Comparative Analysis of Catabolic and Anabolic Dehydroshikimate Dehydratases for 3,4-DHBA Production in Escherichia coli
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Plasmids, and Growth Conditions
2.2. DNA Manipulation
2.3. Protein Production and Purification
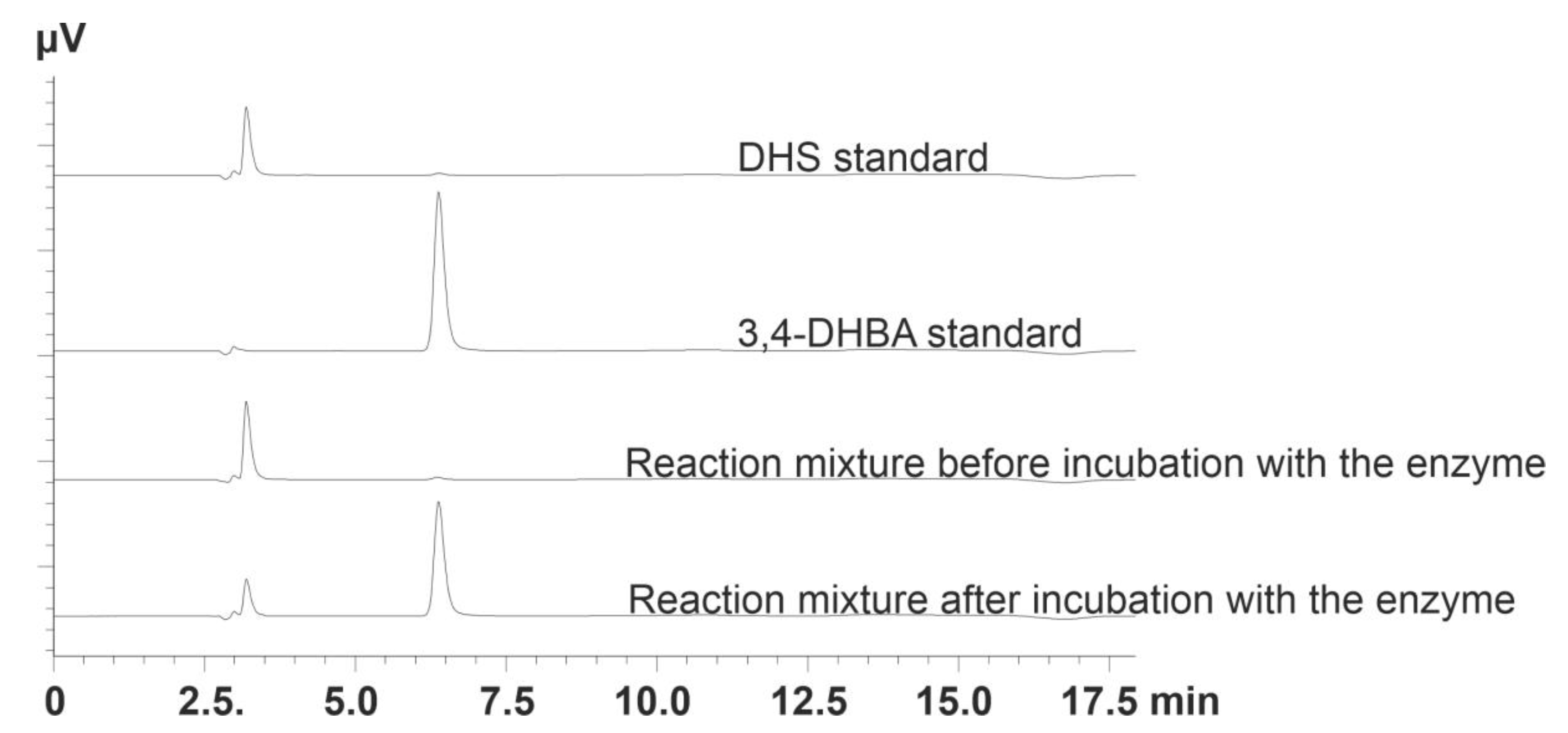
2.4. Measurement of DSD Activity
2.5. HPLC Analysis
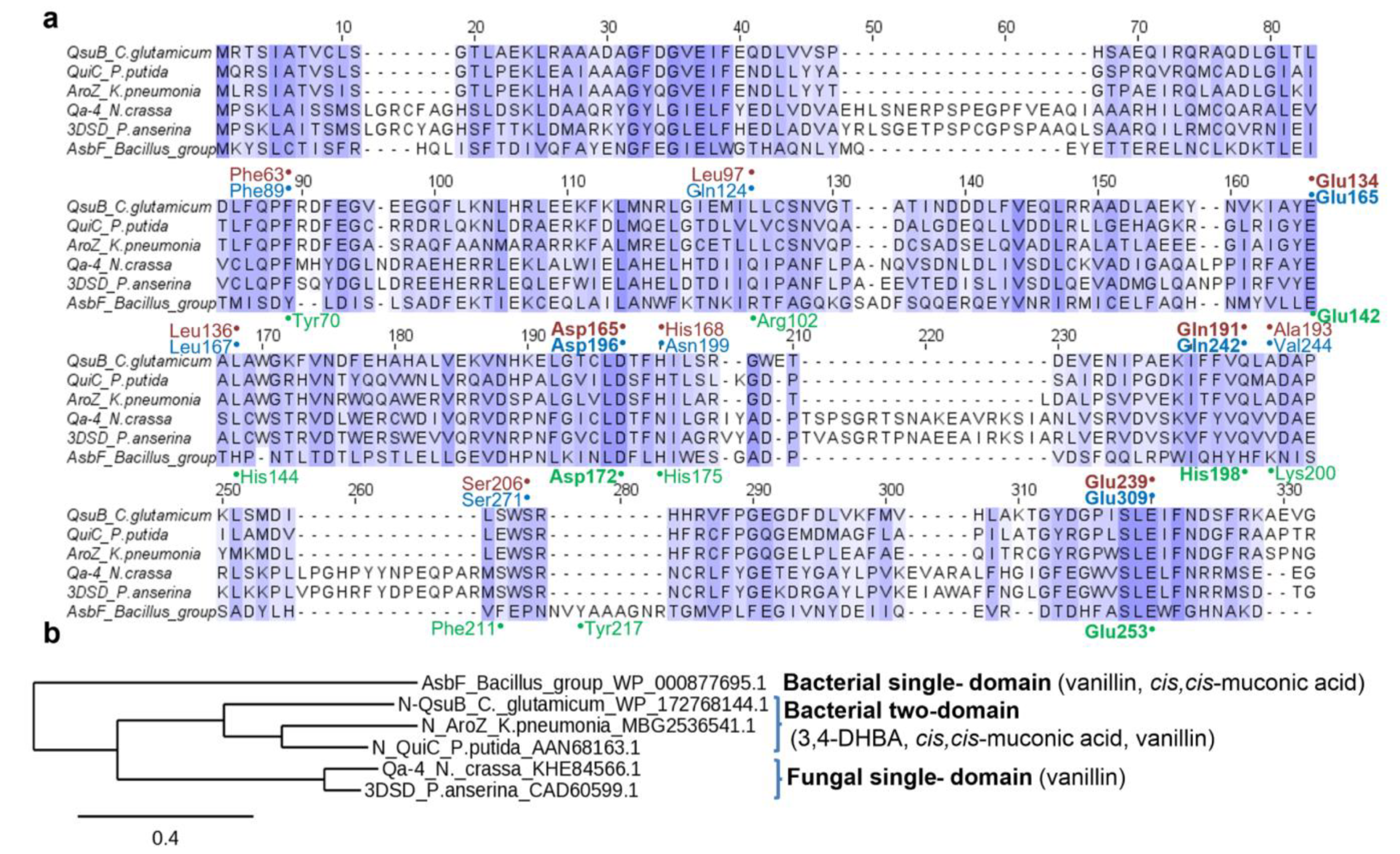
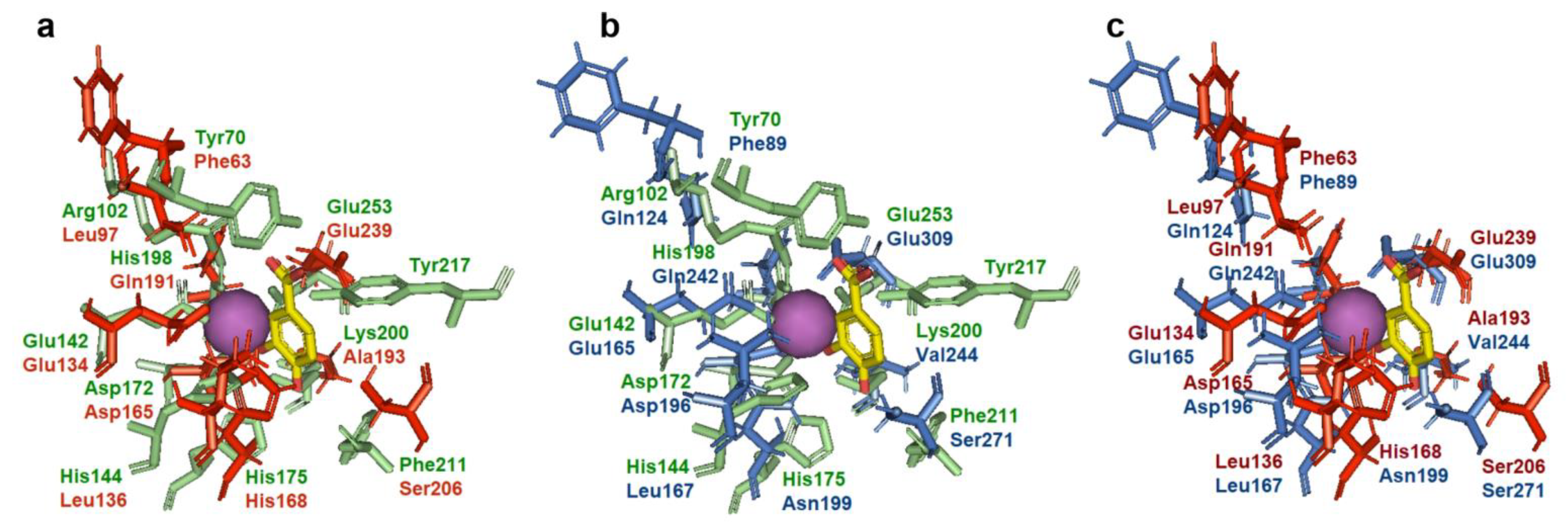
2.6. Sequence Alignment and 3D Structural Analysis
2.7. Statistical Analysis
3. Results
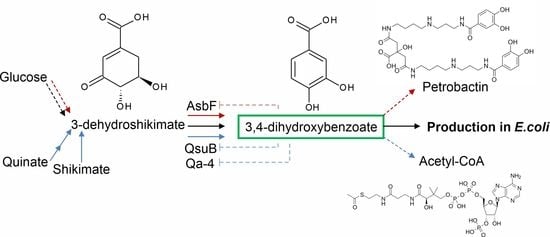
3.1. Selection of the Enzymes for Comparative Analysis
3.2. Overexpression of DSDs in E. coli Using the T7 RNA Polymerase-Based System
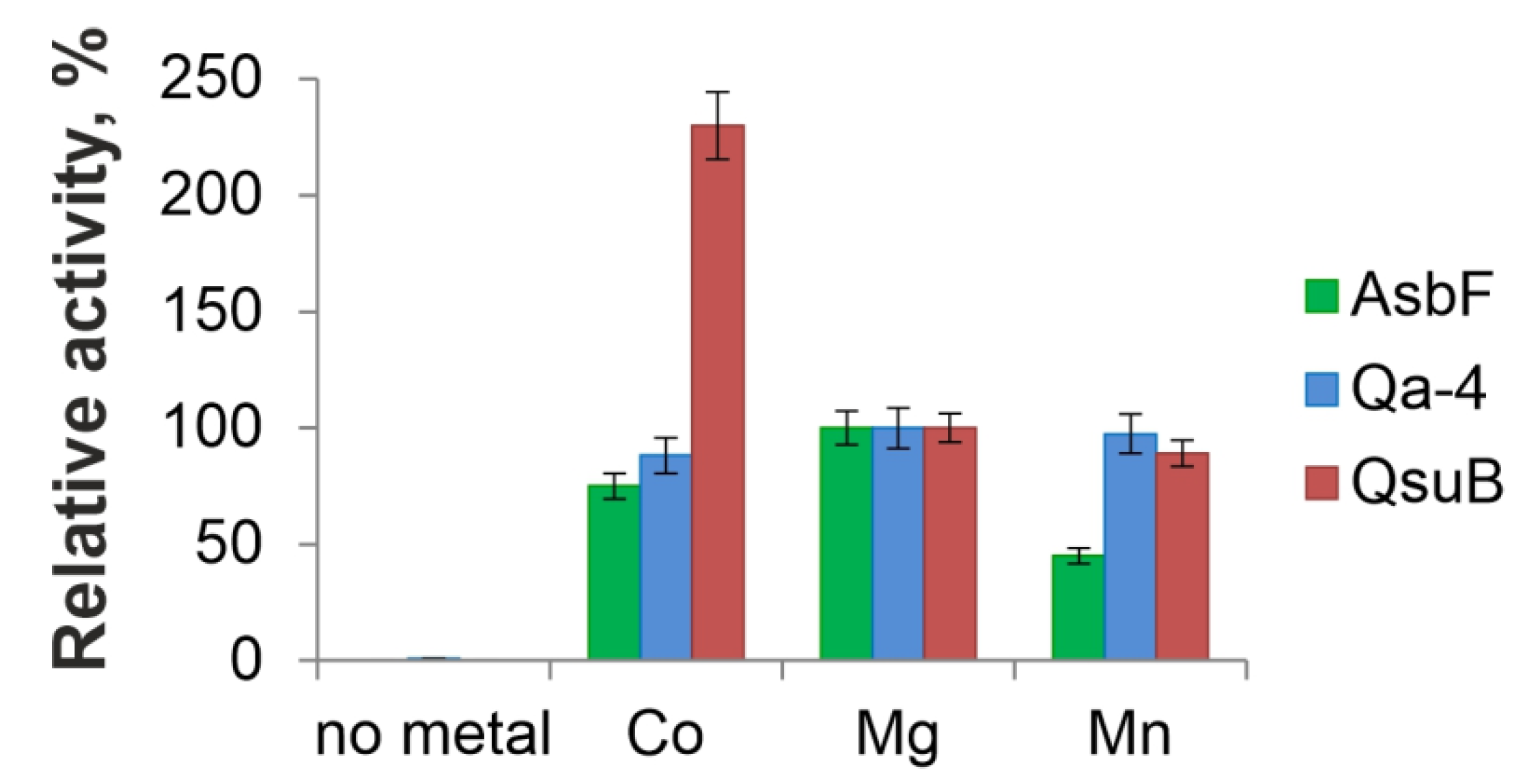
3.3. Metal-Dependence Comparison of AsbF, Qa-4, and QsuB Enzymes
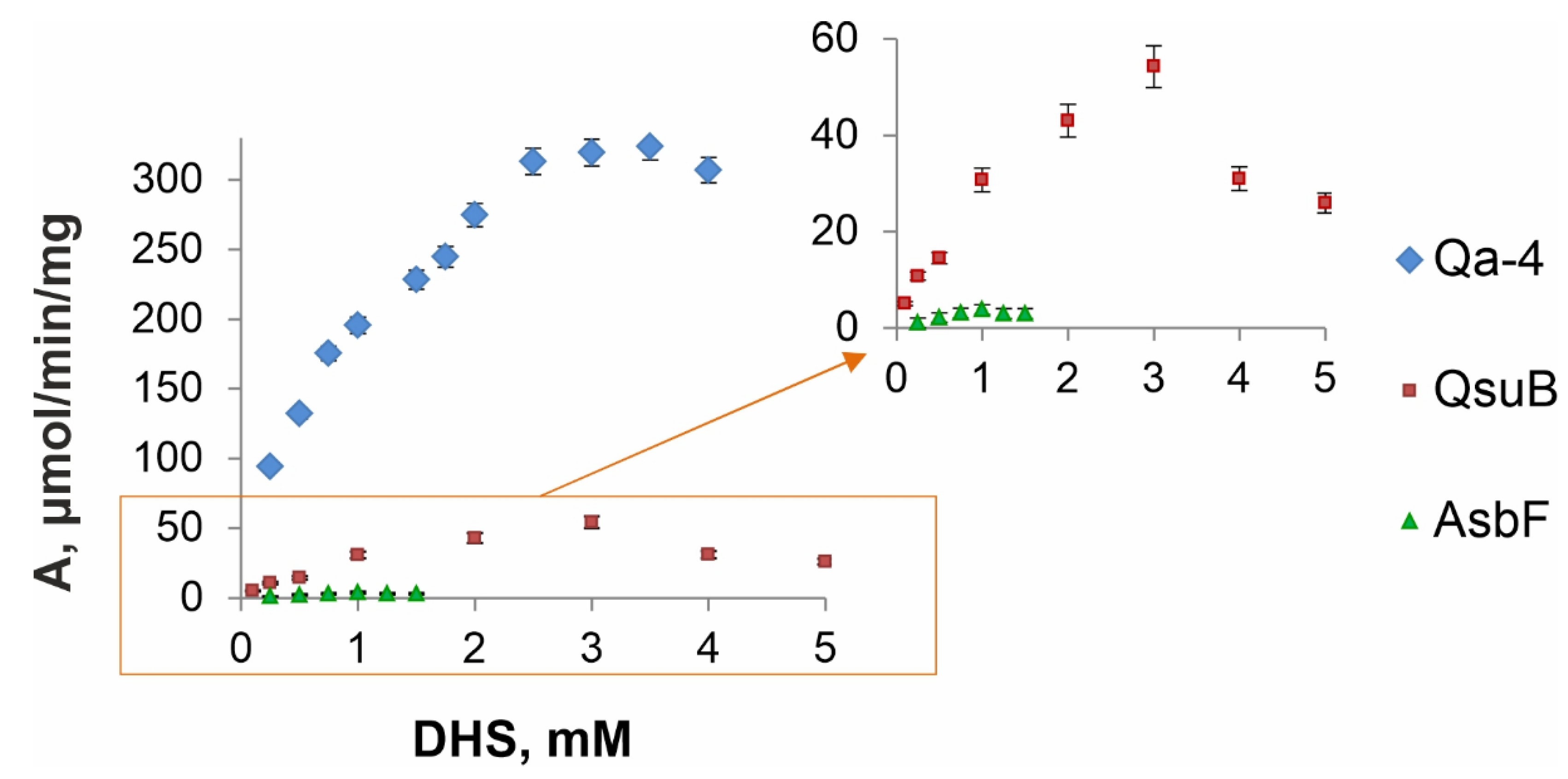
3.4. Comparative Kinetic Analysis of DSDs
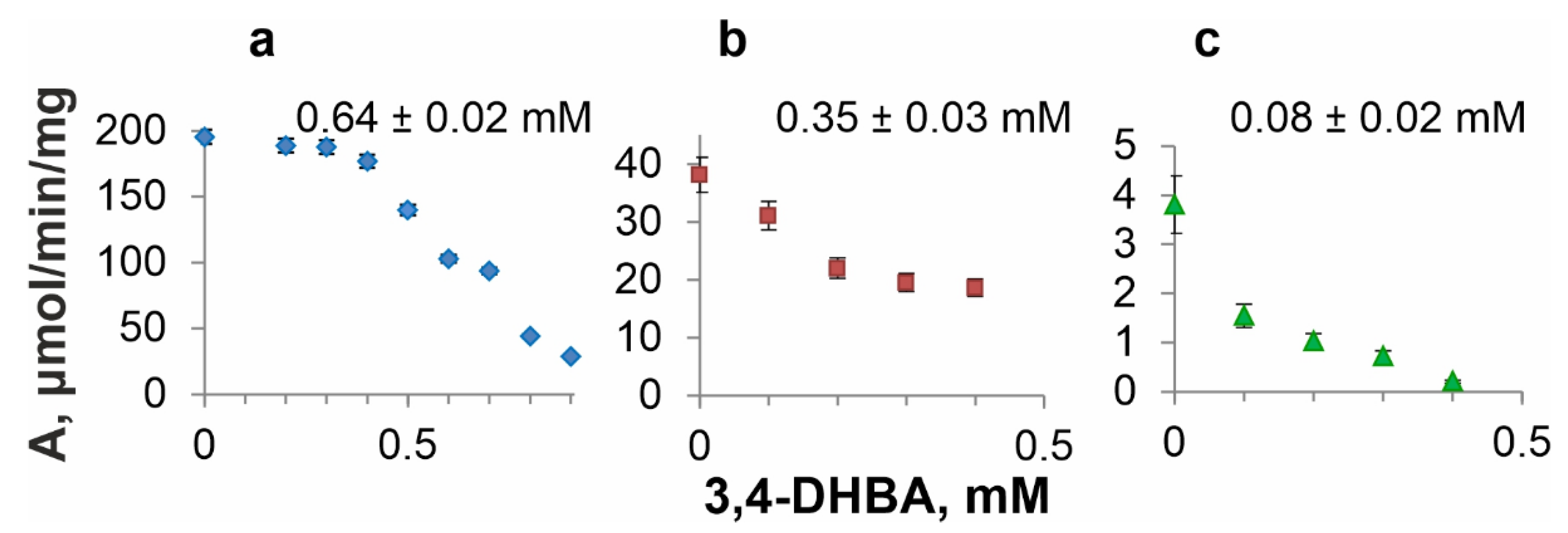
3.5. DSD Inhibition by 3,4-DHBA
3.6. 3,4-DHBA Production Using AsbF, Qa-4, QsuB in E. coli
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Metsämuuronen, S.; Sirén, H. Bioactive phenolic compounds, metabolism and properties: A review on valuable chemical compounds in Scots pine and Norway spruce. Phytochem. Rev. 2019, 18, 623–666. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Wang, X.; Chen, D.; Chen, S. Antioxidant activity and mechanism of protocatechuic acid in vitro. Funct. Foods Health Dis. 2011, 1, 232–244. [Google Scholar] [CrossRef]
- Zhou, Z.; Zhang, Y.; Ding, X.R.; Chen, S.H.; Yang, J.; Wang, X.J.; Jia, G.L.; Chen, H.S.; Bo, X.C.; Wang, S.Q. Protocatechuic aldehyde inhibits hepatitis B virus replication both in vitro and in vivo. Antivir. Res. 2007, 74, 59–64. [Google Scholar] [CrossRef] [PubMed]
- Lende, A.B.; Kshirsagar, A.D.; Deshpande, A.D.; Muley, M.M.; Patil, R.R.; Bafna, P.A.; Naik, S.R. Anti-inflammatory and analgesic activity of protocatechuic acid in rats and mice. Inflammopharmacology 2011, 19, 255–263. [Google Scholar] [CrossRef]
- Kakkar, S.; Bais, S. A review on protocatechuic acid and its pharmacological potential. ISRN Pharmacol. 2014, 2014, 952943. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krzysztoforska, K.; Piechal, A.; Blecharz-Klin, K.; Pyrzanowska, J.; Joniec-Maciejak, I.; Mirowska-Guzel, D.; Widy-Tyszkiewicz, E. Administration of protocatechuic acid affects memory and restores hippocampal and cortical serotonin turnover in rat model of oral D-galactose-induced memory impairment. Behav. Brain. Res. 2019, 368, 111896. [Google Scholar] [CrossRef]
- Lee, J.-H.; Wendisch, V.F. Biotechnological production of aromatic compounds of the extended shikimate pathway from renewable biomass. J. Biotechnol. 2017, 257, 211–221. [Google Scholar] [CrossRef]
- Kim, H.S.; Choi, J.A.; Kim, B.Y.; Ferrer, L.; Choi, J.M.; Wendisch, V.F.; Lee, J.H. Engineered Corynebacterium glutamicum as the Platform for the Production of Aromatic Aldehydes. Front. Bioeng. Biotechnol. 2022, 10, 880277. [Google Scholar] [CrossRef]
- Peek, J.; Roman, J.; Moran, G.; Christendat, D. Structurally diverse dehydroshikimate dehydratase variants participate in microbial quinate catabolism. Mol. Microbiol. 2017, 103, 39–54. [Google Scholar] [CrossRef] [Green Version]
- Fox, D.T.; Hotta, K.; Kim, C.Y.; Koppisch, A.T. The missing link in petrobactin biosynthesis: asbF encodes a (−)-3-dehydroshikimate dehydratase. Biochemistry 2008, 47, 12251–12253. [Google Scholar] [CrossRef]
- Pfleger, B.F.; Kim, Y.; Nusca, T.D.; Maltseva, N.; Lee, J.Y.; Rath, C.M.; Scaglione, J.B.; Janes, B.K.; Anderson, E.C.; Bergman, N.H.; et al. Structural and functional analysis of AsbF: Origin of the stealth 3,4-dihydroxybenzoic acid subunit for petrobactin biosynthesis. Proc. Natl. Acad. Sci. USA 2008, 105, 17133–17138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hawkins, A.R.; Giles, N.H.; Kinghorn, J.R. Genetical and biochemical aspects of quinate breakdown in the filamentous fungus Aspergillus nidulans. Biochem. Genet. 1982, 20, 271–286. [Google Scholar] [CrossRef]
- Shmonova, E.A.; Voloshina, O.V.; Ovsienko, M.V.; Smirnov, S.V.; Nolde, D.E.; Doroshenko, V.G. Characterization of the Corynebacterium glutamicum dehydroshikimate dehydratase QsuB and its potential for microbial production of protocatechuic acid. PLoS ONE 2020, 15, e0231560. [Google Scholar] [CrossRef]
- Draths, K.M.; Frost, J.W. Environmentally Compatible Synthesis of Catechol from D-Glucose. J. Am. Chem. Soc. 1995, 117, 2395–2400. [Google Scholar] [CrossRef]
- Li, K.; Frost, J.W. Synthesis of Vanillin from Glucose. J. Am. Chem. Soc. 1998, 120, 10545–10546. [Google Scholar] [CrossRef]
- Niu, W.; Draths, K.M.; Frost, J.W. Benzene-free synthesis of adipic acid. Biotechnol. Prog. 2002, 18, 201–211. [Google Scholar] [CrossRef]
- Hansen, E.H.; Møller, B.L.; Kock, G.R.; Bünner, C.M.; Kristensen, C.; Jensen, O.R.; Okkels, F.T.; Olsen, C.E.; Motawia, M.S.; Hansen, J. De novo biosynthesis of vanillin in fission yeast (Schizosaccharomyces pombe) and baker’s yeast (Saccharomyces cerevisiae). Appl. Environ. Microbiol. 2009, 75, 2765–2774. [Google Scholar] [CrossRef] [Green Version]
- Weber, C.; Brückner, C.; Weinreb, S.; Lehr, C.; Essl, C.; Boles, E. Biosynthesis of cis,cis-muconic acid and its aromatic precursors, catechol and protocatechuic acid, from renewable feedstocks by Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2012, 78, 8421–8430. [Google Scholar] [CrossRef] [Green Version]
- Örn, O.E.; Sacchetto, S.; van Niel, E.W.J.; Hatti-Kaul, R. Enhanced Protocatechuic Acid Production From Glucose Using Pseudomonas putida 3-Dehydroshikimate Dehydratase Expressed in a Phenylalanine-Overproducing Mutant of Escherichia coli. Front. Bioeng. Biotechnol. 2021, 9, 695704. [Google Scholar] [CrossRef]
- Kogure, T.; Suda, M.; Hiraga, K.; Inui, M. Protocatechuate overproduction by Corynebacterium glutamicum via simultaneous engineering of native and heterologous biosynthetic pathways. Metab. Eng. 2021, 65, 232–242. [Google Scholar] [CrossRef]
- Sambrook, J.R.D. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2001. [Google Scholar]
- Datsenko, K.A.; Wanner, B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 2000, 97, 6640–6645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Minaeva, N.I.; Gak, E.R.; Zimenkov, D.V.; Skorokhodova, A.Y.; Biryukova, I.V.; Mashko, S.V. Dual-In/Out strategy for genes integration into bacterial chromosome: A novel approach to step-by-step construction of plasmid-less marker-less recombinant E. coli strains with predesigned genome structure. BMC Biotechnol. 2008, 8, 63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smirnov, S.V.; Kodera, T.; Samsonova, N.N.; Kotlyarova, V.A.; Rushkevich, N.Y.; Kivero, A.D.; Sokolov, P.M.; Hibi, M.; Ogawa, J.; Shimizu, S. Metabolic engineering of Escherichia coli to produce (2S, 3R, 4S)-4-hydroxyisoleucine. Appl. Microbiol. Biotechnol. 2010, 88, 719–726. [Google Scholar] [CrossRef] [PubMed]
- Katashkina, Z.; Skorokhodova, A.; Zimenkov, D.V.; Gulevich, A.; Minaeva, N.I.; Doroshenko, V.G.; Biriukova, I.V.; Mashko, S.V. Tuning of expression level of the genes of interest located in the bacterial chromosome. Mol. Biol. (Moscow Russ. Fed. Engl. Ed.) 2005, 39, 823–831. [Google Scholar]
- Quan, J.; Tian, J. Circular polymerase extension cloning of complex gene libraries and pathways. PLoS ONE 2009, 4, e6441. [Google Scholar] [CrossRef]
- Strøman, P.; Reinert, W.R.; Giles, N.H. Purification and characterization of 3-dehydroshikimate dehydratase, an enzyme in the inducible quinic acid catabolic pathway of Neurospora crassa. J. Biol. Chem. 1978, 253, 4593–4598. [Google Scholar] [CrossRef]
- Notredame, C.; Higgins, D.G.; Heringa, J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000, 302, 205–217. [Google Scholar] [CrossRef] [Green Version]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview Version 2-a multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef] [Green Version]
- Dereeper, A.; Guignon, V.; Blanc, G.; Audic, S.; Buffet, S.; Chevenet, F.; Dufayard, J.F.; Guindon, S.; Lefort, V.; Lescot, M.; et al. Phylogeny.fr: Robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008, 36, W465–W469. [Google Scholar] [CrossRef]
- Roy, A.; Kucukural, A.; Zhang, Y. I-TASSER: A unified platform for automated protein structure and function prediction. Nat. Protoc. 2010, 5, 725–738. [Google Scholar] [CrossRef] [Green Version]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kunjapur, A.M.; Hyun, J.C.; Prather, K.L.J. Deregulation of S-adenosylmethionine biosynthesis and regeneration improves methylation in the E. coli de novo vanillin biosynthesis pathway. Microb. Cell Fact. 2016, 15, 61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harrington, L.B.; Jha, R.K.; Kern, T.L.; Schmidt, E.N.; Canales, G.M.; Finney, K.B.; Koppisch, A.T.; Strauss, C.E.; Fox, D.T. Rapid Thermostabilization of Bacillus thuringiensis Serovar Konkukian 97–27 Dehydroshikimate Dehydratase through a Structure-Based Enzyme Design and Whole Cell Activity Assay. ACS Synth. Biol. 2017, 6, 120–129. [Google Scholar] [CrossRef]
- Gross, S.R. The enzymatic conversion of 5-dehydroshikimic acid to protocatechuic acid. J. Biol. Chem. 1958, 233, 1146–1151. [Google Scholar] [CrossRef]
- Olins, P.O.; Rangwala, S.H. A novel sequence element derived from bacteriophage T7 mRNA acts as an enhancer of translation of the lacZ gene in Escherichia coli. J. Biol. Chem. 1989, 264, 16973–16976. [Google Scholar] [CrossRef]
- Seo, S.W.; Yang, J.S.; Kim, I.; Yang, J.; Min, B.E.; Kim, S.; Jung, G.Y. Predictive design of mRNA translation initiation region to control prokaryotic translation efficiency. Metab. Eng. 2013, 15, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Draths, K.M.; Knop, D.R.; Frost, J.W. Shikimic Acid and Quinic Acid: Replacing Isolation from Plant Sources with Recombinant Microbial Biocatalysis. J. Am. Chem. Soc. 1999, 121, 1603–1604. [Google Scholar] [CrossRef]
- Kubota, T.; Tanaka, Y.; Hiraga, K.; Inui, M.; Yukawa, H. Characterization of shikimate dehydrogenase homologues of Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 2013, 97, 8139–8149. [Google Scholar] [CrossRef]
- Xue, K.; Prezioso, S.M.; Christendat, D. QuiC2 represents a functionally distinct class of dehydroshikimate dehydratases identified in Listeria species including Listeria monocytogenes. Environ. Microbiol. 2020, 22, 2680–2692. [Google Scholar] [CrossRef]
- Wei, K.; Long, L.; Lin, Q.; Ding, S. Functional characterization of a new 3-dehydroshikimate dehydratase from Eupenicillium parvum and its potential for protocatechuic acid production. Biosci. Biotechnol. Biochem. 2022, zbac078, Published online ahead of print, 24 May 2022. [Google Scholar] [CrossRef]


| Strain or Plasmid | Relevant Characteristics | Source and Description |
|---|---|---|
| E. coli strains | ||
| MG1655 | Laboratory strain E. coli K12 F− λ− ilvG− rfb-50 rph-1 | VKPM a B6195 |
| BL21(DE3) | The strain was used for the expression of genes cloned in the pET22b vector. F–ompT gal dcm lon hsdSB(rB–mB–) λ(DE3 [lacI lacUV5-T7p07 ind1 sam7 nin5]) [malB+]K-12(λS) | Novagen (Merck Millipore, Darmstadt, Germany) |
| MG1655 ∆aroE | The DHS accumulating strain containing in-frame deletion of the aroE gene | [13] |
| MG1655 ∆aroE PlacUV5-qsuBattφ80 | 3,4-DHBA producing strains containing DSD genes integrated into the bacterial chromosome and expressed using the IPTG-inducible promoter PlacUV5 | [13] |
| MG1655 ∆aroE PlacUV5-asbFattφ80 | The integration of PlacUV5-asbF and PlacUV5-qa-4 was carried out using “Dual In/Out” method [23] followed by marker excision. | |
| MG1655 ∆aroE PlacUV5-qa-4attφ80 | ||
| Plasmids | ||
| pAH162-λattL-TcR-λattR | The integrative vector for the “Dual In/Out” method | [23] |
| pELAC | A template for the PlacUV5 promoter amplification, ApR | [24] |
| pAH123 | The helper plasmid containing phage φ80 integrase for the “Dual In/Out” method, ApR | [23] |
| pMW-int-xis | The helper plasmid for marker removal: oriR101, repA101ts, λcIts857, λP R → λxis-int, ApR | [25] |
| pET22b | The vector for protein expression, ApR | Novagen (Merck Millipore, Darmstadt, Germany) |
| pET22b-qsuB | The plasmid was used for the production of His-tagged QsuB | [13] |
| pET22b- asbF | The plasmid was used for the production of His-tagged AsbF | The DNA fragment containing pET22b was amplified using the primers P1/P2. The DNA fragments of asbF and qa-4 were amplified using synthetic DNA fragments as templates and the primers P3/P4 and P5/P6, respectively. |
| pET22b- qa-4 | The plasmid was used for the production of His-tagged Qa-4 | |
| pAH162-λattL-TcR-λattR- Plac-asbF | The integrative plasmids containing the PlacUV5-asbF modification | The DNA fragment containing PlacUV5-DSD was cloned into pAH162-λattL-TcR-λattR between the SalI and SacI restriction sites. PlacUV5-asbF and PlacUV5-qa-4 were obtained by overlapping PCR (the primers P7/P10 and P7/P12, respectively) of DNA fragments containing the promoter PlacUV5 and DSD coding region. These DNA fragments were amplified by PCR with the primers P7/P8 for PlacUV5 and P9/P10 for asbF, P11/P12 for qa-4 using pELAC and pET22b-DSD plasmids as templates, respectively. |
| pAH162-λattL-TcR-λattR-Plac-qa-4 | The integrative plasmids containing the PlacUV5-qa-4 modification | |
| Primer | Oligonucleotide Sequence |
|---|---|
| P1 | ATGTATATCTCCTTCTTAAAGTTAAACAAAA |
| P2 | CACCACCACCACCACCACTGAGATCCGGCTGCTAACAAAG |
| P3 | TTTTGTTTAACTTTAAGAAGGAGATATACATATGAAATATTCGCTTTGTACTATTAGCT |
| P4 | GTGGTGGTGGTGGTGGTGCGAAGTAACAACTTCCAGTTTGCGA |
| P5 | TTTTGTTTAACTTTAAGAAGGAGATATACATATGCCATCGAAACTAGCAATATCGAGCA |
| P6 | GTGGTGGTGGTGGTGGTGTAACGACGCGGAAACTCGC |
| P7 | TTTTTGTCGACTCTAGAGGATCTGCGGGCAG |
| P8 | ATGTATATCTCCTTCTTAAATCTAGATCCTGTGTGAAATTGTTATCC |
| P9 | TTTAAGAAGGAGATATACATATGAAATATTCG |
| P10 | TCACGAAGTAACAACTTCCAGTTTGCGA |
| P11 | TTTAAGAAGGAGATATACATATGCCATCGAA |
| P12 | TCATGTAACGACGCGGAAACTCGC |
| Enzyme | Km (DHS), µM | kcat, s−1 | kcat/Km, (103 μM−1 s−1) |
|---|---|---|---|
| Qa-4 | 598 ± 16 | 218.6 ± 1.1 | 365.6 ± 68.8 |
| QsuB | 961 ± 77 | 60.8 ± 0.9 | 63.3 ± 11.7 |
| AsbF | 36 ± 7 | 1.1 ± 0.1 | 29.0 ± 7.1 |
| MG1655 ∆aroE Strain | OD540 | DHS, g/L | 3.4-DHBA, g/L | Residual Glucose, g/L | 1 mM IPTG |
|---|---|---|---|---|---|
| - | 31 ± 1 | 3.3 ± 0.1 | <0.1 | 10.0 ± 0.3 | + |
| 31 ± 1 | 3.4 ± 0.2 | <0.1 | 9.5 ± 0.2 | - | |
| PlacUV5-asbFattφ80 | 31 ± 1 | 2.3 ± 0.1 | 0.20 ± 0.01 | 9.5 ± 0.1 | + |
| 30 ± 1 | 2.3 ± 0.1 | <0.1 | 9.9 ± 0.3 | - | |
| PlacUV5-qsuBattφ80 | 30 ± 1 | 0.2 ± 0.1 | 2.7 ± 0.2 | 11.0 ± 1.5 | + |
| 30 ± 1 | 2.1 ± 0.1 | 1.0 ± 0.1 | 10.2 ± 0.2 | - | |
| PlacUV5-qa-4attφ80 | 29 ± 1 | 0.2 ± 0.1 | 2.7 ± 0.1 | 12.0 ± 0.8 | + |
| 29 ± 1 | 0.8 ± 0.1 | 2.1 ± 0.1 | 11.0 ± 0.4 | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shmonova, E.A.; Savrasova, E.A.; Fedorova, E.N.; Doroshenko, V.G. Comparative Analysis of Catabolic and Anabolic Dehydroshikimate Dehydratases for 3,4-DHBA Production in Escherichia coli. Microorganisms 2022, 10, 1357. https://doi.org/10.3390/microorganisms10071357
Shmonova EA, Savrasova EA, Fedorova EN, Doroshenko VG. Comparative Analysis of Catabolic and Anabolic Dehydroshikimate Dehydratases for 3,4-DHBA Production in Escherichia coli. Microorganisms. 2022; 10(7):1357. https://doi.org/10.3390/microorganisms10071357
Chicago/Turabian StyleShmonova, Ekaterina A., Ekaterina A. Savrasova, Elizaveta N. Fedorova, and Vera G. Doroshenko. 2022. "Comparative Analysis of Catabolic and Anabolic Dehydroshikimate Dehydratases for 3,4-DHBA Production in Escherichia coli" Microorganisms 10, no. 7: 1357. https://doi.org/10.3390/microorganisms10071357
APA StyleShmonova, E. A., Savrasova, E. A., Fedorova, E. N., & Doroshenko, V. G. (2022). Comparative Analysis of Catabolic and Anabolic Dehydroshikimate Dehydratases for 3,4-DHBA Production in Escherichia coli. Microorganisms, 10(7), 1357. https://doi.org/10.3390/microorganisms10071357