathway. Biochem. Biophys. Res. Commun. 2017, 486, 774–780. [Google Scholar] [CrossRef] [PubMed]
- Ramsundar, B.; Eastman, P.; Walters, P.; Pande, V. Deep Learning for the Life Sciences: Applying Deep Learning to Genomics, Microscopy, Drug Discovery, and More; O’Reilly Media: Sebastopol, CA, USA, 2019. [Google Scholar]
- Ribeiro, M.; Singh, S.; Guestrin, C. “Why Should I Trust You?”: Explaining the Predictions of Any Classifier. In Proceedings of the 2016 Conference of the North American Chapter of the Association for Computational Linguistics: Demonstrations, San Diego, CA, USA, June 2016; DeNero, J., Finlayson, M., Reddy, S., Eds.; Association for Computational Linguistics: Stroudsburg, PA, USA, 2016; pp. 1135–1144. [Google Scholar] [CrossRef]
- Harrington, R.A.; Adhikari, V.; Rayner, M.; Scarborough, P. Nutrient composition databases in the age of big data: foodDB, a comprehensive, real-time database infrastructure. BMJ Open 2019, 9, e026652. [Google Scholar] [CrossRef]
- Micah, H.; Claire, F.-L.; Rob, K. The Human Microbiome Project: Exploring the microbial part of ourselves in a changing world. Nature 2007, 449, 804–810. [Google Scholar]
- The Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 2012, 486, 207–214. [Google Scholar] [CrossRef] [Green Version]
- Huson, D.H.; Auch, A.F.; Qi, J.; Schuster, S.C. MEGAN analysis of metagenomic data. Genome Res. 2007, 17, 377–386. [Google Scholar] [CrossRef] [Green Version]
- Keegan, K.P.; Glass, E.M.; Meyer, F. MG-RAST, a metagenomics service for analysis of microbial community structure and function. In Microbial Environmental Genomics (MEG); Humana Press, Springer Science + Business Media: New York, NY, USA, 2016; pp. 207–233. [Google Scholar]
- Davis, J.J.; Wattam, A.R.; Aziz, R.K.; Brettin, T.; Butler, R.; Butler, R.M.; Chlenski, P.; Conrad, N.; Dickerman, A.; Dietrich, E.M.; et al. The PATRIC Bioinformatics Resource Center: Expanding data and analysis capabilities. Nucleic Acids Res. 2020, 48, D606–D612. [Google Scholar] [CrossRef] [Green Version]

| Plant Name | Type of Tannins | Reference | |
|---|---|---|---|
| Hydrolysable Tannins | |||
| Pomegranate (Punica granatum) | Punicalagin | Ellagitannins | [10] |
| Casuarictin | |||
| Pedunculagin | |||
| Strawberry (Fragaria ananassa) | Sanguiin | [11] | |
| Oak (Quercus sp.) and Chestnut (Castanea sp.) | Vescalagin | [12,13] | |
| Castalagin | |||
| Sumac (Rhus semialata) | Pentagalloyl-glucoside | Gallotannins | [13,14] |
| Hexagalloyl-glucoside | |||
| Heptagalloyl-glucoside | |||
| Octagalloyl-glucoside | |||
| Nonagalloyl-glucoside | |||
| Decagalloyl-glucoside | |||
| Condensed Tannins | |||
| Tea (Camellia sinensis) | (-)-epicatechin (+)-catechin (-)-epigallocatechin gallate | Monomers | [15] |
| Cocoa (Theobroma cacao) | |||
| Apple (Malus pumila) | |||
| Grapes (Vitis vinifera) | |||
| Berries (Vaccinium sp.) | |||
| Peanut (Arachis hypogaea) | |||
| Persimmon (Diospyros lotus) | |||
| Plums (Prunus sp.) | Proanthocyanidin B-Type | Dimers | [16] |
| Avocado (Persea americana) | Proanthocyanidin A-type | ||
| Cinnamon (Cinnamomum sp.) | Procyanidin C1, C2 | Trimers | |
| Arecatanin A2 Cinnamtannin A2 | Tetramers | ||
| Biological Effect | Substructure | Metabolite |
|---|---|---|
| Anti-inflammatory | coc(=O)c(c)c | Ellagic acid |
| cC(=O)O | Gallic acid | |
| cc(=O)oc(c)c | Urolithin B | |
| ccc(cc)C(=O)O | Vanillic acid | |
| cc(=O)oc(c)c | Urolithin A | |
| Antioxident | coc(=O)c(c)c | Ellagic acid |
| cc(O)c(CC)c(c)O | Gallocatechin | |
| ccc(cc)C(=O)O | Gallic acid | |
| ccc | Vanillic acid | |
| Anticancer | cc(O)cc(c)c | Ellagic acid |
| cc(C)cc(c)O | 5-(3′,5′-dihydroxyphenyl)-gamma-valerolactone | |
| cc(C)cc(c)O | 5-(3′,4′,5′-trihydroxyphenyl)-gamma-valerolactone | |
| ccc(cc)C(=O)O | Gallic acid | |
| cc(C)cc(c)O | 4-Hydroxy-5-(3,4,5-trihydroxyphenyl)valeric ac... | |
| cc(=O)oc(c)c | Urolithin C | |
| cccc(c)C | Vanillic acid | |
| cc(=O)oc(c)c | Urolithin A | |
| Anti-atherosclerotic | cc(O)cc(c)c | Urolithin A |
| CCCC(=O)O | 5-(3′,4′-Dihydroxyphenyl)-gamma-valerolactone |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sallam, I.E.; Abdelwareth, A.; Attia, H.; Aziz, R.K.; Homsi, M.N.; von Bergen, M.; Farag, M.A. Effect of Gut Microbiota Biotransformation on Dietary Tannins and Human Health Implications. Microorganisms 2021, 9, 965. https://doi.org/10.3390/microorganisms9050965
Sallam IE, Abdelwareth A, Attia H, Aziz RK, Homsi MN, von Bergen M, Farag MA. Effect of Gut Microbiota Biotransformation on Dietary Tannins and Human Health Implications. Microorganisms. 2021; 9(5):965. https://doi.org/10.3390/microorganisms9050965
Chicago/Turabian StyleSallam, Ibrahim E., Amr Abdelwareth, Heba Attia, Ramy K. Aziz, Masun Nabhan Homsi, Martin von Bergen, and Mohamed A. Farag. 2021. "Effect of Gut Microbiota Biotransformation on Dietary Tannins and Human Health Implications" Microorganisms 9, no. 5: 965. https://doi.org/10.3390/microorganisms9050965
APA StyleSallam, I. E., Abdelwareth, A., Attia, H., Aziz, R. K., Homsi, M. N., von Bergen, M., & Farag, M. A. (2021). Effect of Gut Microbiota Biotransformation on Dietary Tannins and Human Health Implications. Microorganisms, 9(5), 965. https://doi.org/10.3390/microorganisms9050965










