Characterization of the Soil Bacterial Community from Selected Boxwood Gardens across the United States
Abstract
:1. Introduction
2. Materials and Methods
2.1. Boxwood Gardens
2.2. Soil Sampling
2.3. DNA Extraction and PCR Amplification
2.4. Nanopore Library Preparation and Sequencing
2.5. Database Construction
2.6. Bioinformatics
2.7. Data Analyses and Statistics
2.7.1. Community Diversity and Structure
2.7.2. Identification of Major Bacterial Orders
2.7.3. Identification of Bacterial Species with Biological Control Potential
3. Results
3.1. Sequencing Summary
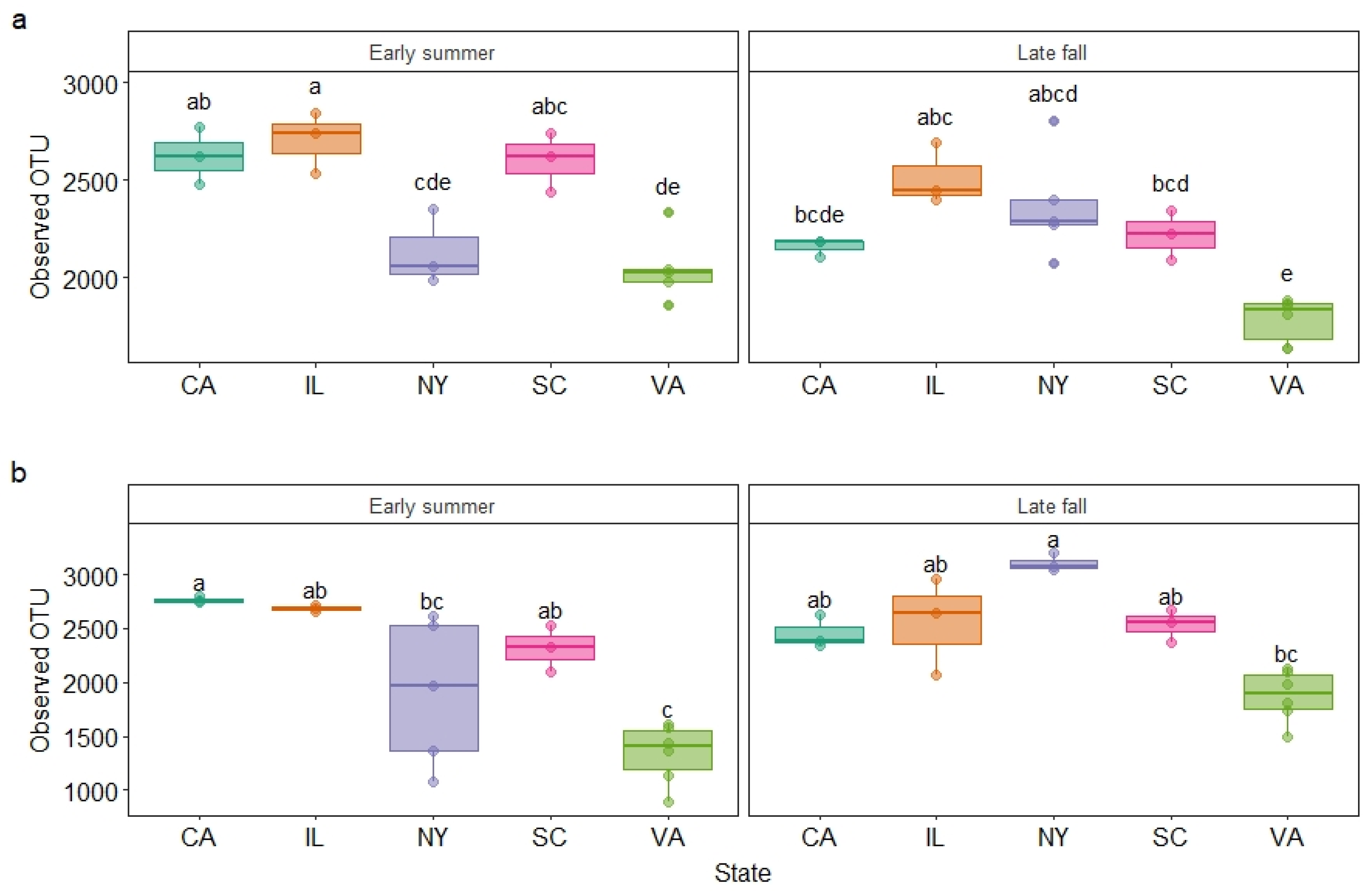
3.2. Garden Soil Bacterial Community Diversity
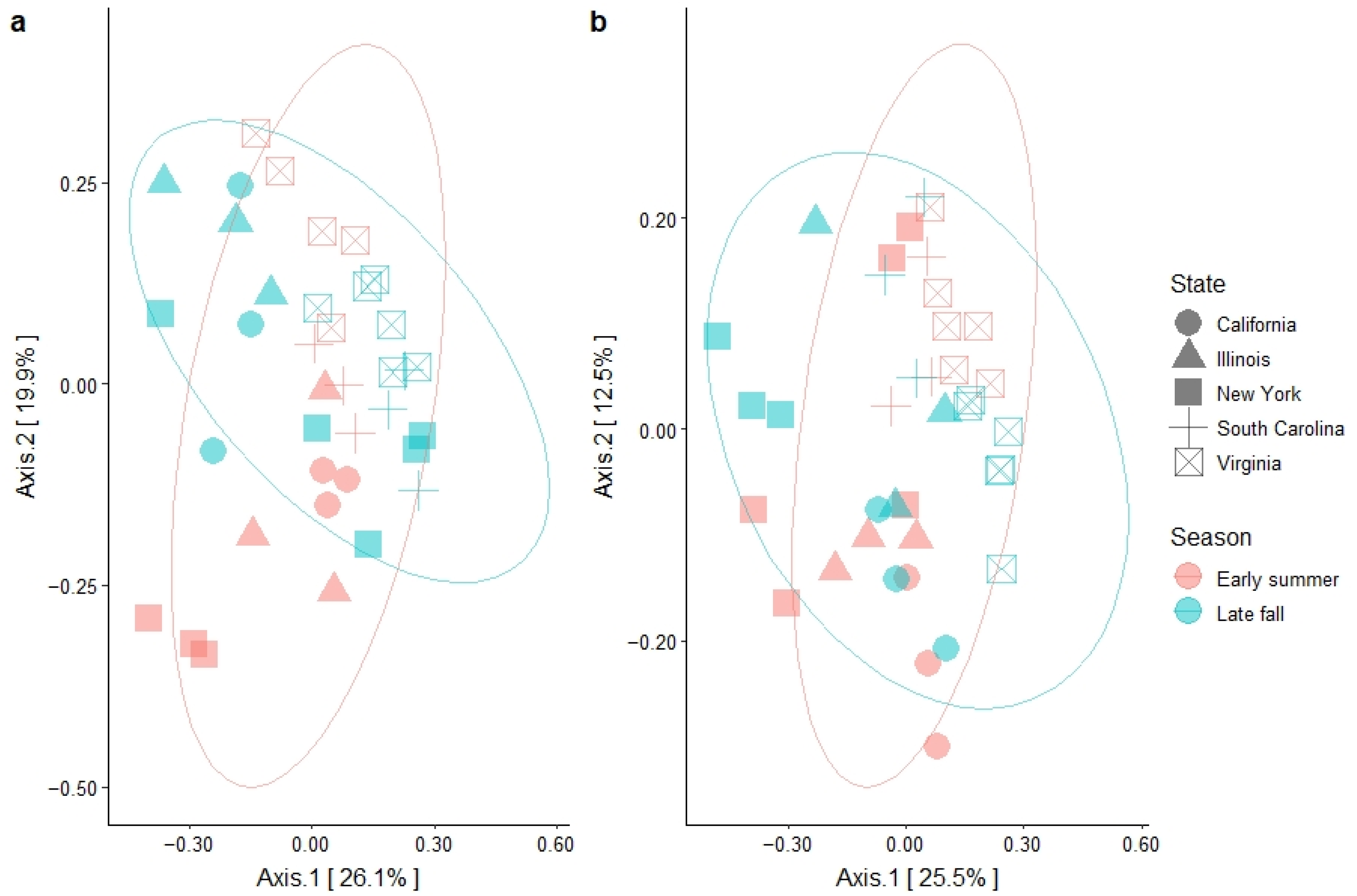
3.3. Garden Soil Bacterial Community Structure
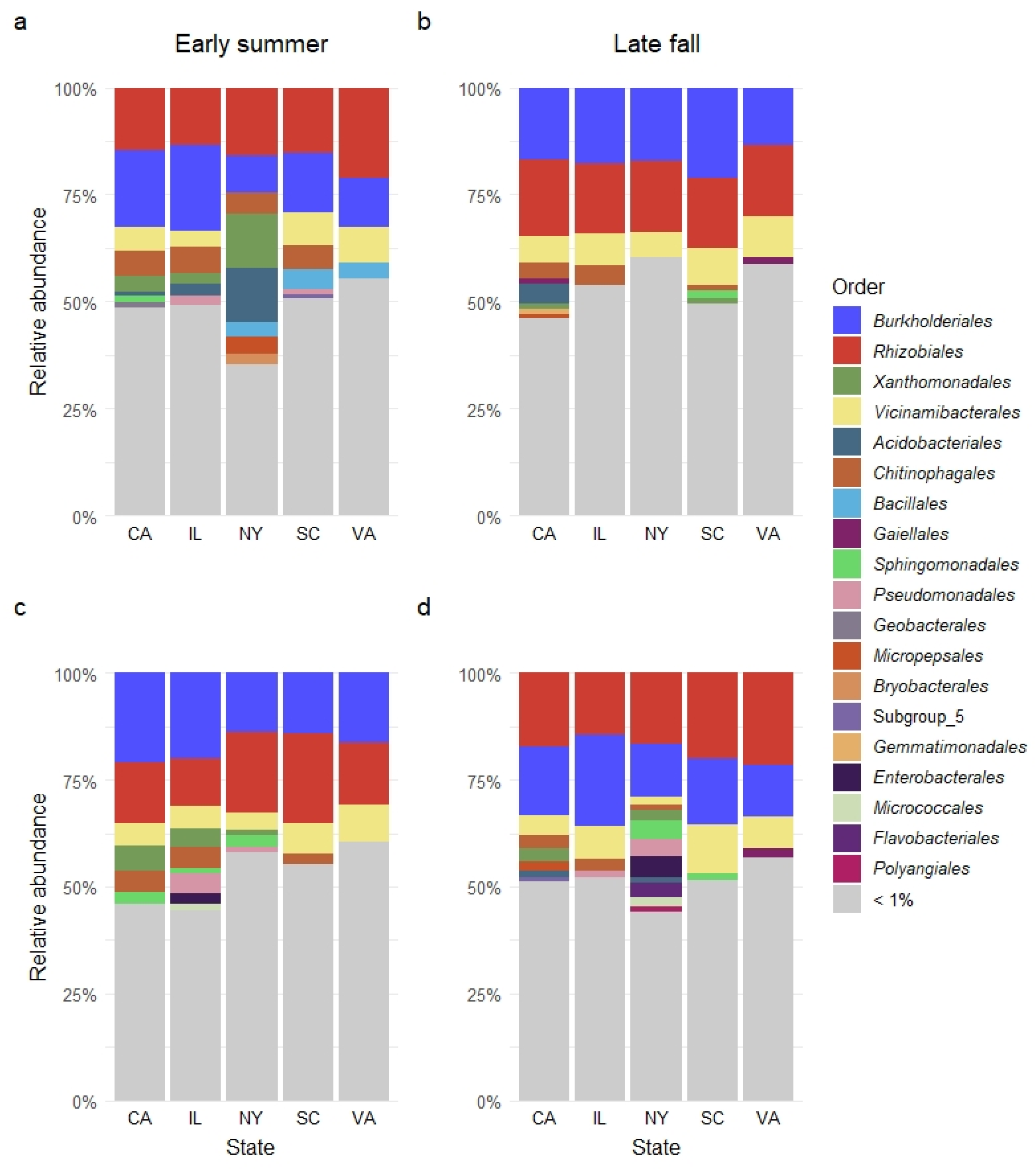
3.4. Relative Abundance of Bacterial Orders in Sampled Garden Soil
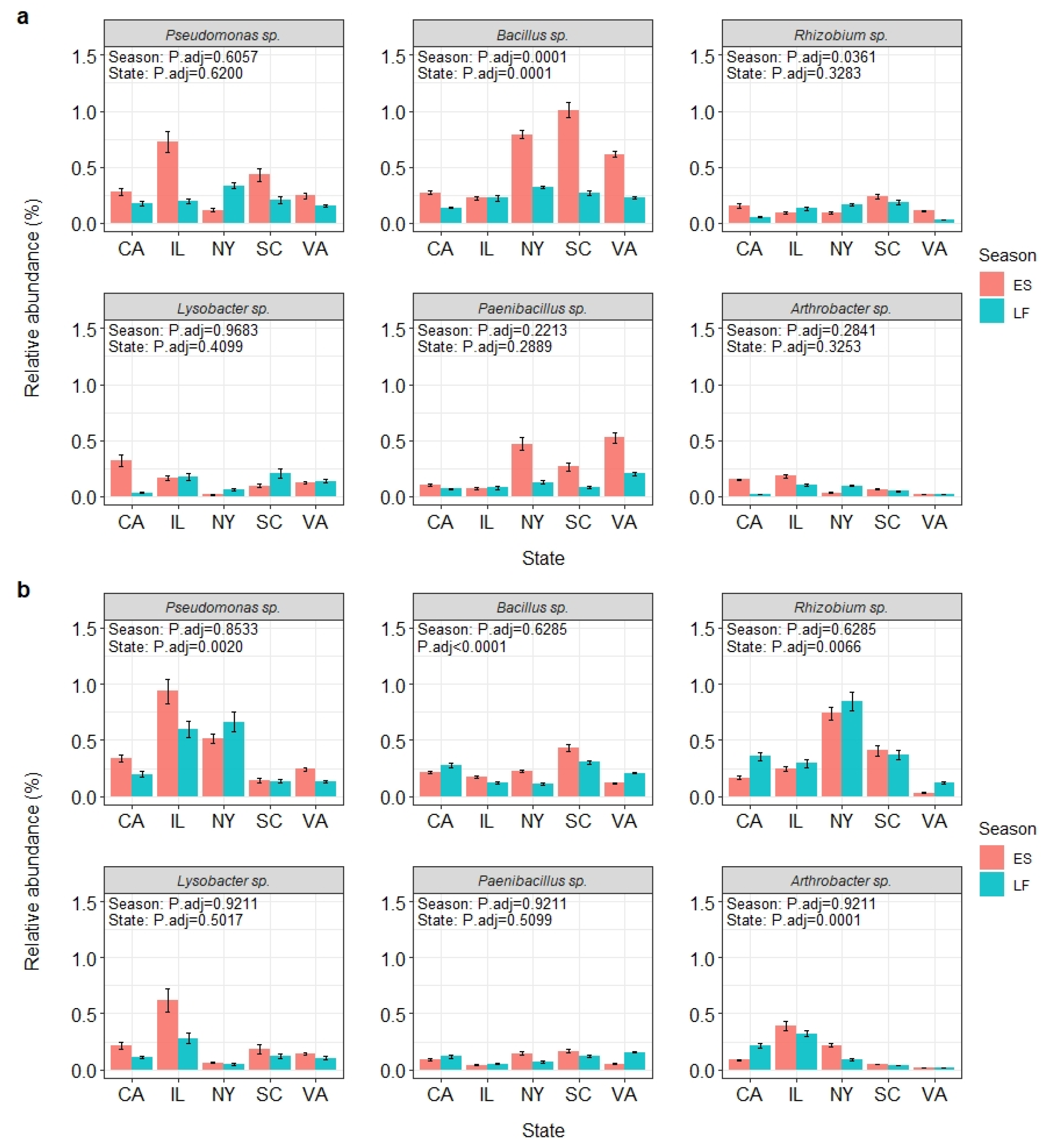
3.5. Bacterial Species with Biological Control Potential
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Baker, K.F.; Cook, R.J. Biological Control of Plant Pathogens; W. H. Freeman: San Francisco, CA, USA, 1974. [Google Scholar]
- Jayaraman, S.; Naorem, A.K.; Lal, R.; Dalal, R.C.; Sinha, N.K.; Patra, A.K.; Chaudhari, S.K. Disease-Suppressive Soils—Beyond Food Production: A Critical Review. J. Soil Sci. Plant Nutr. 2021, 21, 1437–1465. [Google Scholar] [CrossRef] [PubMed]
- Janvier, C.; Villeneuve, F.-O.; Alabouvette, C.; Véronique Edel-Hermann, V.; Mateille, T.; Steinberg, C. Soil Health through Soil Disease Suppression: Which Strategy from Descriptors to Indicators? Soil Biol. Biochem. 2007, 39, 1–23. [Google Scholar] [CrossRef]
- Wei, Z.; Gu, Y.; Friman, V.P.; Kowalchuk, G.A.; Xu, Y.; Shen, Q.; Jousset, A. Initial Soil Microbiome Composition and Functioning Predetermine Future Plant Health. Sci. Adv. 2019, 5, 759–784. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Larkin Robert, P.; Hopkins Donald, L.; Martin Frank, N. Suppression of Fusarium Wilt of Watermelon by Nonpathogenic Fusarium oxysporum and Other Microorganisms Recovered from a Disease-Suppressive Soil. Phytopathology 1996, 86, 812–819. [Google Scholar] [CrossRef]
- Henry, A.W. The Natural Microflora of the Soil in Relation to the Foot-Rot Problem of Wheat. Can. J. Res. 1931, 4, 69–77. [Google Scholar] [CrossRef]
- Goh, Y.K.; Zarul, M.; Zoqratt, H.M.; Goh, Y.K.; Ayub, Q.; Su, A.; Ting, Y. Discovering Naturally-Occurring Microbiota in Disease Suppressive Soil: Potential Role of Biological Elements in Suppressing Ganoderma boninense. Biol. Control 2022, 165, 104787. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, Q.; Cui, H.; Li, Y.; Xu, N.; Lu, T.; Chen, J.; Penuelas, J.; Hu, B.; Qian, H. Composition Identification and Functional Verification of Bacterial Community in Disease-Suppressive Soils by Machine Learning. Environ. Microbiol. 2022. [Google Scholar] [CrossRef]
- Mendes, R.; Kruijt, M.; de Bruijn, I.; Dekkers, E.; van der Voort, M.; Schneider, J.H.M.; Piceno, Y.M.; DeSantis, T.Z.; Andersen, G.L.; Bakker, P.A.H.M.; et al. Deciphering the Rhizosphere Microbiome for Disease-Suppressive Bacteria. Science 2011, 332, 1097–1100. [Google Scholar] [CrossRef]
- Schlatter, D.; Kinkel, L.; Thomashow, L.; Weller, D.; Paulitz, T. Disease Suppressive Soils: New Insights from the Soil Microbiome. Phytopathology 2017, 107, 1284–1297. [Google Scholar] [CrossRef] [Green Version]
- Jamil, F.N.; Hashim, A.M.; Yusof, M.T.; Saidi, N.B. Analysis of Soil Bacterial Communities and Physicochemical Properties Associated with Fusarium Wilt Disease of Banana in Malaysia. Sci. Rep. 2022, 12, 999. [Google Scholar] [CrossRef]
- Raaijmakers, J.M.; Mazzola, M. Soil Immune Responses. Science 2016, 352, 1392–1393. [Google Scholar] [CrossRef]
- Garbeva, P.; Postma, J.; van Veen, J.A.; van Elsas, J.D. Effect of Above-Ground Plant Species on Soil Microbial Community Structure and Its Impact on Suppression of Rhizoctonia solani AG3. Environ. Microbiol. 2006, 8, 233–246. [Google Scholar] [CrossRef]
- Weller, D.M.; Raaijmakers, J.M.; McSpadden Gardener, B.B.; Thomashow, L.S. Microbial Populations Responsible for Specific Soil Suppressiveness to Plant Pathogens. Annu. Rev. Phytopathol. 2002, 40, 309–348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Köhl, J.; Kolnaar, R.; Ravensberg, W.J. Mode of Action of Microbial Biological Control Agents against Plant Diseases: Relevance beyond Efficacy. Front. Plant Sci. 2019, 10, 845. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, X.; Zhang, S.; Jiang, Q.; Bai, Y.; Shen, G.; Li, S.; Ding, W. Using Community Analysis to Explore Bacterial Indicators for Disease Suppression of Tobacco Bacterial Wilt. Sci. Rep. 2016, 6, 36773. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Bruggen, A.H.C.; Semenov, A.M. A New Approach to the Search for Indicators of Root Disease Suppression. Australas. Plant Pathol. 1999, 28, 4–10. [Google Scholar] [CrossRef]
- de Boer, M.; van der Sluis, I.; van Loon, L.C.; Bakker, P.A.H.M. Combining Fluorescent Pseudomonas spp. Strains to Enhance Suppression of Fusarium Wilt of Radish. Eur. J. Plant Pathol. 1999, 105, 201–210. [Google Scholar] [CrossRef]
- Kumar, P.; Dubey, R.C.; Maheshwari, D.K. Bacillus Strains Isolated from Rhizosphere Showed Plant Growth Promoting and Antagonistic Activity against Phytopathogens. Microbiol. Res. 2012, 167, 493–499. [Google Scholar] [CrossRef]
- Cordovez, V.; Carrion, V.J.; Etalo, D.W.; Mumm, R.; Zhu, H.; van Wezel, G.P.; Raaijmakers, J.M. Diversity and Functions of Volatile Organic Compounds Produced by Streptomyces from a Disease-Suppressive Soil. Front. Microbiol. 2015, 6, 01081. [Google Scholar] [CrossRef] [Green Version]
- Expósito, R.G.; de Bruijn, I.; Postma, J.; Raaijmakers, J.M. Current Insights into the Role of Rhizosphere Bacteria in Disease Suppressive Soils. Front. Microbiol. 2017, 8, 2529. [Google Scholar] [CrossRef]
- Albayrak, Ç.B. Bacillus Species as Biocontrol Agents for Fungal Plant Pathogens. In Bacilli and Agrobiotechnology: Phytostimulation and Biocontrol: Volume 2; Islam, M.T., Rahman, M.M., Pandey, P., Boehme, M.H., Haesaert, G., Eds.; Springer International Publishing: Cham, Switzerland, 2019; pp. 239–265. [Google Scholar] [CrossRef]
- Postma, J.; Schilder, M.T.; Bloem, J.; van Leeuwen-Haagsma, W.K. Soil Suppressiveness and Functional Diversity of the Soil Microflora in Organic Farming Systems. Soil Biol. Biochem. 2008, 40, 2394–2406. [Google Scholar] [CrossRef] [Green Version]
- Peralta, A.L.; Sun, Y.; McDaniel, M.D.; Lennon, J.T. Crop Rotational Diversity Increases Disease Suppressive Capacity of Soil Microbiomes. Ecosphere 2018, 9, e02235. [Google Scholar] [CrossRef]
- Bonanomi, G.; de Filippis, F.; Zotti, M.; Idbella, M.; Cesarano, G.; Al-Rowaily, S.; Abd-Elgawad, A. Repeated Applications of Organic Amendments Promote Beneficial Microbiota, Improve Soil Fertility and Increase Crop Yield. Appl. Soil Ecol. 2020, 156, 103714. [Google Scholar] [CrossRef]
- Bonanomi, G.; Lorito, M.; Vinale, F.; Woo, S.L. Organic Amendments, Beneficial Microbes, and Soil Microbiota: Toward a Unified Framework for Disease Suppression. Annu. Rev. Phytopathol. 2018, 56, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Raaijmakers, J.M.; Weller, D.M. Natural Plant Protection by 2,4-Diacetylphloroglucinol-Producing Pseudomonas spp. in Take-All Decline Soils. Mol. Plant-Microbe Interact. 1998, 11, 144–152. [Google Scholar] [CrossRef] [Green Version]
- Kwak, M.J.; Kong, H.G.; Choi, K.; Kwon, S.K.; Song, J.Y.; Lee, J.; Lee, P.A.; Choi, S.Y.; Seo, M.; Lee, H.J.; et al. Rhizosphere Microbiome Structure Alters to Enable Wilt Resistance in Tomato. Nat. Biotechnol. 2018, 36, 1100–1116. [Google Scholar] [CrossRef] [PubMed]
- Mazzola, M. Mechanisms of Natural Soil Suppressiveness to Soilborne Diseases. Antonie Van Leeuwenhoek 2002, 81, 557–564. [Google Scholar] [CrossRef]
- LeBlanc, N.; Crouch, J.A. Prokaryotic Taxa Play Keystone Roles in the Soil Microbiome Associated with Woody Perennial Plants in the Genus Buxus. Ecol. Evol. 2019, 9, 11102–11111. [Google Scholar] [CrossRef] [Green Version]
- Daughtrey, M.L. Boxwood Blight: Threat to Ornamentals. Annu. Rev. Phytopathol. 2019, 57, 189–209. [Google Scholar] [CrossRef]
- Hong, C. Saving American Gardens from Boxwood Blight. J. Am. Boxwood Soc. 2019, 58, 4–10. [Google Scholar]
- Hong, C.; Daughtrey, M.; Howle, M.; Schirmer, S.; Kosta, K.; Kong, P.; Pychlik, P.; Williamson, M.; Mirmow, N.; Suslow, K. Rapid Decline of Calonectria pseudonaviculata Soil Population in Selected Gardens across the United States. Plant Dis. 2022. [Google Scholar] [CrossRef]
- Kinkel, L.L.; Bakker, M.G.; Schlatter, D.C. A Coevolutionary Framework for Managing Disease-Suppressive Soils. Annu. Rev. Phytopathol. 2011, 49, 47–67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of General 16S Ribosomal RNA Gene PCR Primers for Classical and Next-Generation Sequencing-Based Diversity Studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 2013, 41, 590–596. [Google Scholar] [CrossRef]
- Yilmaz, P.; Parfrey, L.W.; Yarza, P.; Gerken, J.; Pruesse, E.; Quast, C.; Schweer, T.; Peplies, J.; Ludwig, W.; Glöckner, F.O. The SILVA and “All-Species Living Tree Project (LTP)” Taxonomic Frameworks. Nucleic Acids Res. 2014, 42, D643–D648. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science Using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Robeson, M.S.; O’Rourke, D.R.; Kaehler, B.D.; Ziemski, M.; Dillon, M.R.; Foster, J.T.; Bokulich, N.A. RESCRIPt: Reproducible Sequence Taxonomy Reference Database Management. PLOS Comput. Biol. 2021, 17, e1009581. [Google Scholar] [CrossRef]
- Kim, D.; Song, L.; Breitwieser, F.P.; Salzberg, S.L. Centrifuge: Rapid and Sensitive Classification of Metagenomic Sequences. Genome Res. 2016, 26, 1721–1729. [Google Scholar] [CrossRef] [Green Version]
- De Coster, W.; D’Hert, S.; Schultz, D.T.; Cruts, M.; Van Broeckhoven, C. NanoPack: Visualizing and Processing Long-Read Sequencing Data. Bioinformatics 2018, 34, 2666–2669. [Google Scholar] [CrossRef]
- Li, H. Minimap2: Pairwise Alignment for Nucleotide Sequences. Bioinformatics 2018, 34, 3094–3100. [Google Scholar] [CrossRef]
- Marijon, P.; Chikhi, R.; Varré, J.S. Yacrd and Fpa: Upstream Tools for Long-Read Genome Assembly. Bioinformatics 2020, 36, 3894–3896. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical: Vienna, Austria, 2020. [Google Scholar]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mikryukov, V. MetagMisc: Miscellaneous Functions for Metagenomic Analysis. R Version 0.0.4. 2021. Available online: https://github.com/vmikk/metagMisc/ (accessed on 18 May 2022).
- Chao, A.; Jost, L. Coverage-Based Rarefaction and Extrapolation: Standardizing Samples by Completeness Rather than Size. Ecology 2012, 93, 2533–2547. [Google Scholar] [CrossRef] [PubMed]
- Daniel, L. sjstats: Statistical Functions for Regression Models. Version 0.18.1. 2021. Available online: https://doi.org/10.5281/zenodo.1284472 (accessed on 27 June 2022).
- Cohen, J. Statistical Power Analysis for the Behavioral Sciences, 2nd ed.; Routledge: New York, NY, USA, 1988. [Google Scholar] [CrossRef]
- Bray, J.R.; Curtis, J.T. An Ordination of the Upland Forest Communities of Southern Wisconsin. Ecol. Monogr. 1957, 27, 325–349. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package. R Package Version 2.5-7. 2013. Available online: https://CRAN.R-project.org/package=vegan (accessed on 9 May 2022).
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Kruskal, W.H.; Wallis, W.A. Use of Ranks in One-Criterion Variance Analysis. J. Am. Stat. Assoc. 1952, 47, 583–621. [Google Scholar] [CrossRef]
- Morales-Cedeño, L.R.; Orozco-Mosqueda, M.d.C.; Loeza-Lara, P.D.; Parra-Cota, F.I.; de los Santos-Villalobos, S.; Santoyo, G. Plant Growth-Promoting Bacterial Endophytes as Biocontrol Agents of Pre- and Post-Harvest Diseases: Fundamentals, Methods of Application and Future Perspectives. Microbiol. Res. 2021, 242, 126612. [Google Scholar] [CrossRef]
- Deketelaere, S.; Tyvaert, L.; França, S.C.; Hofte, M. Desirable Traits of a Good Biocontrol Agent against Verticillium Wilt. Front. Microbiol. 2017, 8, 1186. [Google Scholar] [CrossRef] [Green Version]
- Islam, M.T. Potentials for Biological Control of Plant Diseases by Lysobacter spp., with Special Reference to Strain SB-K88. In Bacteria in Agrobiology: Plant Growth Responses; Maheshwari, D.K., Ed.; Springer: Berlin/Heidelberg, Germany, 2011; pp. 335–363. [Google Scholar] [CrossRef]
- Khan, N.I.; Filonow, A.B.; Singleton, L.L. Augmentation of Soil with Sporangia of Actinoplanes spp. for Biological Control of Pythium Damping-Off. Biocontrol Sci. Technol. 1997, 7, 11–22. [Google Scholar] [CrossRef]
- Seo, H.J.; Park, A.R.; Kim, S.; Yeon, J.; Yu, N.H.; Ha, S.; Chang, J.Y.; Park, H.W.; Kim, J.C. Biological Control of Root-Knot Nematodes by Organic Acid-Producing Lactobacillus brevis Wikim0069 Isolated from Kimchi. Plant Pathol. J. 2019, 35, 662–673. [Google Scholar] [CrossRef]
- Shen, Z.; Ruan, Y.; Xue, C.; Zhong, S.; Li, R.; Shen, Q. Soils Naturally Suppressive to Banana Fusarium Wilt Disease Harbor Unique Bacterial Communities. Plant Soil 2015, 393, 21–33. [Google Scholar] [CrossRef]
- Inderbitzin, P.; Ward, J.; Barbella, A.; Solares, N.; Izyumin, D.; Burman, P.; Chellemi, D.O.; Subbarao, K.V. Soil Microbiomes Associated with Verticillium Wilt-Suppressive Broccoli and Chitin Amendments Are Enriched with Potential Biocontrol Agents. Phytopathology 2018, 108, 31–43. [Google Scholar] [CrossRef] [Green Version]
- Lazcano, C.; Boyd, E.; Holmes, G.; Hewavitharana, S.; Pasulka, A.; Ivors, K. The Rhizosphere Microbiome Plays a Role in the Resistance to Soil-Borne Pathogens and Nutrient Uptake of Strawberry Cultivars under Field Conditions. Sci. Rep. 2021, 11, 3188. [Google Scholar] [CrossRef]
- Nisrina, L.; Effendi, Y.; Pancoro, A. Revealing the Role of Plant Growth Promoting Rhizobacteria in Suppressive Soils against Fusarium oxysporum f. sp. cubense Based on Metagenomic Analysis. Heliyon 2021, 7, e07636. [Google Scholar] [CrossRef] [PubMed]
- Chaparro, J.M.; Sheflin, A.M.; Manter, D.K.; Vivanco, J.M. Manipulating the Soil Microbiome to Increase Soil Health and Plant Fertility. Biol. Fertil. Soils 2012, 48, 489–499. [Google Scholar] [CrossRef]
- Larkin, R.P. Soil Health Paradigms and Implications for Disease Management. Annu. Rev. Phytopathol. 2015, 53, 199–221. [Google Scholar] [CrossRef] [PubMed]
- Kong, P.; Hong, C. Endophytic Burkholderia sp. SSG as a Potential Biofertilizer Promoting Boxwood Growth. PeerJ 2020, 8, e9547. [Google Scholar] [CrossRef] [PubMed]
- Kong, P.; Richardson, P.; Hong, C. Burkholderia sp. SSG Is a Broad-Spectrum Antagonist against Plant Diseases Caused by Diverse Pathogens. Biol. Control 2020, 151, 104380. [Google Scholar] [CrossRef]
- Kong, P.; Li, X.; Gouker, F.; Hong, C. cDNA Transcriptome of Arabidopsis Reveals Various Defense Priming Induced by a Broad-Spectrum Biocontrol Agent Burkholderia sp. SSG. Int. J. Mol. Sci. 2022, 23, 3151. [Google Scholar] [CrossRef] [PubMed]
- Kong, P.; Hong, C. Complete Genome Sequence of a Boxwood Endophyte Burkholderia sp. SSG with Broad Biotechnological Application Potential. Biotechnol. Rep. 2020, 26, e00455. [Google Scholar] [CrossRef]
- Wang, Q.; Sheng, J.; Pan, L.; Cao, H.; Li, C.; Lambers, H.; Wang, X. Soil Property Determines the Ability of Rhizobial Inoculation to Enhance Nitrogen Fixation and Phosphorus Acquisition in Soybean. Appl. Soil Ecol. 2022, 171, 104346. [Google Scholar] [CrossRef]
- Garrido-Oter, R.; Nakano, R.T.; Dombrowski, N.; Ma, K.W.; McHardy, A.C.; Schulze-Lefert, P. Modular Traits of the Rhizobiales Root Microbiota and Their Evolutionary Relationship with Symbiotic Rhizobia. Cell Host Microbe 2018, 24, 155–167.e5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abbas, A.; Khan, S.U.; Khan, W.U.; Saleh, T.A.; Khan, M.H.U.; Ullah, S.; Ali, A.; Ikram, M. Antagonist Effects of Strains of Bacillus spp. against Rhizoctonia solani for Their Protection against Several Plant Diseases: Alternatives to Chemical Pesticides. Comptes Rendus Biol. 2019, 342, 124–135. [Google Scholar] [CrossRef] [PubMed]
- Ashwini, N.; Srividya, S. Potentiality of Bacillus subtilis as Biocontrol Agent for Management of Anthracnose Disease of Chilli Caused by Colletotrichum gloeosporioides OGC1. 3 Biotech 2014, 4, 127–136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, P.; Quan, C.; Wang, Y.; Wang, J.; Fan, S. Bacillus amyloliquefaciens Q-426 as a Potential Biocontrol Agent against Fusarium oxysporum f. sp. spinaciae. J. Basic Microbiol. 2014, 54, 448–456. [Google Scholar] [CrossRef]
- Cartieaux, F.; Thibaud, M.C.; Zimmerli, L.; Lessard, P.; Sarrobert, C.; David, P.; Gerbaud, A.; Robaglia, C.; Somerville, S.; Nussaume, L. Transcriptome Analysis of Arabidopsis Colonized by a Plant-Growth Promoting Rhizobacterium Reveals a General Effect on Disease Resistance. Plant J. 2003, 36, 177–188. [Google Scholar] [CrossRef]
- Bano, N.; Musarrat, J. Characterization of a New Pseudomonas aeruginosa Strain NJ-15 as a Potential Biocontrol Agent. Curr. Microbiol. 2003, 46, 324–328. [Google Scholar] [CrossRef]
- Chen, D.M.; Yang, H.J.; Huang, J.G.; Yuan, L. Lysobacter enzymogenes LE16 Autolysates Have Potential as Biocontrol Agents—Lysobacter sp. Autolysates as Biofungicide. J. Appl. Microbiol. 2020, 129, 1684–1692. [Google Scholar] [CrossRef]
- Hao, Z.; Van Tuinen, D.; Wipf, D.; Fayolle, L.; Chataignier, O.; Li, X.; Chen, B.; Gianinazzi, S.; Gianinazzi-Pearson, V.; Adrian, M. Biocontrol of Grapevine Aerial and Root Pathogens by Paenibacillus sp. Strain B2 and Paenimyxin in vitro and in planta. Biol. Control 2017, 109, 42–50. [Google Scholar] [CrossRef]
- Ramlawi, S.; Abusharkh, S.; Carroll, A.; McMullin, D.R.; Avis, T.J. Biological and Chemical Characterization of Antimicrobial Activity in Arthrobacter spp. Isolated from Disease-Suppressive Compost. J. Basic Microbiol. 2021, 61, 745–756. [Google Scholar] [CrossRef]
- Das, K.; Prasanna, R.; Kumar Saxena, A. Rhizobia: A Potential Biocontrol Agent for Soilborne Fungal Pathogens. Folia Microbiol. 2017, 62, 425–435. [Google Scholar] [CrossRef]
- Antonopoulos, D.F.; Tjamos, S.E.; Antoniou, P.P.; Rafeletos, P.; Tjamos, E.C. Effect of Paenibacillus alvei, Strain K165, on the Germination of Verticillium dahliae Microsclerotia in planta. Biol. Control 2008, 46, 166–170. [Google Scholar] [CrossRef]
- Ni, H.; Kong, W.L.; Zhang, Y.; Wu, X.Q. Effects of Volatile Organic Compounds Produced by Pseudomonas aurantiaca ST-TJ4 against Verticillium dahliae. J. Fungi 2022, 8, 697. [Google Scholar] [CrossRef]
- Meschke, H.; Walter, S.; Schrempf, H. Characterization and Localization of Prodiginines from Streptomyces lividans Suppressing Verticillium dahliae in the Absence or Presence of Arabidopsis thaliana. Environ. Microbiol. 2012, 14, 940–952. [Google Scholar] [CrossRef]
- Fan, R.; Gong, X.; Gao, L.; Shang, W.; Hu, X.; Xu, X. Temporal Dynamics of the Survival of Verticillium dahliae Microsclerotia with or without Melanin in Soils Amended with Biocontrol Agents. Eur. J. Plant Pathol. 2020, 157, 521–531. [Google Scholar] [CrossRef]
- Dart, N.; Hong, C.; Craig, C.A.; Fry, J.T.; Hu, X. Soil Inoculum Production, Survival, and Infectivity of the Boxwood Blight Pathogen, Calonectria pseudonaviculata. Plant Dis. 2015, 99, 1689–1694. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berg, G.; Roskot, N.; Steidle, A.; Eberl, L.; Zock, A.; Smalla, K. Plant-Dependent Genotypic and Phenotypic Diversity of Antagonistic Rhizobacteria Isolated from Different Verticillium Host Plants. Appl. Environ. Microbiol. 2002, 68, 3328–3338. [Google Scholar] [CrossRef] [Green Version]
- Cernava, T.; Müller, H.; Aschenbrenner, I.A.; Grube, M.; Berg, G. Analyzing the Antagonistic Potential of the Lichen Microbiome against Pathogens by Bridging Metagenomic with Culture Studies. Front. Microbiol. 2015, 6, 620. [Google Scholar] [CrossRef] [Green Version]
- van Bruggen, A.H.C.; Semenov, A.M. In Search of Biological Indicators for Soil Health and Disease Suppression. Appl. Soil Ecol. 2000, 15, 13–24. [Google Scholar] [CrossRef]
- Lebeis, S.L.; Paredes, S.H.; Lundberg, D.S.; Breakfield, N.; Gehring, J.; McDonald, M.; Malfatti, S.; del Rio, T.G.; Jones, C.D.; Tringe, S.G.; et al. Salicylic Acid Modulates Colonization of the Root Microbiome by Specific Bacterial Taxa. Science 2015, 349, 860–864. [Google Scholar] [CrossRef] [Green Version]
- de Souza, R.S.C.; Armanhi, J.S.L.; Arruda, P. From Microbiome to Traits: Designing Synthetic Microbial Communities for Improved Crop Resiliency. Front. Plant Sci. 2020, 11, 1179. [Google Scholar] [CrossRef]
- Berg, G.; Smalla, K. Plant Species and Soil Type Cooperatively Shape the Structure and Function of Microbial Communities in the Rhizosphere. FEMS Microbiol. Ecol. 2009, 68, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Burns, J.H.; Anacker, B.L.; Strauss, S.Y.; Burke, D.J. Soil Microbial Community Variation Correlates Most Strongly with Plant Species Identity, Followed by Soil Chemistry, Spatial Location and Plant Genus. AoB Plants 2015, 7, plv030. [Google Scholar] [CrossRef] [Green Version]
- Ehrenfeld, J.G.; Ravit, B.; Elgersma, K. Feedback in the Plant-Soil System. Annu. Rev. Environ. Resour. 2005, 30, 75–115. [Google Scholar] [CrossRef] [Green Version]
- Saunders, R.; Saunders, T.; Saunders, B.; Saunders, J. Boxwood Guide, 5th ed.; Saunders Brothers: Piney River, VA, USA, 2018. [Google Scholar]
- Zhou, Y.; Wei, Y.; Zhao, Z.; Li, J.; Li, H.; Yang, P.; Tian, S.; Ryder, M.; Toh, R.; Yang, H.; et al. Microbial Communities along the Soil-Root Continuum Are Determined by Root Anatomical Boundaries, Soil Properties, and Root Exudation. Soil Biol. Biochem. 2022, 171, 108721. [Google Scholar] [CrossRef]
- Xue, P.P.; Carrillo, Y.; Pino, V.; Minasny, B.; McBratney, A.B. Soil Properties Drive Microbial Community Structure in a Large Scale Transect in South Eastern Australia. Sci. Rep. 2018, 8, 11725. [Google Scholar] [CrossRef] [Green Version]
- Batdorf, L.R. Boxwood Handbook: A Practical Guide to Knowing and Growing Boxwood, 3rd ed.; The American Boxwood Society: Boyce, VA, USA, 1995. [Google Scholar]
- Deng, X.; Zhang, N.; Li, Y.; Zhu, C.; Qu, B.; Liu, H.; Li, R.; Bai, Y.; Shen, Q.; Falcao Salles, J. Bio-organic Soil Amendment Promotes the Suppression of Ralstonia solanacearum by Inducing Changes in the Functionality and Composition of Rhizosphere Bacterial Communities. New Phytol. 2022, 235, 1558–1574. [Google Scholar] [CrossRef] [PubMed]
- van Bruggen, A.H.C.; Sharma, K.; Kaku, E.; Karfopoulos, S.; Zelenev, V.V.; Blok, W.J. Soil Health Indicators and Fusarium Wilt Suppression in Organically and Conventionally Managed Greenhouse Soils. Appl. Soil Ecol. 2014, 86, 192–201. [Google Scholar] [CrossRef]
- Palojärvi, A.; Kellock, M.; Parikka, P.; Jauhiainen, L.; Alakukku, L. Tillage System and Crop Sequence Affect Soil Disease Suppressiveness and Carbon Status in Boreal Climate. Front. Microbiol. 2020, 11, 4786. [Google Scholar] [CrossRef] [PubMed]
- Stirling, G.R.; Smith, M.K.; Smith, J.P.; Stirling, A.M.; Hamill, S.D. Organic Inputs, Tillage and Rotation Practices Influence Soil Health and Suppressiveness to Soilborne Pests and Pathogens of Ginger. Australas. Plant Pathol. 2012, 41, 99–112. [Google Scholar] [CrossRef]
- Islam, W.; Noman, A.; Naveed, H.; Huang, Z.; Chen, H.Y.H. Role of Environmental Factors in Shaping the Soil Microbiome. Environ. Sci. Pollut. Res. 2020, 27, 41225–41247. [Google Scholar] [CrossRef]
- Pershina, E.V.; Ivanova, E.A.; Korvigo, I.O.; Chirak, E.L.; Sergaliev, N.H.; Abakumov, E.V.; Provorov, N.A.; Andronov, E.E. Investigation of the Core Microbiome in Main Soil Types from the East European Plain. Sci. Total Environ. 2018, 631–632, 1421–1430. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Zeng, J.; Zhu, Q.; Zhang, Z.; Lin, X. pH Is the Primary Determinant of the Bacterial Community Structure in Agricultural Soils Impacted by Polycyclic Aromatic Hydrocarbon Pollution. Sci. Rep. 2017, 7, 40093. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xue, D.; Christenson, R.; Genger, R.; Gevens, A.; Lankau, R.A. Soil Microbial Communities Reflect Both Inherent Soil Properties and Management Practices in Wisconsin Potato Fields. Am. J. Potato Res. 2018, 95, 696–708. [Google Scholar] [CrossRef]
- Naidoo, Y.; Valverde, A.; Pierneef, R.E.; Cowan, D.A. Differences in Precipitation Regime Shape Microbial Community Composition and Functional Potential in Namib Desert Soils. Microb. Ecol. 2021, 83, 689–701. [Google Scholar] [CrossRef]
- Donhauser, J.; Niklaus, P.A.; Rousk, J.; Larose, C.; Frey, B. Temperatures beyond the Community Optimum Promote the Dominance of Heat-Adapted, Fast Growing and Stress Resistant Bacteria in Alpine Soils. Soil Biol. Biochem. 2020, 148, 107873. [Google Scholar] [CrossRef]
| Year | Season | Sequence Reads | Sample Coverage |
|---|---|---|---|
| 2017 | Early summer | 1,788,962 | 0.991 |
| Late Fall | 2,127,058 | 0.991 | |
| 2018 | Early summer | 1,871,473 | 0.995 |
| Late Fall | 1,732,272 | 0.990 | |
| ∑ | 7,519,765 | Avg = 0.992 | |
| Year | Variable | Observed OTU Richness | Shannon Index | ||
|---|---|---|---|---|---|
| F | p-Value | F | p-Value | ||
| 2017 | State | 20.60 | <0.0001 | 27.72 | <0.0001 |
| Season | 12.97 | 0.0013 | 5.45 | 0.0273 | |
| Season × State | 4.53 | 0.0063 | 8.56 | 0.0001 | |
| 2018 | State | 15.06 | <0.0001 | 31.35 | <0.0001 |
| Season | 11.72 | 0.0019 | 0.54 | 0.4704 | |
| Season × State | 5.42 | 0.0023 | 4.23 | 0.0084 | |
| Year | Variable | PERMANOVA † | Dispersion ‡ | ||
|---|---|---|---|---|---|
| R2 | p-Value | F | p-Value | ||
| 2017 | State | 0.33 | <0.0001 | 5.07 | 0.0028 |
| Season | 0.06 | 0.0006 | 0.41 | 0.5268 | |
| State × Season | 0.22 | <0.0001 | - § | - | |
| 2018 | State | 0.38 | <0.0001 | 4.89 | 0.0033 |
| Season | 0.05 | 0.0026 | 0.45 | 0.5063 | |
| State × Season | 0.13 | 0.0004 | - | - | |
| Order | Relative Abundance (%) | Season | State | ||||||
|---|---|---|---|---|---|---|---|---|---|
| 2017 | 2018 | 2017 | 2018 | ||||||
| χ2 | p-Adj | χ2 | p-Adj | χ2 | p-Adj | χ2 | p-Adj | ||
| Rhizobiales | 16.8 | 1.64 | 0.5751 | 3.84 | 0.7027 | 9.59 | 0.0741 | 15.63 | 0.0117 |
| Burkholderiales | 16.0 | 0.02 | 0.9482 | 0.31 | 0.9372 | 21.70 | 0.0020 | 22.51 | 0.0022 |
| Vicinamibacterales | 6.9 | 1.34 | 0.6096 | 0.003 | 0.9893 | 15.52 | 0.0103 | 19.41 | 0.0041 |
| Chitinophagales | 3.7 | 5.80 | 0.2555 | 2.78 | 0.7787 | 18.55 | 0.0043 | 20.48 | 0.0032 |
| Xanthomonadales | 2.8 | 2.81 | 0.4531 | 1.17 | 0.8771 | 18.23 | 0.0048 | 26.25 | 0.0000 |
| Gaiellales | 1.9 | 0.06 | 0.9288 | 9.24 | 0.2309 | 21.51 | 0.0021 | 9.56 | 0.0812 |
| Sphingomonadales | 1.8 | 0.68 | 0.6814 | 1.30 | 0.8643 | 25.69 | 0.0011 | 24.69 | 0.0015 |
| Bacillales | 1.8 | 10.44 | 0.0993 | 0.09 | 0.9836 | 1.65 | 0.8147 | 14.05 | 0.0184 |
| Acidobacteriales | 1.7 | 3.92 | 0.3257 | 0.77 | 0.9372 | 13.58 | 0.0197 | 13.40 | 0.0224 |
| Gemmatimonadales | 1.6 | 0.004 | 0.9715 | 0.01 | 0.9893 | 14.99 | 0.0124 | 15.26 | 0.0131 |
| Year | Season | State | Total BCA Sequence Reads | Total Sequence Reads | BCA Abundance (%) |
|---|---|---|---|---|---|
| 2017 | Early summer | California | 6893 | 409,612 | 1.68 |
| Illinois | 8048 | 396,132 | 2.03 | ||
| New York | 5529 | 315,309 | 1.75 | ||
| South Carolina | 8077 | 315,116 | 2.56 | ||
| Virginia | 7514 | 352,793 | 2.13 | ||
| Late fall | California | 1266 | 174,912 | 0.72 | |
| Illinois | 1503 | 136,271 | 1.10 | ||
| New York | 8937 | 668,827 | 1.34 | ||
| South Carolina | 5590 | 467,264 | 1.20 | ||
| Virginia | 7254 | 679,784 | 1.07 | ||
| 2018 | Early Summer | California | 7313 | 429,281 | 1.70 |
| Illinois | 13,630 | 346,078 | 3.94 | ||
| New York | 9419 | 386,764 | 2.44 | ||
| South Carolina | 4166 | 249,726 | 1.67 | ||
| Virginia | 4330 | 459,624 | 0.94 | ||
| Late fall | California | 6020 | 349,274 | 1.72 | |
| Illinois | 5644 | 251,778 | 2.24 | ||
| New York | 4282 | 143,548 | 2.98 | ||
| South Carolina | 2933 | 215,495 | 1.36 | ||
| Virginia | 8226 | 772,177 | 1.07 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.; Kong, P.; Daughtrey, M.; Kosta, K.; Schirmer, S.; Howle, M.; Likins, M.; Hong, C. Characterization of the Soil Bacterial Community from Selected Boxwood Gardens across the United States. Microorganisms 2022, 10, 1514. https://doi.org/10.3390/microorganisms10081514
Li X, Kong P, Daughtrey M, Kosta K, Schirmer S, Howle M, Likins M, Hong C. Characterization of the Soil Bacterial Community from Selected Boxwood Gardens across the United States. Microorganisms. 2022; 10(8):1514. https://doi.org/10.3390/microorganisms10081514
Chicago/Turabian StyleLi, Xiaoping, Ping Kong, Margery Daughtrey, Kathleen Kosta, Scott Schirmer, Matthew Howle, Michael Likins, and Chuanxue Hong. 2022. "Characterization of the Soil Bacterial Community from Selected Boxwood Gardens across the United States" Microorganisms 10, no. 8: 1514. https://doi.org/10.3390/microorganisms10081514
APA StyleLi, X., Kong, P., Daughtrey, M., Kosta, K., Schirmer, S., Howle, M., Likins, M., & Hong, C. (2022). Characterization of the Soil Bacterial Community from Selected Boxwood Gardens across the United States. Microorganisms, 10(8), 1514. https://doi.org/10.3390/microorganisms10081514







