The Microbiome of the ‘Williams’ Pear Variety Grown in the Organic Orchard and Antifungal Activity by the Autochthonous Bacterial and Yeast Isolates
Abstract
:1. Introduction
2. Material and Methods
2.1. Plant Material and Culturable Microbiota
2.2. Identification of Bacterial, Yeast, and Fungal Isolates
2.3. Antifungal Activity of Autochthonous Bacterial and Yeast Isolates
2.4. Amplicon Sequencing of Unculturable Microbiota
2.5. Reprocessing, Sequence Inference, Taxonomy Annotation, and Data Availability
3. Results
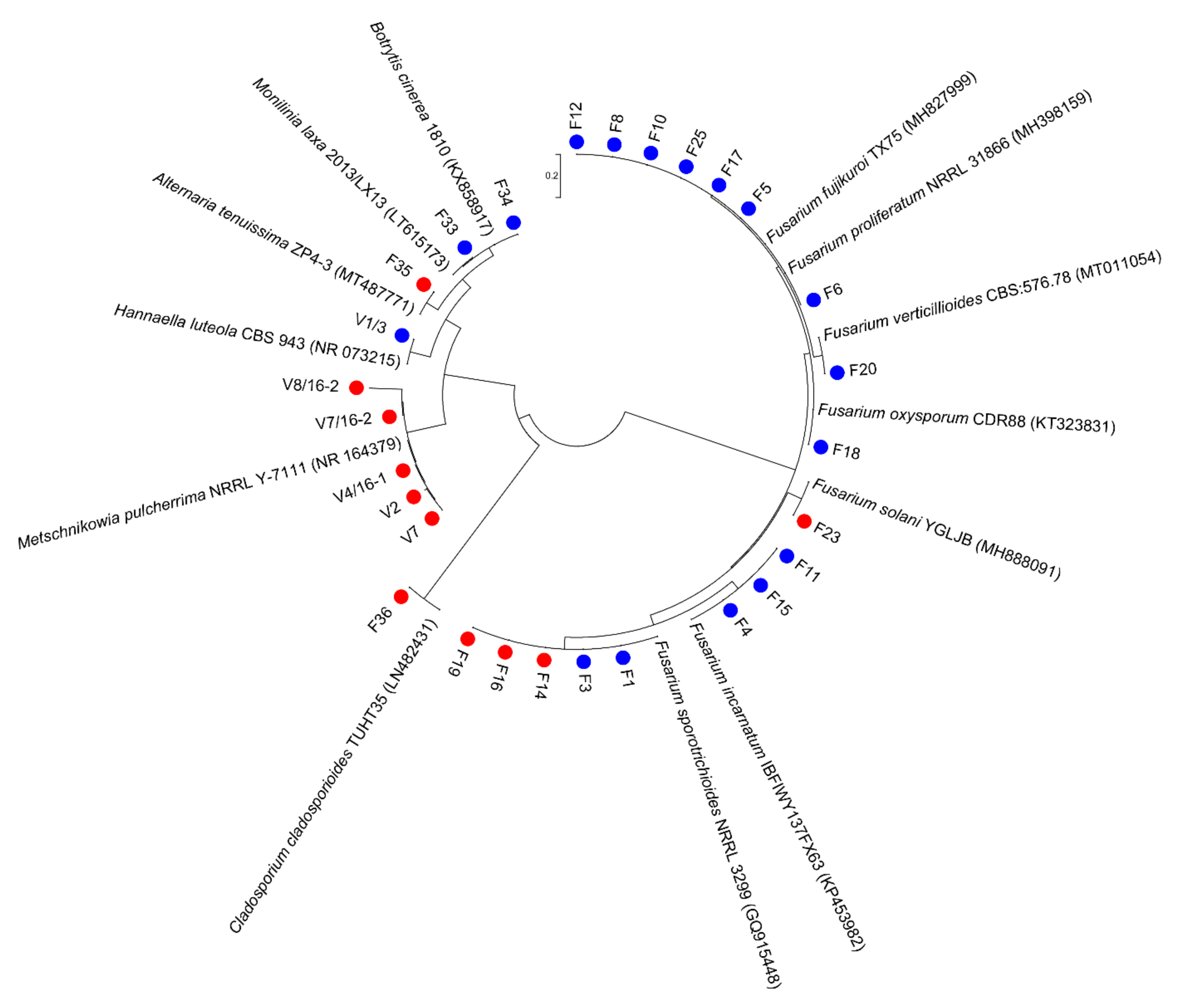
3.1. Culturable Microbiota
3.2. Antifungal Activity
3.3. Metabarcoding Data and Alpha Diversity of Microbial Communities
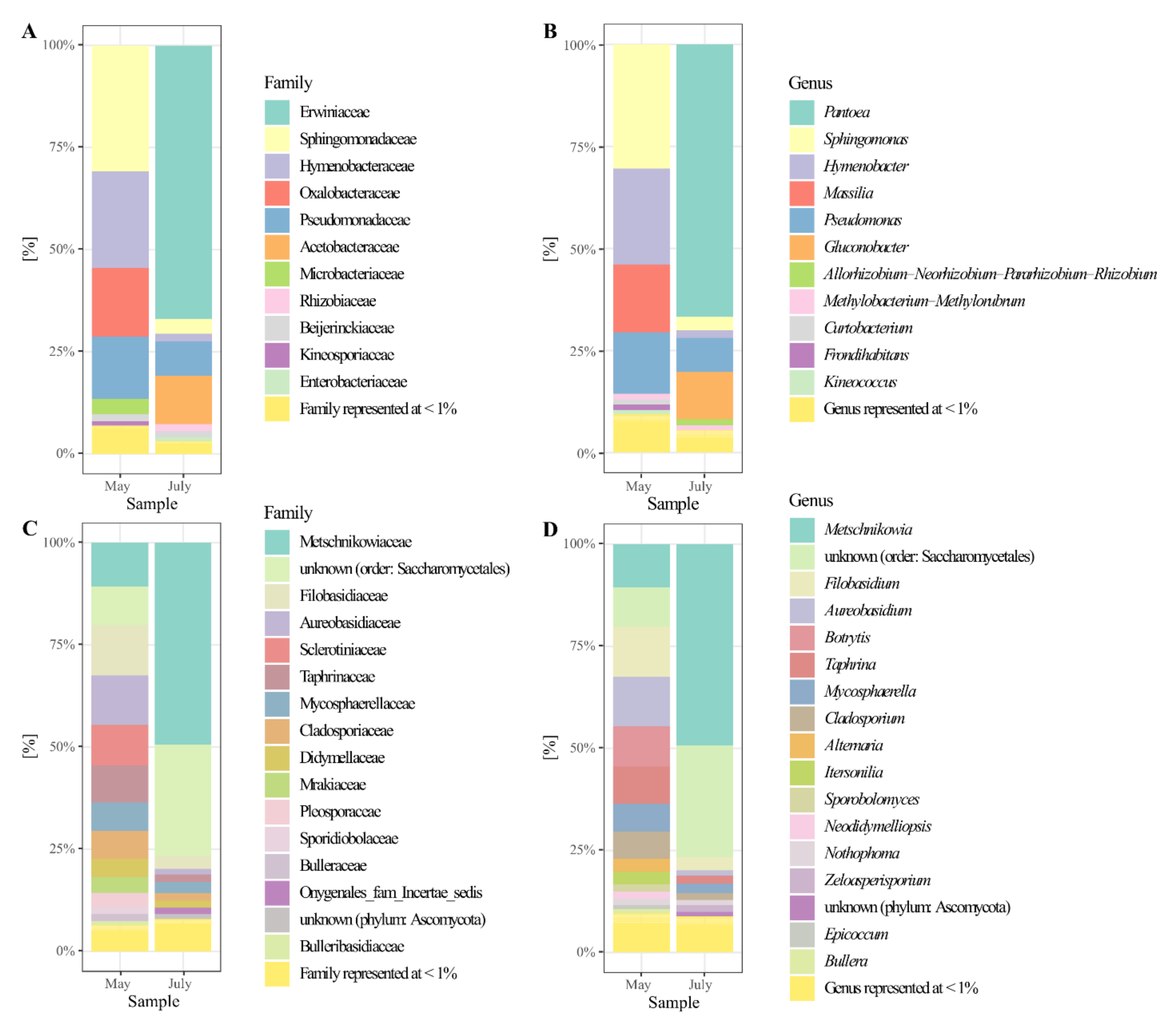
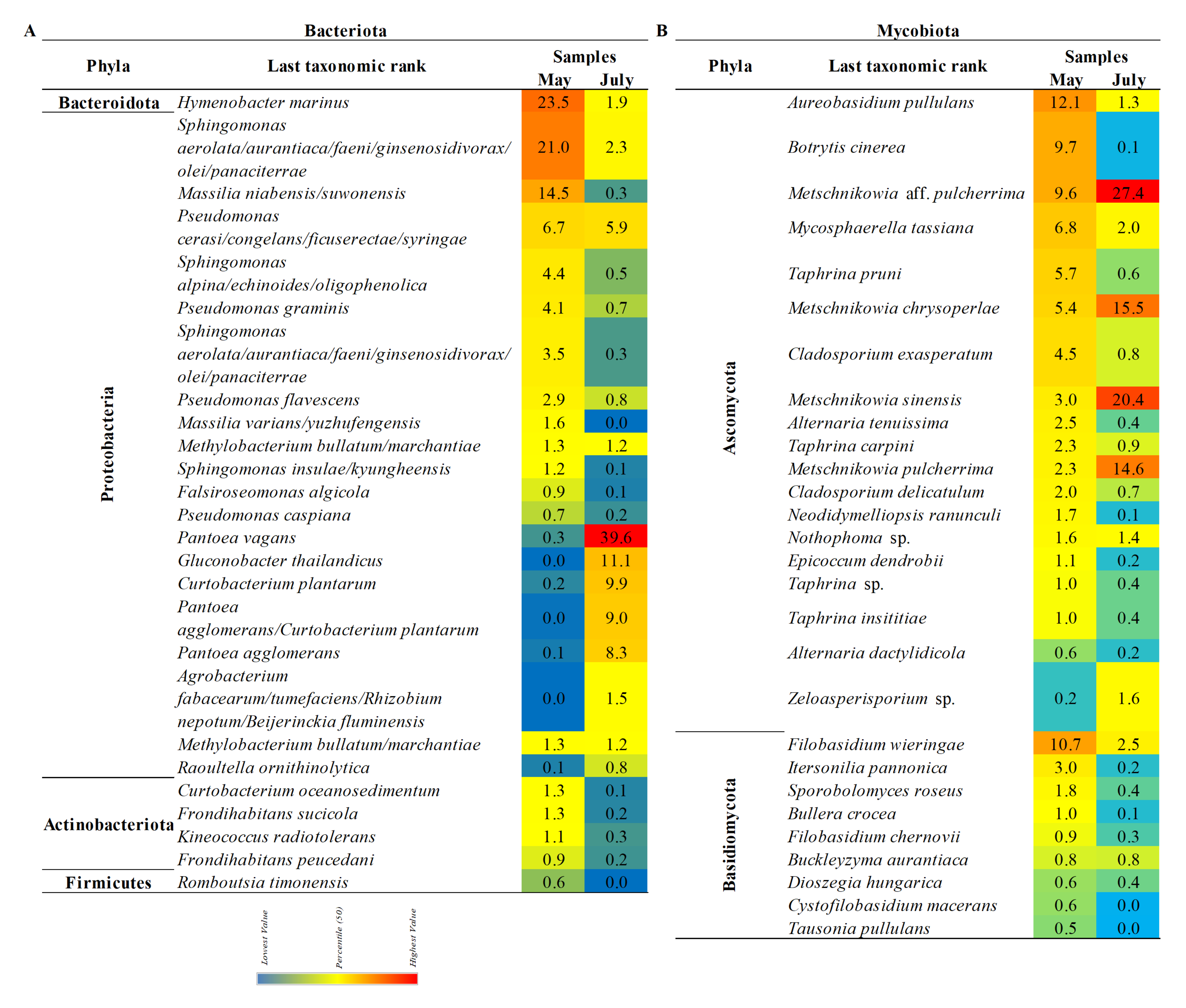
3.4. The Composition of the Pear’S Bacteriota
3.5. The Composition of the Pear’s Mycobiota
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Hancock, J.F.; Lobos, G.A. Pears. In Temperate Fruit Crop Breeding: Germplasm to Genomics; Hancock, J., Ed.; Springer Kluwer Academic Publishers: Dordrecht, The Netherlands, 2008; pp. 299–336. [Google Scholar]
- Kocsisné, G.M.; Bolla, D.; Anhalt-Brüderl, U.C.; Forneck, A.; Taller, J.; Kocsis, L. Genetic diversity and similarity of pear (Pyrus communis L.) cultivars in Central Europe revealed by SSR markers. Genet. Resour. Crop Evol. 2020, 67, 1755–1763. [Google Scholar] [CrossRef]
- Nikićević, N. Effects of some production factors on chemical composition and sensory qualities of Williams pear brandy. J. Agric. Sci. 2005, 50, 193–206. [Google Scholar]
- Urošević, I.; Nikićević, N.; Stanković, L.; Andjelković, B.; Urošević, T.; Krstić, G.; Tešević, V. Influence of yeast and nutrients on the quality of apricot brandy. J. Serb. Chem. Soc. 2014, 79, 1223–1234. [Google Scholar] [CrossRef]
- Moragrega, C.; Llorente, I.; Manceau, C.; Montesinos, E. Susceptibility of European pear cultivars to Pseudomonas syringae pv. syringae using immature fruit and detached leaf assays. Eur. J. Plant Pathol. 2003, 109, 319–326. [Google Scholar] [CrossRef]
- Sardella, D.; Muscat, A.; Brincat, J.P.; Gatt, R.; Decelis, S.; Valdramidis, V. A comprehensive review of the pear fungal diseases. Int. J. Fruit Sci. 2016, 16, 351–377. [Google Scholar] [CrossRef]
- Smessaert, J.; van Geel, M.; Verreth, C.; Crauwels, S.; Honnay, O.; Keulemans, W.; Lievens, B. Temporal and spatial variation in bacterial communities of “Jonagold” apple (Malus × domestica Borkh.) and “Conference” pear (Pyrus communis L.) floral nectar. Microbiologyopen 2019, 8, e918. [Google Scholar] [CrossRef] [Green Version]
- Zambounis, A.; Osathanunkul, M.; Madesis, P. Metagenome data of bacterial diversity in pear (Pyrus communis L.) rhizospheres associated with Phytophthora infection and amino acid treatment. Data Br. 2019, 26, 104396. [Google Scholar] [CrossRef]
- Martínez-Alonso, M.; Escolano Sánchez, J.; Montesinos Seguí, E.; Gaju, N. Diversity of the bacterial community in the surface soil of a pear orchard based on 16S rRNA gene analysis. Int. Microbiol. 2010, 13, 123–134. [Google Scholar]
- Sessa, L.; Abreo, E.; Lupo, S. Diversity of fungal latent pathogens and true endophytes associated with fruit trees in Uruguay. Phytopathology 2018, 166, 633–647. [Google Scholar] [CrossRef]
- Zambounis, A.; Ganopoulos, I.; Tsaftaris, A.; Valasiadis, D.; Madesis, P. Metagenomics analysis of fungal communities associated with postharvest diseases in pear fruits under the effect of management practices. Arch. Microbiol. 2020, 202, 2391–2400. [Google Scholar] [CrossRef]
- Bashir, I.; War, A.F.; Rafiq, I.; Reshi, Z.A.; Rashid, I.; Shouche, Y.S. Phyllosphere microbiome: Diversity and functions. Microbiol. Res. 2022, 254, 126888. [Google Scholar] [CrossRef] [PubMed]
- Dimkić, I.; Janakiev, T.; Petrović, M.; Degrassi, G.; Fira, D. Plant-associated Bacillus and Pseudomonas antimicrobial activities in plant disease suppression via biological control mechanisms-A review. Physiol. Mol. 2022, 117, 101754. [Google Scholar] [CrossRef]
- Hulot, J.F.; Hiller, N. Exploring the Benefits of Biocontrol for Sustainable Agriculture–A Literature Review on Biocontrol in Light of the European Green Deal; Institute for European Environmental Policy: Brussels, Belgium, 2021. [Google Scholar]
- Kong, W.L.; Li, P.S.; Wu, X.Q.; Wu, T.Y.; Sun, X.R. Forest tree-associated bacterial diffusible and volatile organic compounds against various phytopathogenic fungi. Microorganisms 2020, 8, 590. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wenneker, M. Fungal Pathogens in Pome Fruit Orchards and Causal Agents of Postharvest Decay. Ph.D. Thesis, Wageningen University, Wageningen, The Netherlands, 2019. [Google Scholar]
- Perincherry, L.; Lalak-Kańczugowska, J.; Stępień, Ł. Fusarium-produced mycotoxins in plant-pathogen interactions. Toxins 2019, 11, 664. [Google Scholar] [CrossRef] [Green Version]
- Zhu, X.Q.; Niu, C.W.; Chen, X.Y.; Guo, L.Y. Monilinia species are associated with brown rot of cultivated apple and pear fruit in China. Plant Dis. 2016, 100, 2240–2250. [Google Scholar] [CrossRef] [Green Version]
- Janakiev, T.; Dimkić, I.Z.; Unković, N.; Ljaljević Grbić, M.; Opsenica, D.M.; Gašić, U.M.; Stanković, S.; Berić, T. Phyllosphere fungal communities of plum and antifungal activity of indigenous phenazine-producing Pseudomonas synxantha against Monilinia laxa. Front. Microbiol. 2019, 10, 2287. [Google Scholar] [CrossRef] [Green Version]
- Janakiev, T.; Dimkić, I.; Bojić, S.; Fira, D.; Stanković, S.; Berić, T. Bacterial communities of plum phyllosphere and characterization of indigenous antagonistic Bacillus thuringiensis R3/3 isolate. J. Appl. Microbiol. 2020, 128, 528–543. [Google Scholar] [CrossRef]
- Dimkić, I.; Berić, T.; Stević, T.; Pavlović, S.; Šavikin, K.; Fira, D.; Stanković, S. Additive and synergistic effects of Bacillus spp. isolates and essential oils on the control of phytopathogenic and saprophytic fungi from medicinal plants and marigold seeds. Biol Control 2015, 87, 6–13. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.J.W.T.; Taylor, J.L. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols—A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: London, UK, 1990; Volume 18, pp. 315–322. [Google Scholar]
- O’Donnell, K.; Cigelnik, E. Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Mol. Phylogenet. Evol. 1997, 7, 103–116. [Google Scholar] [CrossRef]
- Kim, J.J.; Kim, S.H.; Lee, S.; Breuil, C. Distinguishing Ophiostoma ips and Ophiostoma montium, two bark beetle-associated sapstain fungi. FEMS Microbiol. Lett. 2003, 222, 187–192. [Google Scholar] [CrossRef] [Green Version]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef] [PubMed]
- Toju, H.; Tanabe, A.S.; Yamamoto, S.; Sato, H. High-coverage ITS primers for the DNA-based identification of ascomycetes and basidiomycetes in environmental samples. PLoS ONE 2012, 7, e40863. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bushnell, B.; Rood, J.; Singer, E. BBMerge–accurate paired shotgun read merging via overlap. PLoS ONE 2017, 12, e0185056. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [Green Version]
- McMurdie, P.J.; Holmes, S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [Green Version]
- Oksanen, J.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’hara, R.B.; Wagner, H. Community Ecology Package; R Package Version; The R Foundation: Vienna, Austria, 2013; pp. 321–326. [Google Scholar]
- Haas, J.C.; Street, N.R.; Sjödin, A.; Lee, N.M.; Högberg, M.N.; Näsholm, T.; Hurry, V. Microbial community response to growing season and plant nutrient optimisation in a boreal Norway spruce forest. Soil Biol. Biochem. 2018, 125, 197–209. [Google Scholar] [CrossRef]
- Bao, L.; Gu, L.; Sun, B.; Cai, W.; Zhang, S.; Zhuang, G.; Bai, Z.; Zhuang, X. Seasonal variation of epiphytic bacteria in the phyllosphere of Gingko biloba, Pinus bungeana and Sabina chinensis. FEMS Microbiol. Ecol. 2020, 96, fiaa017. [Google Scholar] [CrossRef]
- Schlechter, R.O.; Miebach, M.; Remus-Emsermann, M.N. Driving factors of epiphytic bacterial communities: A review. J. Adv. Res. 2019, 19, 57–65. [Google Scholar] [CrossRef]
- Rastogi, G.; Coaker, G.L.; Leveau, J.H. New insights into the structure and function of phyllosphere microbiota through high-throughput molecular approaches. FEMS Microbiol. Lett. 2013, 348, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Ofek, M.; Hadar, Y.; Minz, D. Ecology of root colonizing Massilia (Oxalobacteraceae). PLoS ONE 2012, 7, e40117. [Google Scholar] [CrossRef]
- Luziatelli, F.; Gatti, L.; Ficca, A.G.; Medori, G.; Silvestri, C.; Melini, F.; Mule, R.; Ruzzi, M. Metabolites secreted by a plant-growth-promoting Pantoea agglomerans strain improved rooting of Pyrus communis L. cv Dar Gazi cuttings. Front. Microbiol. 2020, 11, 2412. [Google Scholar] [CrossRef]
- Janisiewicz, W.J.; Buyer, J.S. Culturable bacterial microflora associated with nectarine fruit and their potential for control of brown rot. Can. J. Microbiol. 2010, 56, 480–486. [Google Scholar] [CrossRef]
- Ding, T.; Melcher, U. Influences of plant species, season and location on leaf endophytic bacterial communities of non-cultivated plants. PLoS ONE 2016, 11, e0150895. [Google Scholar] [CrossRef] [Green Version]
- Ruiz-Pérez, C.A.; Restrepo, S.; Zambrano, M.M. Microbial and functional diversity within the phyllosphere of Espeletia species in an Andean high-mountain ecosystem. Appl. Environ. Microbiol. 2016, 82, 1807–1817. [Google Scholar] [CrossRef] [Green Version]
- Bowsher, A.W.; Benucci, G.M.N.; Bonito, G.; Shade, A. Seasonal dynamics of core fungi in the switchgrass phyllosphere, and co-occurrence with leaf bacteria. Phytobiomes J. 2020, 5, 60–68. [Google Scholar] [CrossRef]
- Zhang, Y.; Gao, C.; Masum, M.; Islam, M.; Cheng, Y.; Wei, C.; Guan, Y.; Guan, J. Dynamic Microbiome Changes Reveal the Effect of 1-Methylcyclopropene Treatment on Reducing Post-harvest Fruit Decay in “Doyenne du Comice” Pear. Front. Microbiol. 2021, 12, 729014. [Google Scholar] [CrossRef]
- Gupta, A.; Singh, V.K.; Qazi, G.N.; Kumar, A. Gluconobacter oxydans: Its biotechnological applications. J. Mol. Microbiol. Biotechnol. 2001, 3, 445–456. [Google Scholar]
- Bevardi, M.; Frece, J.; Mesarek, D.; Bošnir, J.; Mrvčić, J.; Delaš, F.; Markov, K. Antifungal and antipatulin activity of Gluconobacter oxydans isolated from apple surface. Arh. Hig. Rada Toksikol. 2013, 64, 279–283. [Google Scholar] [CrossRef] [Green Version]
- Vacher, C.; Hampe, A.; Porté, A.J.; Sauer, U.; Compant, S.; Morris, C.E. The phyllosphere: Microbial jungle at the plant–climate interface. Annu. Rev. Ecol. Evol. 2016, 47, 1–24. [Google Scholar] [CrossRef]
- Singh, P.; Santoni, S.; This, P.; Péros, J.P. Genotype-environment interaction shapes the microbial assemblage in grapevine’s phyllosphere and carposphere: An NGS approach. Microorganisms 2018, 6, 96. [Google Scholar] [CrossRef] [Green Version]
- Milazzo, C.; Zulak, K.G.; Muria-Gonzalez, M.J.; Jones, D.; Power, M.; Bransgrove, K.; Bunce, M.; Lopez-Ruiz, F.J. High-Throughput Metabarcoding Characterizes Fungal Endophyte Diversity in the Phyllosphere of a Barley Crop. Phytobiomes J. 2021, 5, 316–325. [Google Scholar] [CrossRef]
- Zhu, S.; Lei, Y.; Wang, C.; Wei, Y.; Wang, C.; Sun, Y. Patterns of yeast diversity distribution and its drivers in rhizosphere soil of Hami melon orchards in different regions of Xinjiang. BMC Microbiol. 2021, 21, 170. [Google Scholar] [CrossRef]
- Glushakova, A.M.; Kachalkin, A.V. Endophytic yeasts in Malus domestica and Pyrus communis fruits under anthropogenic impact. Microbiology 2017, 86, 128–135. [Google Scholar] [CrossRef]
- Mari, M.; Martini, C.; Guidarelli, M.; Neri, F. Postharvest biocontrol of Monilinia laxa, Monilinia fructicola and Monilinia fructigena on stone fruit by two Aureobasidium pullulans strains. Biol. Control 2012, 60, 132–140. [Google Scholar] [CrossRef]
- Liu, J.; Sui, Y.; Wisniewski, M.; Droby, S.; Liu, Y. Utilization of antagonistic yeasts to manage postharvest fungal diseases of fruit. Int. J. Food Microbiol. 2013, 167, 153–160. [Google Scholar] [CrossRef]
- Banani, H.; Spadaro, D.; Zhang, D.; Matic, S.; Garibaldi, A.; Gullino, M.L. Postharvest application of a novel chitinase cloned from Metschnikowia fructicola and overexpressed in Pichia pastoris to control brown rot of peaches. Int. J. Food Microbiol. 2015, 199, 54–61. [Google Scholar] [CrossRef]
- Parafati, L.; Vitale, A.; Restuccia, C.; Cirvilleri, G. Biocontrol ability and action mechanism of food-isolated yeast strains against Botrytis cinerea causing post-harvest bunch rot of table grape. Food Microbiol. 2015, 47, 85–92. [Google Scholar] [CrossRef]
- Agarbati, A.; Canonico, L.; Pecci, T.; Romanazzi, G.; Ciani, M.; Comitini, F. Biocontrol of non-Saccharomyces yeasts in vineyard against the gray mold disease agent Botrytis cinerea. Microorganisms 2022, 10, 200. [Google Scholar] [CrossRef]
- Barata, A.; Malfeito-Ferreira, M.; Loureiro, V. The microbial ecology of wine grape berries. Int. J. Food Microbiol. 2012, 153, 243–259. [Google Scholar] [CrossRef]
- Gschaedler, A. Contribution of non-conventional yeasts in alcoholic beverages. Curr. Opin. Food Sci. 2017, 13, 73–77. [Google Scholar] [CrossRef]
- Lee, H.B.; Patriarca, A.; Magan, N. Alternaria in food: Ecophysiology, mycotoxin production and toxicology. Mycobiology 2015, 43, 93–106. [Google Scholar] [CrossRef] [Green Version]
- Rupp, S.; Weber, R.W.; Rieger, D.; Detzel, P.; Hahn, M. Spread of Botrytis cinerea strains with multiple fungicide resistance in German horticulture. Front. Microbiol. 2017, 7, 2075. [Google Scholar] [CrossRef] [Green Version]
- Cissé, O.H.; Almeida, J.M.; Fonseca, Á.; Kumar, A.A.; Salojärvi, J.; Overmyer, K.; Hauser, P.M.; Pagni, M. Genome sequencing of the plant pathogen Taphrina deformans, the causal agent of peach leaf curl. mBio 2013, 4, e00055-13. [Google Scholar] [CrossRef] [Green Version]
- Wessels, B.A.; Linde, C.C.; Fourie, P.H.; Mostert, L. Genetic population structure and fungicide resistance of Botrytis cinerea in pear orchards in the Western Cape of South Africa. Plant Pathol. 2016, 65, 1473–1483. [Google Scholar] [CrossRef] [Green Version]
- Bensch, K.; Braun, U.; Groenewald, J.Z.; Crous, P.W. The genus Cladosporium. Stud. Mycol. 2012, 72, 1–401. [Google Scholar] [CrossRef] [Green Version]
- Amanelah Baharvandi, H.; Zafari, D. Identification of Cladosporium delicatulum as a mycoparasite of Taphrina pruni. Arch. Phytopathol. Pflanzenschutz 2015, 48, 688–697. [Google Scholar] [CrossRef]
- Thambugala, K.M.; Daranagama, D.A.; Phillips, A.J.; Kannangara, S.D.; Promputtha, I. Fungi vs. fungi in biocontrol: An overview of fungal antagonists applied against fungal plant pathogens. Front. Cell. Infect. Microbiol. 2020, 10, 718. [Google Scholar] [CrossRef]
- Temperini, C.V.; Pardo, A.G.; Pose, G.N. Diversity of airborne Cladosporium species isolated from agricultural environments of northern Argentinean Patagonia: Molecular characterization and plant pathogenicity. Aerobiologia 2018, 34, 227–239. [Google Scholar] [CrossRef]
- Pitkaranta, M.; Meklin, T.; Hyvarinen, A.; Paulin, L.; Auvinen, P.; Nevalainen, A.; Rintala, H. Analysis of fungal flora in indoor dust by ribosomal DNA sequence analysis, quantitative PCR, and culture. Appl. Environ. Microbiol. 2008, 74, 233–244. [Google Scholar] [CrossRef] [Green Version]
- Rojas, E.C.; Sapkota, R.; Jensen, B.; Jørgensen, H.J.; Henriksson, T.; Jørgensen, L.N.; Nicolaisen, M.; Collinge, D.B. Fusarium head blight modifies fungal endophytic communities during infection of wheat spikes. Microb. Ecol. 2020, 79, 397–408. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duvenage, F.J.; Duvenage, S.; Du Plessis, E.M.; Volschenk, Q.; Korsten, L. Viable bacterial population and persistence of foodborne pathogens on the pear carpoplane. J. Sci. Food Agric. 2017, 97, 1185–1192. [Google Scholar] [CrossRef]
- Brady, C.; Hunter, G.; Kirk, S.; Arnold, D.; Denman, S. Rahnella victoriana sp. nov., Rahnella bruchi sp. nov., Rahnella woolbedingensis sp. nov., classification of Rahnella genomospecies 2 and 3 as Rahnella variigena sp. nov. and Rahnella inusitata sp. nov., respectively and emended description of the genus Rahnella. Syst. Appl. Microbiol. 2014, 37, 545–552. [Google Scholar] [PubMed]
- Vadkertiová, R.; Molnárová, J.; Vránová, D.; Sláviková, E. Yeasts and yeast-like organisms associated with fruits and blossoms of different fruit trees. Can. J. Microbiol. 2012, 58, 1344–1352. [Google Scholar] [CrossRef]
- Bourret, T.B.; Grove, G.G.; Vandemark, G.J.; Henick-Kling, T.; Glawe, D.A. Diversity and molecular determination of wild yeasts in a central Washington State vineyard. North Am. Fungi 2013, 8, 1–32. [Google Scholar] [CrossRef] [Green Version]
- Puławska, J.; Willems, A.; De Meyer, S.E.; Süle, S. Rhizobium nepotum sp. nov. isolated from tumors on different plant species. Syst. Appl. Microbiol. 2012, 35, 215–220. [Google Scholar] [CrossRef]
- Sipiczki, M. Metschnikowia pulcherrima and related pulcherrimin-producing yeasts: Fuzzy species boundaries and complex antimicrobial antagonism. Microorganisms 2020, 8, 1029. [Google Scholar] [CrossRef]
- Spadaro, D.; Vola, R.; Piano, S.; Gullino, M.L. Mechanisms of action and efficacy of four isolates of the yeast Metschnikowia pulcherrima active against postharvest pathogens on apples. Postharvest Biol. Technol. 2002, 24, 123–134. [Google Scholar] [CrossRef]
- Matić, S.; Spadaro, D.; Garibaldi, A.; Gullino, M.L. Antagonistic yeasts and thermotherapy as seed treatments to control Fusarium fujikuroi on rice. Biol. Control 2014, 73, 59–67. [Google Scholar] [CrossRef]
- Papp, L.A.; Horváth, E.; Peles, F.; Pócsi, I.; Miklós, I. Insight into Yeast–Mycotoxin Relations. Agriculture 2021, 11, 1291. [Google Scholar] [CrossRef]
- Polat, Z.; Gültekin, M.A.; Palacıoğlu, G.; Akçay, M.E.; Bayraktar, H. First report of Fusarium avenaceum causing branch canker on pear in Turkey. J. Plant Pathol. 2022, 104, 815. [Google Scholar] [CrossRef]
- Janisiewicz, W.J.; Jurick II, W.M.; Vico, I.; Peter, K.A.; Buyer, J.S. Culturable bacteria from plum fruit surfaces and their potential for controlling brown rot after harvest. Postharvest Biol. Technol. 2013, 76, 145–151. [Google Scholar] [CrossRef]

| Phenophases | Total Number of Species | Identified Species |
|---|---|---|
| May | 13 | Phoma sp., Nigrospora sp., Trichoderma harzianum, Penicillium sp., Botrytis cinerea, Pseudopithomyces sp., Fusarium incarnatum, Phoma putaminum, Epicoccum sp., Trichothecium roseum, Macrophomina phaseolina, Fusarium verticillioides, Fusarium proliferatum |
| July | 5 | Penicillium corylophilum, Alternaria tenuissima, Cladosporium cladosporioides, Stemphylium botryosum, Curvularia sp. |
| May and July | 10 | Aspergillus niger, Aspergillus flavus, Trichoderma viride, Alternaria sp., Fusarium oxysporum, Fusarium solani, Cladosporium sp., Phoma betae, Fusarium sporotrichioides, Rhizopus sp. |
| Antagonists | Fusarium proliferatum/Fujikuroi | Fusarium verticillioides | Fusarium sporotrichioides | Fusarium solani | Fusarium oxysporum | Fusarium incarnatum |
| Bacteria | ||||||
| Pseudomonas graminis V2/1 | 39.8 a ± 2.00 | 39.0 ab ± 1.99 | 65.1 ab ± 1.04 | 28.0 bc ± 3.18 | 42.5 ab ± 1.88 | 21.6 c ± 6.43 |
| Pseudomonas graminis V2/2 | 11.4 cd ± 2.40 | 11.7 e ± 0.75 | 69.4 a ± 1.80 | 12.6 cd ± 1.46 | 13.0 cd ± 0.50 | 16.1 c ± 2.63 |
| Pseudomonas graminis V3/1 | 12.6 bcd ± 0.84 | 19.9 de ± 2.41 | 52.9 b ± 1.36 | 10.9 cd ± 1.57 | 18.9 cd ± 2.00 | 8.8 c ± 3.13 |
| Pseudomonas putida V8 | 19.3 bc ± 2.31 | 9.1 e ± 1.50 | 65.5 ab ± 2.08 | 3.5 d ± 1.10 | 7.4 d ± 2.29 | 6.4 c ± 2.04 |
| Frigoribacterium endophyticum/faeni V3/2 | 4.5 d ± 0.80 | 15.2 de ± 2.29 | 65.1 ab ± 3.42 | 0.0 d ± 0.00 | 14.6 cd ± 2.13 | 15.7 c ± 2.03 |
| Yeasts | ||||||
| Metschnikowia pulcherrima V2 | 21.6 b ± 2.01 | 22.9 cd ± 3.03 | 63.1 ab ± 5.28 | 46.3 ab ± 4.76 | 24.8 bcd ± 2.67 | 27.3 bc ± 6.22 |
| Metschnikowia pulcherrima V7 | 45.6 a ± 2.44 | 33.6 bc ± 3.00 | 52.7 b ± 5.96 | 52.4 a ± 2.34 | 26.4 bc ± 7.95 | 50.9 ab ± 3.21 |
| Hannaella luteola V1/3 | 42.3 a ± 2.01 | 45.5 a ± 1.98 | 70.2 a ± 1.03 | 56.9 a ± 8.56 | 53.2 a ± 1.98 | 56.9 a ± 8.56 |
| Monilinia laxa | Botrytis cinerea | Alternaria tenuissima | Cladosporium cladosporioides | |||
| Bacteria | ||||||
| Pseudomonas graminis V2/1 | 63.2 a ± 0.44 | 42.6 ab ± 1.29 | 40.5 bc ± 0.88 | 52.2 ab ± 2.51 | ||
| Pseudomonas graminis V2/2 | 0.0 b ± 0.00 | 35.4 bc ± 8.31 | 34.4 c ± 0.88 | 43.4 ab ± 2.51 | ||
| Pseudomonas graminis V3/1 | 17.8 b ± 1.49 | 36.6 bc ± 8.75 | 43.6 b ± 2.64 | 45.6 ab ± 6.26 | ||
| Pseudomonas putida V8 | 15.7 b ± 3.46 | 38.5 ab ± 3.97 | 34.4 c ± 0.88 | 14.4 c ± 1.43 | ||
| Frigoribacterium endophyticum/faeni V3/2 | 5.7 b ± 1.54 | 12.8 c ± 3.44 | 21.7 d ± 1.02 | 36.9 b ± 6.26 | ||
| Yeasts | ||||||
| Metschnikowia pulcherrima V2 | 52.2 a ± 0.77 | 46.3 ab ± 3.86 | 60.5 a ± 1.50 | 52.2 ab ± 2.51 | ||
| Metschnikowia pulcherrima V7 | 54.0 a ± 6.92 | 61.7 a ± 2.56 | 64.9 a ± 0.88 | 58.0 a ± 1.47 | ||
| Hannaella luteola V1/3 | 65.0 a ± 6.43 | 45.5 ab ± 2.22 | 65.8 a ± 3.03 | 60.9 a ± 2.51 | ||
| 16S | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Sample | OBS | Chao1 | SE.Chao1 | ACE | SE.ACE | Shannon | Simpson | InvSimpson | Level |
| May | 12.00 | 12.00 | 0.00 | 12.00 | 1.71 | 0.90 | 0.48 | 1.93 | Phylum |
| July | 8.00 | 8.00 | 0.00 | 8.00 | 1.37 | 0.16 | 0.06 | 1.06 | |
| May | 61.00 | 61.00 | 0.00 | 61.00 | 3.89 | 1.90 | 0.80 | 4.88 | Family |
| July | 32.00 | 32.00 | 0.00 | 32.00 | 2.62 | 1.28 | 0.53 | 2.12 | |
| May | 99.00 | 99.08 | 0.34 | 99.67 | 4.94 | 2.05 | 0.80 | 5.02 | Genus |
| July | 66.00 | 66.00 | 0.08 | 66.41 | 3.38 | 1.37 | 0.53 | 2.14 | |
| May | 644.00 | 694.04 | 14.44 | 693.57 | 12.86 | 4.23 | 0.94 | 17.29 | ASV |
| July | 432.00 | 454.00 | 8.83 | 456.30 | 10.55 | 2.48 | 0.80 | 5.06 | |
| ITS | |||||||||
| Sample | OBS | Chao1 | SE.Chao1 | ACE | SE.ACE | Shannon | Simpson | InvSimpson | Level |
| May | 3.00 | 3.00 | 0.00 | – | – | 0.59 | 0.38 | 1.61 | Phylum |
| July | 3.00 | 3.00 | 0.00 | – | – | 0.29 | 0.15 | 1.18 | Phylum |
| May | 72.00 | 72.00 | 0.00 | 72.00 | 3.26 | 2.73 | 0.92 | 11.99 | Family |
| July | 67.00 | 67.00 | 0.00 | 67.00 | 2.92 | 1.76 | 0.68 | 3.11 | |
| May | 109.00 | 109.00 | 0.00 | 109.00 | 3.79 | 2.86 | 0.92 | 12.39 | Genus |
| July | 93.00 | 93.00 | 0.00 | 93.00 | 3.55 | 1.81 | 0.68 | 3.11 | |
| May | 285.00 | 285.00 | 0.00 | 285.00 | 5.86 | 3.78 | 0.95 | 20.81 | ASV |
| July | 276.00 | 276.00 | 0.00 | 276.00 | 5.46 | 4.27 | 0.97 | 39.68 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Janakiev, T.; Berić, T.; Stević, T.; Stanković, S.; Bačić, J.; Majstorović, H.; Fira, D.; Dimkić, I. The Microbiome of the ‘Williams’ Pear Variety Grown in the Organic Orchard and Antifungal Activity by the Autochthonous Bacterial and Yeast Isolates. Microorganisms 2022, 10, 1282. https://doi.org/10.3390/microorganisms10071282
Janakiev T, Berić T, Stević T, Stanković S, Bačić J, Majstorović H, Fira D, Dimkić I. The Microbiome of the ‘Williams’ Pear Variety Grown in the Organic Orchard and Antifungal Activity by the Autochthonous Bacterial and Yeast Isolates. Microorganisms. 2022; 10(7):1282. https://doi.org/10.3390/microorganisms10071282
Chicago/Turabian StyleJanakiev, Tamara, Tanja Berić, Tatjana Stević, Slaviša Stanković, Jasmina Bačić, Helena Majstorović, Djordje Fira, and Ivica Dimkić. 2022. "The Microbiome of the ‘Williams’ Pear Variety Grown in the Organic Orchard and Antifungal Activity by the Autochthonous Bacterial and Yeast Isolates" Microorganisms 10, no. 7: 1282. https://doi.org/10.3390/microorganisms10071282
APA StyleJanakiev, T., Berić, T., Stević, T., Stanković, S., Bačić, J., Majstorović, H., Fira, D., & Dimkić, I. (2022). The Microbiome of the ‘Williams’ Pear Variety Grown in the Organic Orchard and Antifungal Activity by the Autochthonous Bacterial and Yeast Isolates. Microorganisms, 10(7), 1282. https://doi.org/10.3390/microorganisms10071282








