An In Situ Study to Understand Community Structure of Estuarine Microbes on the Plastisphere
Abstract
:1. Introduction
2. Materials and Methods
2.1. Selection of Microplastics
2.2. Preparation and Sterilization of Beads
2.3. In Situ Incubation
2.4. Subsampling for Microbial Community Analysis
2.5. 16S Sequencing and Bioinformatic Pipeline
2.6. Diversity Measures and PERMANOVA
2.7. Ecological Interpretation of 16S Marker Data
2.8. Hydrological Data
3. Results
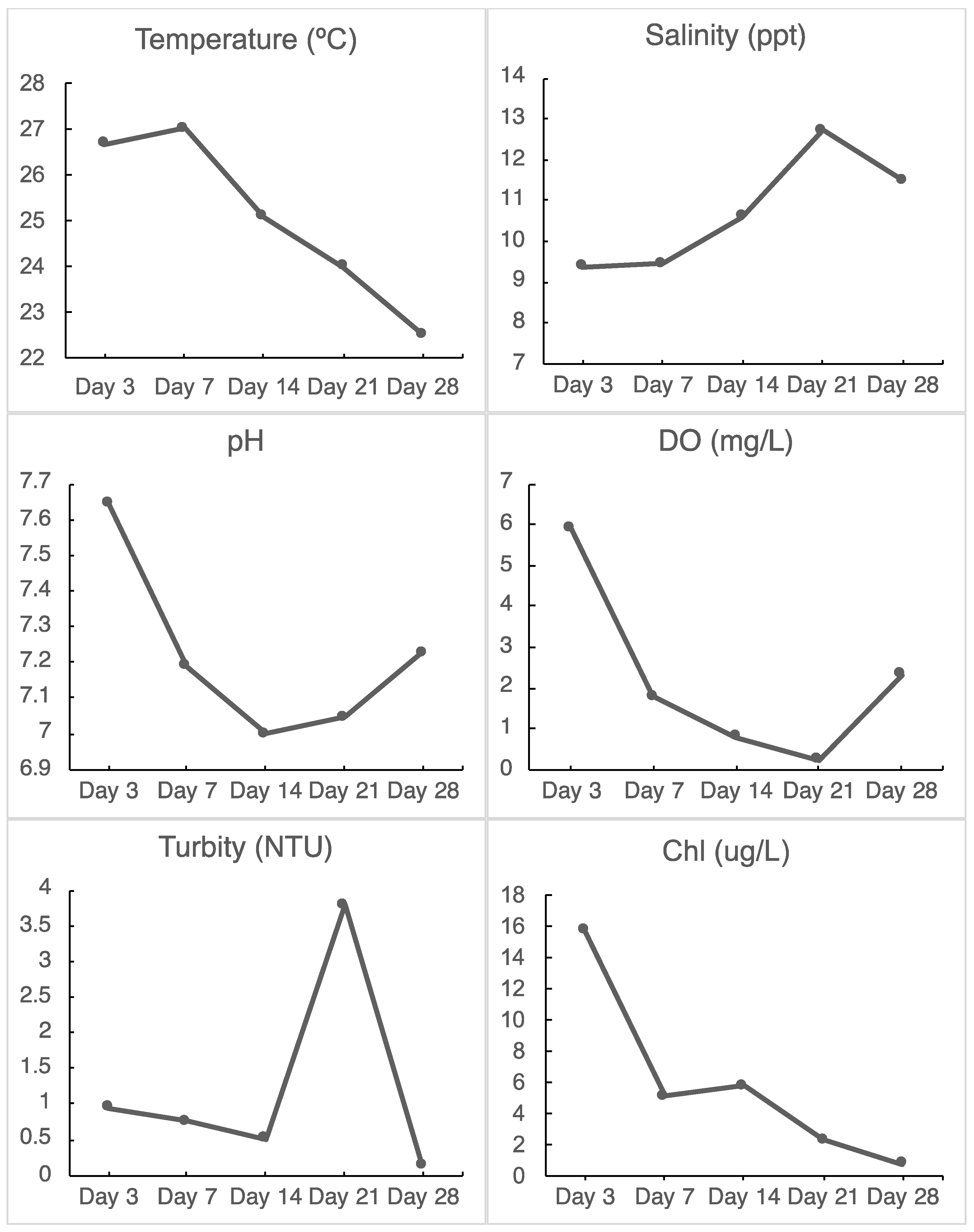
3.1. Hydrological Data
3.2. DNA Sequencing and Bioinformatic Pipeline
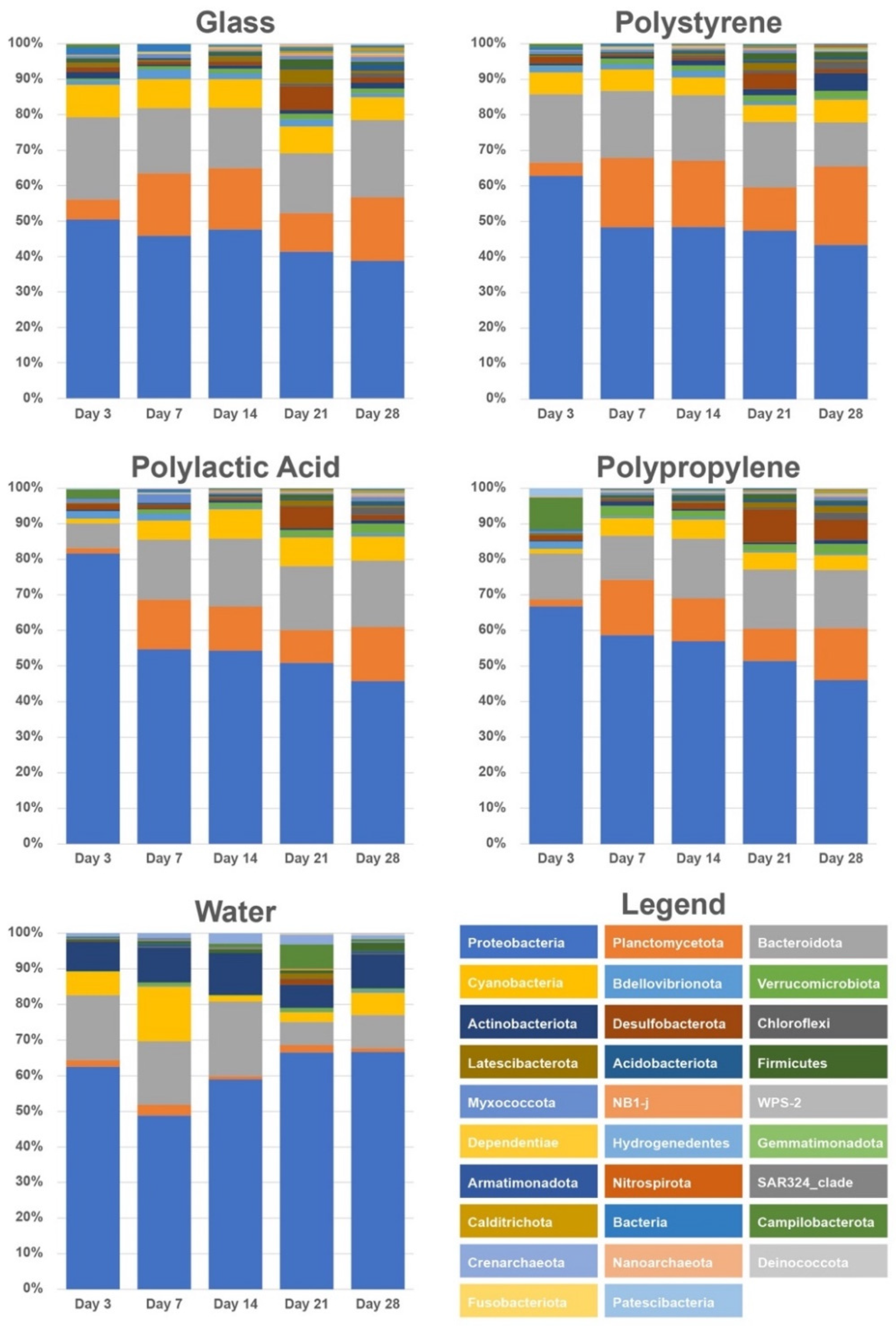
3.3. Microbial Community Analysis
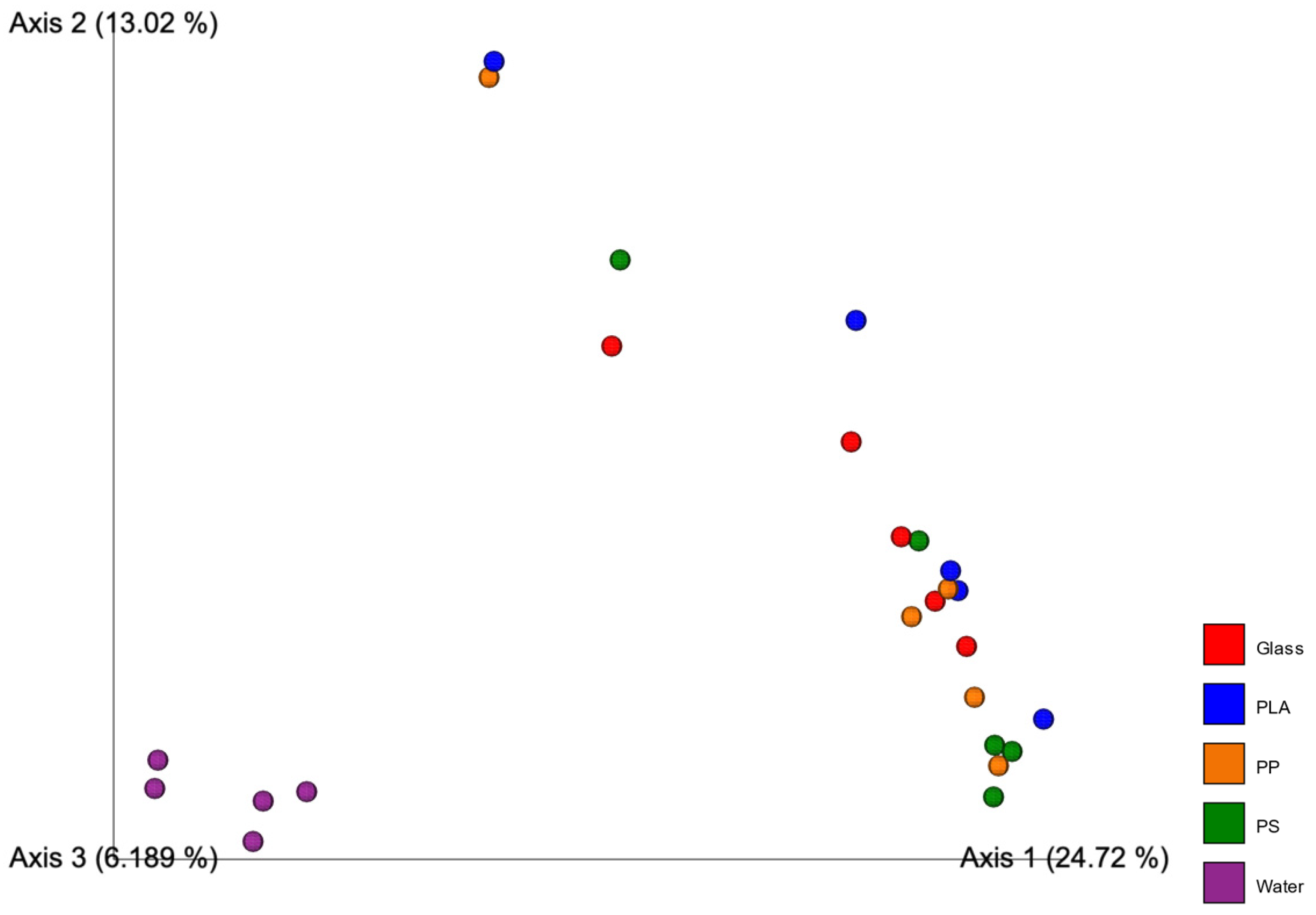
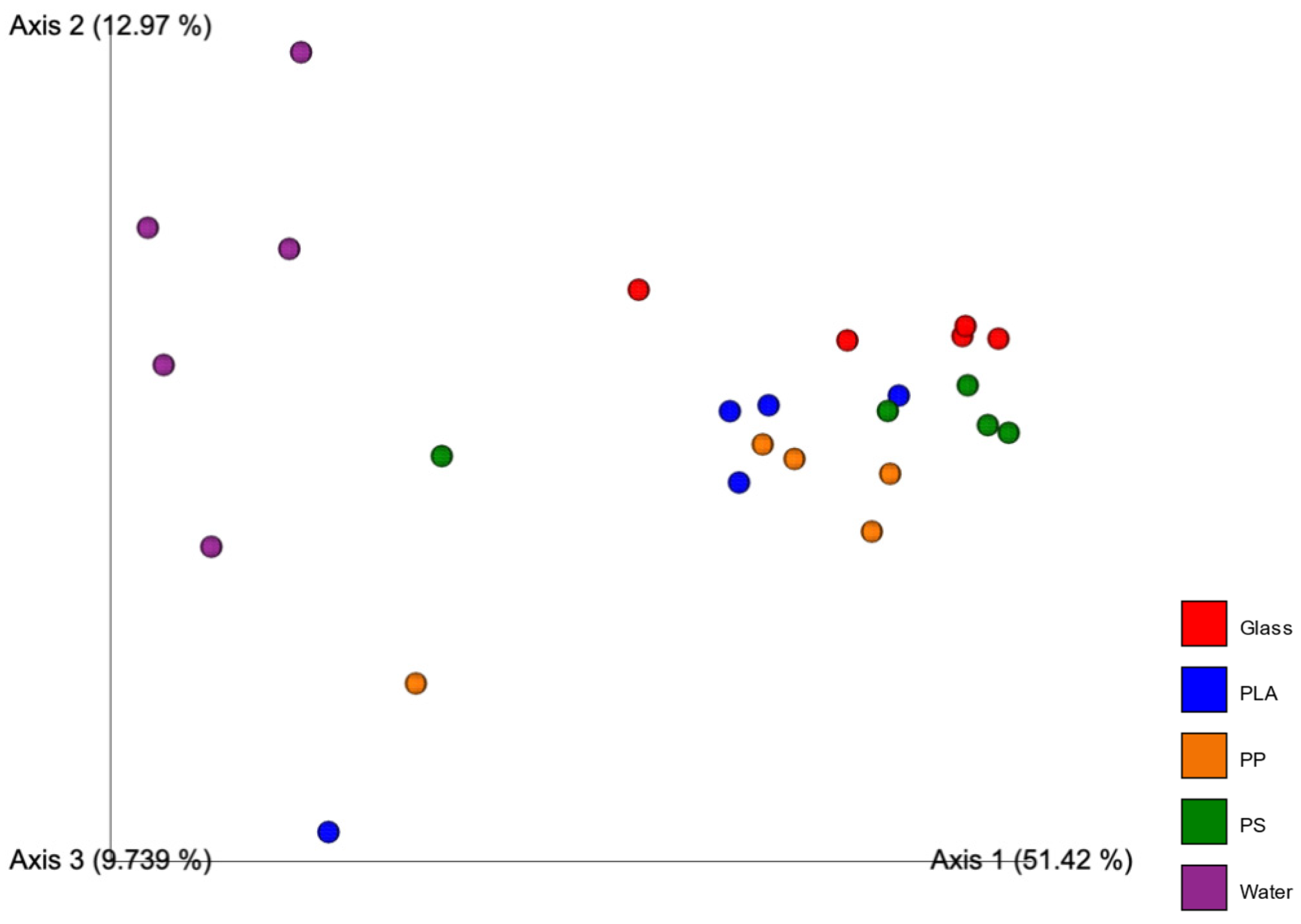
3.4. Diversity Measures and PERMANOVA
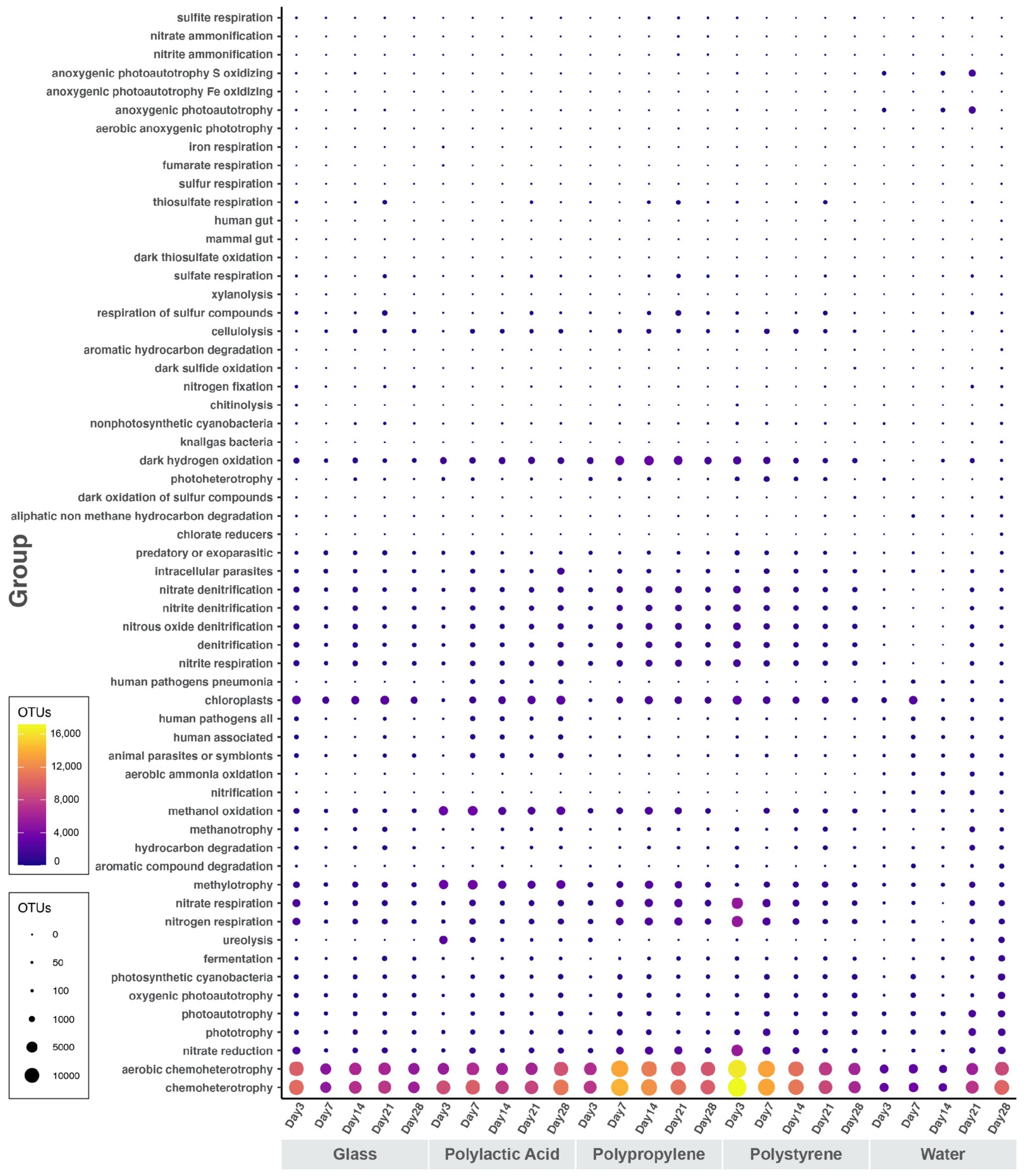
3.5. Ecological Interpretation of 16S Marker Data
4. Discussion
4.1. Low Dissolved Oxygen in the Inner Harbor during the Incubation Period
4.2. Planctomycetes Thrived on Biofilms
4.3. Desulfobacteriota Became Abundant on Day 21
4.4. Succession of Biofilm Communities
4.5. Diversity Measures and PERMANOVA
4.6. Ecological Interpretation of 16S Marker Data
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Barnes, D.K.A.; Galgani, F.; Thompson, R.C.; Barlaz, M. Accumulation and fragmentation of plastic debris in global environments. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2009, 364, 1985–1998. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Betts, K. Why small plastic particles may pose a big problem in the oceans. Environ. Sci. Technol. 2008, 42, 8995. [Google Scholar] [CrossRef] [PubMed]
- A Cooper, D.; Corcoran, P.L. Effects of mechanical and chemical processes on the degradation of plastic beach debris on the island of Kauai, Hawaii. Mar. Pollut. Bull. 2010, 60, 650–654. [Google Scholar] [CrossRef] [PubMed]
- Fendall, L.S.; Sewell, M.A. Contributing to marine pollution by washing your face: Microplastics in facial cleansers. Mar. Pollut. Bull. 2009, 58, 1225–1228. [Google Scholar] [CrossRef]
- Eriksen, M.; Lebreton, L.C.M.; Carson, H.S.; Thiel, M.; Moore, C.J.; Borerro, J.C.; Galgani, F.; Ryan, P.G.; Reisser, J. Plastic Pollution in the World’s Oceans: More than 5 Trillion Plastic Pieces Weighing over 250,000 Tons Afloat at Sea. PLoS ONE 2014, 9, e111913. [Google Scholar] [CrossRef] [Green Version]
- Yonkos, L.T.; Friedel, E.A.; Perez-Reyes, A.C.; Ghosal, S.; Arthur, C.D. Microplastics in Four Estuarine Rivers in the Chesapeake Bay, U.S.A. Environ. Sci. Technol. 2014, 48, 14195–14202. [Google Scholar] [CrossRef]
- Bikker, J.; Lawson, J.; Wilson, S.; Rochman, C. Microplastics and other anthropogenic particles in the surface waters of the Chesapeake Bay. Mar. Pollut. Bull. 2020, 156, 111257. [Google Scholar] [CrossRef]
- Costerton, J.W.; Cheng, K.J.; Geesey, G.G.; Ladd, T.I.; Nickel, J.C.; Dasgupta, M.; Marrie, T.J. Bacterial Biofilms in Nature and Disease. Annu. Rev. Microbiol. 1987, 41, 435–464. [Google Scholar] [CrossRef]
- Zettler, E.R.; Mincer, T.J.; Amaral-Zettler, L.A. Life in the “Plastisphere”: Microbial Communities on Plastic Marine Debris. Environ. Sci. Technol. 2013, 47, 7137–7146. [Google Scholar] [CrossRef]
- Harrison, J.P.; Schratzberger, M.; Sapp, M.; Osborn, A.M. Rapid bacterial colonization of low-density polyethylene microplastics in coastal sediment microcosms. BMC Microbiol. 2014, 14, 232. [Google Scholar] [CrossRef] [Green Version]
- Eerkes-Medrano, D.; Thompson, R.C.; Aldridge, D.C. Microplastics in freshwater systems: A review of the emerging threats, identification of knowledge gaps and prioritisation of research needs. Water Res. 2015, 75, 63–82. [Google Scholar] [CrossRef]
- Bryant, J.A.; Clemente, T.M.; Viviani, D.; Fong, A.A.; Thomas, K.; Kemp, P.; Karl, D.M.; White, A.E.; DeLong, E.F. Diversity and Activity of Communities Inhabiting Plastic Debris in the North Pacific Gyre. mSystems 2016, 1, e00024-16. [Google Scholar] [CrossRef] [Green Version]
- Mincer, T.J.; Zettler, E.R.; Amaral-Zettler, L.A. Biofilms on Plastic Debris and Their Influence on Marine Nutrient Cycling, Productivity, and Hazardous Chemical Mobility. In Handbook of Environmental Chemistry; Springer: Berlin/Heidelberg, Germany, 2016; Volume 78, pp. 221–233. [Google Scholar]
- Lobelle, D.; Cunliffe, M. Early microbial biofilm formation on marine plastic debris. Mar. Pollut. Bull. 2011, 62, 197–200. [Google Scholar] [CrossRef]
- Amaral-Zettler, L.A.; Zettler, E.R.; Mincer, T.J. Ecology of the plastisphere. Nat. Rev. Microbiol. 2020, 18, 139–151. [Google Scholar] [CrossRef]
- Ogonowski, M.; Motiei, A.; Ininbergs, K.; Hell, E.; Gerdes, Z.; Udekwu, K.I.; Bacsik, Z.; Gorokhova, E. Evidence for selective bacterial community structuring on microplastics. Environ. Microbiol. 2018, 20, 2796–2808. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Louca, S.; Parfrey, L.W.; Doebeli, M. Decoupling function and taxonomy in the global ocean microbiome. Science 2016, 353, 1272–1277. [Google Scholar] [CrossRef]
- Oberbeckmann, S.; Labrenz, M. Marine Microbial Assemblages on Microplastics: Diversity, Adaptation, and Role in Degradation. Annu. Rev. Mar. Sci. 2020, 12, 209–232. [Google Scholar] [CrossRef] [Green Version]
- Fuerst, J.A.; Sagulenko, E. Beyond the bacterium: Planctomycetes challenge our concepts of microbial structure and function. Nat. Rev. Genet. 2011, 9, 403–413. [Google Scholar] [CrossRef]
- Tal, Y.; Watts, J.E.M.; Schreier, H.J. Anaerobic Ammonia-Oxidizing Bacteria and Related Activity in Baltimore Inner Harbor Sediment. Appl. Environ. Microbiol. 2005, 71, 1816–1821. [Google Scholar] [CrossRef] [Green Version]
- Capo, E.; Bravo, A.G.; Soerensen, A.L.; Bertilsson, S.; Pinhassi, J.; Feng, C.; Andersson, A.F.; Buck, M.; Björn, E. Deltaproteobacteria and Spirochaetes-Like Bacteria Are Abundant Putative Mercury Methylators in Oxygen-Deficient Water and Marine Particles in the Baltic Sea. Front. Microbiol. 2020, 11, 574080. [Google Scholar] [CrossRef]
- Dussud, C.; Hudec, C.; George, M.; Fabre, P.; Higgs, P.; Bruzaud, S.; Delort, A.-M.; Eyheraguibel, B.; Meistertzheim, A.-L.; Jacquin, J.; et al. Colonization of Non-biodegradable and Biodegradable Plastics by Marine Microorganisms. Front. Microbiol. 2018, 9, 1571. [Google Scholar] [CrossRef]
- Costerton, J.W.; Stewart, P.S.; Greenberg, E.P. Bacterial Biofilms: A Common Cause of Persistent Infections. Science 1999, 284, 1318–1322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rüthi, J.; Bölsterli, D.; Pardi-Comensoli, L.; Brunner, I.; Frey, B. The “Plastisphere” of Biodegradable Plastics Is Characterized by Specific Microbial Taxa of Alpine and Arctic Soils. Front. Environ. Sci. 2020, 8, 562263. [Google Scholar] [CrossRef]
- Ranjani, A.; Dhanasekaran, D.; Gopinath, P.M. An Introduction to Actinobacteria. In Actinobacteria—Basics and Biotechnological Applications; IntechOpen: London, UK, 2016. [Google Scholar] [CrossRef] [Green Version]
- Barka, E.A.; Vatsa, P.; Sanchez, L.; Gaveau-Vaillant, N.; Jacquard, C.; Klenk, H.-P.; Clément, C.; Ouhdouch, Y.; van Wezel, G.P. Taxonomy, Physiology, and Natural Products of Actinobacteria. Microbiol. Mol. Biol. Rev. 2016, 80, 1–43. [Google Scholar] [CrossRef] [Green Version]
- Vaksmaa, A.; Knittel, K.; Asbun, A.A.; Goudriaan, M.; Ellrott, A.; Witte, H.J.; Vollmer, I.; Meirer, F.; Lott, C.; Weber, M.; et al. Microbial Communities on Plastic Polymers in the Mediterranean Sea. Front. Microbiol. 2021, 12, 673553. [Google Scholar] [CrossRef] [PubMed]
- Kletzin, A. Metabolism of Inorganic Sulfur Compounds in Archaea. Archaea: Evolution, Physiology, and Molecular Biology; Wiley: Hoboken, NJ, USA, 2006; pp. 261–274. [Google Scholar] [CrossRef]
- Leigh, J.A.; Whitman, W.B. Archaeal Genetics. In Brenner’s Encyclopedia of Genetics, 2nd ed.; Maloy, S., Hughes, K., Eds.; Elsevier: London, UK, 2013; pp. 188–191. ISBN 9780080961569. [Google Scholar]
- Bharti, A.; Velmourougane, K.; Prasanna, R. Phototrophic Biofilms: Diversity, Ecology and Applications. J. Appl. Phycol. 2017, 29, 2729–2744. [Google Scholar] [CrossRef]
- Barnes, D.K.A.; Milner, P. Drifting plastic and its consequences for sessile organism dispersal in the Atlantic Ocean. Mar. Biol. 2004, 146, 815–825. [Google Scholar] [CrossRef]
| Material | Date | Raw Reads | Filtered Reads | Total OTUs |
|---|---|---|---|---|
| PP | September 18 | 89,012 | 87,180 | 51,068 |
| PS | September 7 | 86,722 | 84,806 | 52,429 |
| PP | September 11 | 81,232 | 79,914 | 45,136 |
| PLA | October 2 | 81,198 | 79,646 | 49,963 |
| PLA | September 7 | 79,838 | 78,042 | 20,296 |
| PLA | September 11 | 71,672 | 70,183 | 34,545 |
| PP | September 25 | 71,366 | 70,247 | 44,202 |
| PP | September 7 | 70,822 | 69,493 | 22,443 |
| PP | October 2 | 70,623 | 69,284 | 45,136 |
| PS | September 11 | 66,111 | 64,789 | 44,677 |
| PS | September 18 | 58,716 | 57,441 | 44,557 |
| PLA | September 25 | 58,648 | 57,554 | 35,586 |
| Glass | September 7 | 56,307 | 55,149 | 34,308 |
| PLA | September 18 | 56,272 | 55,293 | 29,932 |
| Glass | September 25 | 56,230 | 55,255 | 44,396 |
| PS | September 25 | 55,547 | 54,522 | 42,135 |
| Water | September 25 | 53,544 | 52,547 | 27,485 |
| Glass | September 18 | 48,332 | 47,495 | 36,619 |
| Water | October 2 | 43,913 | 43,030 | 31,958 |
| Water | October 2 | 42,485 | 41,687 | 33,393 |
| PS | October 2 | 37,252 | 36,368 | 28,003 |
| Glass | September 11 | 31,910 | 31,349 | 24,459 |
| Water | September 11 | 30,615 | 29,882 | 20,690 |
| Water | September 7 | 20,689 | 20,241 | 16,192 |
| Water | September 18 | 16,525 | 16,251 | 12,750 |
| Group 1 | Group 2 | Sample Size | Permutations | Pseudo-F | p-Value | q-Value |
|---|---|---|---|---|---|---|
| Glass | PLA | 10 | 999 | 1.3036273 | 0.138 | 0.18125 |
| Glass | PP | 10 | 999 | 1.4050173 | 0.088 | 0.176 |
| Glass | PS | 10 | 999 | 1.4156289 | 0.115 | 0.18125 |
| Glass | Water | 10 | 999 | 4.6538861 | 0.01 | 0.025 |
| PLA | PP | 10 | 999 | 0.8238911 | 0.492 | 0.492 |
| PLA | PS | 10 | 999 | 1.3589694 | 0.145 | 0.18125 |
| PLA | Water | 10 | 999 | 4.5207674 | 0.01 | 0.025 |
| PP | PS | 10 | 999 | 1.0297202 | 0.448 | 0.492 |
| PP | Water | 10 | 999 | 4.1069582 | 0.006 | 0.025 |
| PS | Water | 10 | 999 | 4.7291459 | 0.009 | 0.025 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sosa, A.; Chen, F. An In Situ Study to Understand Community Structure of Estuarine Microbes on the Plastisphere. Microorganisms 2022, 10, 1543. https://doi.org/10.3390/microorganisms10081543
Sosa A, Chen F. An In Situ Study to Understand Community Structure of Estuarine Microbes on the Plastisphere. Microorganisms. 2022; 10(8):1543. https://doi.org/10.3390/microorganisms10081543
Chicago/Turabian StyleSosa, Ana, and Feng Chen. 2022. "An In Situ Study to Understand Community Structure of Estuarine Microbes on the Plastisphere" Microorganisms 10, no. 8: 1543. https://doi.org/10.3390/microorganisms10081543






