New Insights into Bacillus-Primed Plant Responses to a Necrotrophic Pathogen Derived from the Tomato-Botrytis Pathosystem
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant and Microorganism Materials
2.2. Resistance Induction Assay and Treatment Application
2.3. Evaluation of Disease Severity
2.4. Isolation of Lipopeptides and UHPLC-MS Analysis
2.5. RNA Isolation from Tomato Leaflets
2.6. RNAseq Data Processing and Differential Gene Expression Analysis
2.7. Principal Component Analysis (PCA), Self-Organizing Map (SOM) Clustering and Functional Annotation
2.8. Differential Gene Expression Evaluation by Quantitative PCR
3. Results
3.1. Disease Severity and Surfactin Production
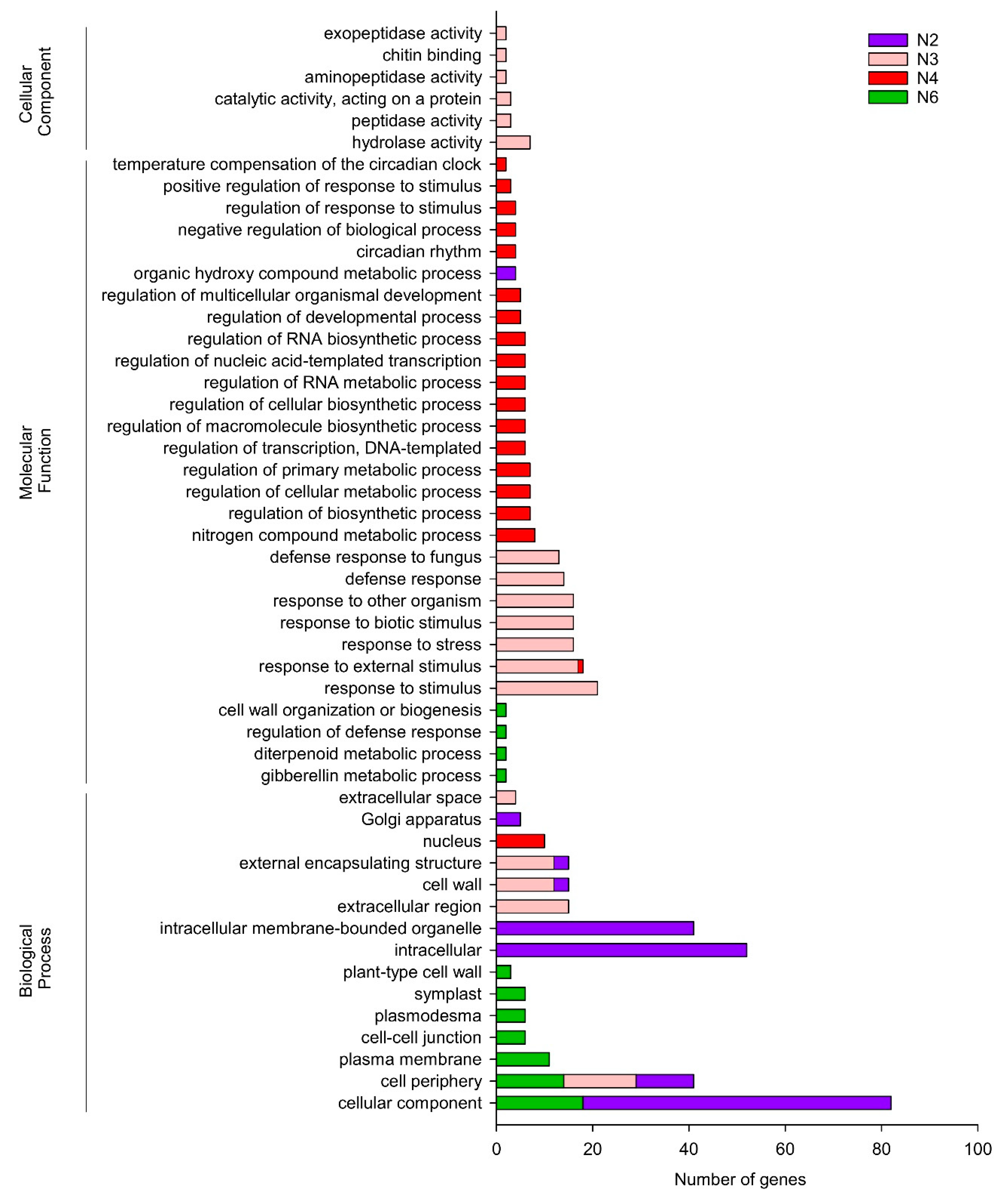
3.2. RNA Sequencing and Differentially Expressed Genes (DEGs) in Response to Priming and Curative Treatments
3.3. Transcript Expression Patterns of Tomato Plant across Interactions with Botrytis cinerea and/or BBC047 Strain
4. Discussion
4.1. Molecular Indicators for ISR
4.2. Gene Expression in the Priming Treatment with BBC047 Strain
4.3. Gene Expression in the Curative Treatment with BBC047 Strain
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- FAOSTAT. 2020. Available online: http://www.fao.org/faostat/en/#data/QC (accessed on 28 December 2021).
- Campos, M.D.; Félix, M.D.R.; Patanita, M.; Materatski, P.; Varanda, C. High throughput sequencing unravels tomato-pathogen interactions towards a sustainable plant breeding. Hortic. Res. 2021, 8, 171. [Google Scholar] [CrossRef] [PubMed]
- Singh, V.K.; Singh, A.K.; Kumar, A. Disease management of tomato through PGPB: Current trends and future perspective. 3 Biotech 2017, 7, 255. [Google Scholar] [CrossRef]
- Panno, S.; Davino, S.; Caruso, A.G.; Bertacca, S.; Crnogorac, A.; Mandic, A.; Noris, E.; Matic, S. A review of the most common and economically important diseases that undermine the cultivation of tomato crop in the Mediterranean basin. Agronomy 2021, 11, 2188. [Google Scholar] [CrossRef]
- Dean, R.; Van Kan, J.A.; Pretorius, Z.A.; Hammond-Kosack, K.E.; Di Pietro, A.; Spanu, P.D.; Rudd, J.J.; Dickman, M.; Kahmann, R.; Ellis, J.; et al. The top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 2012, 13, 414–430. [Google Scholar] [CrossRef] [PubMed]
- Corkley, I.; Fraaije, B.; Hawkins, N. Fungicide resistance management: Maximizing the effective life of plant protection products. Plant Pathol. 2022, 71, 150–169. [Google Scholar] [CrossRef]
- Kloepper, J.W.; Ryu, C.M.; Zhang, S. Induced systemic resistance and promotion of plant growth by Bacillus spp. Phytopathology 2004, 94, 1259–1266. [Google Scholar] [CrossRef] [PubMed]
- Karthika, S.; Varghese, S.; Jisha, M.S. Exploring the efficacy of antagonistic rhizobacteria as native biocontrol agents against tomato plant diseases. 3 Biotech 2020, 10, 320. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.P.; Lee, S.W.; Kim, C.S.; Son, J.H.; Song, J.H.; Lee, K.Y.; Kim, H.J.; Jung, S.J.; Moon, B.J. Evaluation of formulations of Bacillus licheniformis for the biological control of tomato gray mold caused by Botrytis cinerea. Biol. Control 2006, 37, 329–337. [Google Scholar] [CrossRef]
- Dimkić, I.; Janakiev, T.; Petrović, M.; Degrassi, G.; Fira, D. Plant-associated Bacillus and Pseudomonas antimicrobial activities in plant disease suppression via biological control mechanisms—A review. Physiol. Mol. Plant Pathol. 2022, 117, 101754. [Google Scholar] [CrossRef]
- Pieterse, C.M.; Zamioudis, C.; Berendsen, R.L.; Weller, D.M.; Van Wees, S.C.; Bakker, P.A. Induced systemic resistance by beneficial microbes. Annu. Rev. Phytopathol. 2014, 52, 347–375. [Google Scholar] [CrossRef]
- Cawoy, H.; Mariutto, M.; Henry, G.; Fisher, C.; Vasilyeva, N.; Thonart, P.; Dommes, J.; Ongena, M. Plant defense stimulation by natural isolates of Bacillus depends on efficient surfactin production. Mol. Plant-Microbe Interact. 2014, 27, 87–100. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.B.; Shao, D.Y.; Jiang, C.M.; Shi, J.L.; Li, Q.; Huang, Q.S.; Rajoka, M.; Yang, H.; Jin, M.L. Biological activity of lipopeptides from Bacillus. Appl. Microbiol. Biotechnol. 2017, 101, 5951–5960. [Google Scholar] [CrossRef] [PubMed]
- Debois, D.; Fernandez, O.; Franzil, L.; Jourdan, E.; de Brogniez, A.; Willems, L.; Clément, C.; Dorey, S.; De Pauw, E.; Ongena, M. Plant polysaccharides initiate underground crosstalk with bacilli by inducing synthesis of the immunogenic lipopeptide surfactin. Environ. Microbiol. Rep. 2015, 7, 570–582. [Google Scholar] [CrossRef] [PubMed]
- Aleti, G.; Lehner, S.; Bacher, M.; Compant, S.; Nikolic, B.; Plesko, M.; Schuhmacher, R.; Sessitsch, A.; Brader, G. Surfactin variants mediate species-specific biofilm formation and root colonization in Bacillus. Environ. Microbiol. 2016, 18, 2634–2645. [Google Scholar] [CrossRef] [PubMed]
- Al-Ali, A.; Deravel, J.; Krier, F.; Béchet, M.; Ongena, M.; Jacques, P. Biofilm formation is determinant in tomato rhizosphere colonization by Bacillus velezensis FZB42. Environ. Sci. Pollut. Res. Int. 2018, 25, 29910–29920. [Google Scholar] [CrossRef]
- Stoll, A.; Salvatierra-Martínez, R.; González, M.; Araya, M. The role of surfactin production by Bacillus velezensis on colonization, biofilm formation on tomato root and leaf surfaces and subsequent protection (ISR) against Botrytis cinerea. Microorganisms 2021, 9, 2251. [Google Scholar] [CrossRef]
- Nishad, R.; Ahmed, T.; Rahman, V.J.; Kareem, A. Modulation of plant defense system in response to microbial interactions. Front. Microbiol. 2020, 11, 1298. [Google Scholar] [CrossRef]
- Klessig, D.F.; Choi, H.; Dempsey, D.A. Systemic acquired resistance and salicylic acid: Past, present, and future. Mol. Plant-Microbe Interact. 2018, 31, 871–888. [Google Scholar] [CrossRef] [PubMed]
- Vlot, A.C.; Sales, J.H.; Lenk, M.; Bauer, K.; Brambilla, A.; Sommer, A.; Nayem, S. Systemic propagation of immunity in plants. New Phytol. 2021, 229, 1234–1250. [Google Scholar] [CrossRef]
- Ranf, S. Pattern recognition receptors—Versatile genetic tools for engineering broad-spectrum disease resistance in crops. Agronomy 2018, 8, 134. [Google Scholar] [CrossRef]
- Pršić, J.; Ongena, M. Elicitors of plant immunity triggered by beneficial bacteria. Front. Plant Sci. 2020, 11, 594530. [Google Scholar] [CrossRef] [PubMed]
- Beris, D.; Theologidis, I.; Skandalis, N.; Vassilakos, N. Bacillus amyloliquefaciens strain MBI600 induces salicylic acid dependent resistance in tomato plants against tomato spotted wilt virus and potato virus Y. Sci. Rep. 2018, 8, 10320. [Google Scholar] [CrossRef] [PubMed]
- Niu, D.D.; Liu, H.X.; Jiang, C.H.; Wang, Y.P.; Wang, Q.Y.; Jin, H.L.; Guo, J.H. The plant growth-promoting rhizobacterium Bacillus cereus AR156 induces systemic resistance in Arabidopsis thaliana by simultaneously activating salicylate- and jasmonate/ethylene-dependent signaling pathways. Mol. Plant-Microbe Interact. 2011, 24, 533–542. [Google Scholar] [CrossRef] [PubMed]
- Salvatierra-Martinez, R.; Arancibia, W.; Aguilera, S.; Olalde, V.; Bravo, J.; Stoll, A. Colonization ability as an indicator of enhanced biocontrol capacity—An example using two Bacillus amyloliquefaciens strains and Botrytis cinerea infection of tomatoes. J. Phytotahol. 2018, 166, 601–612. [Google Scholar] [CrossRef]
- McKinnon, A.C. Plant tissue preparation for the detection of an endophytic fungus in planta. Methods Mol. Biol. 2016, 1477, 167–173. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, F.; Castro, M.; Príncipe, A.; Borioli, G.; Fischer, S.; Mori, G.; Jofre, E. The plant-associated Bacillus amyloliquefaciens strains MEP218 and ARP23 capable of producing the cyclic lipopeptides iturin or surfactin and fengycin are effective in biocontrol of sclerotinia stem rot disease. J. Appl. Microbiol. 2012, 112, 159–174. [Google Scholar] [CrossRef]
- Koumoutsi, A.; Chen, X.H.; Henne, A.; Liesegang, H.; Hitzeroth, G.; Franke, P.; Vater, J.; Borriss, R. Structural and functional characterization of gene clusters directing nonribosomal synthesis of bioactive cyclic lipopeptides in Bacillus amyloliquefaciens strain FZB42. J. Bacteriol. 2004, 186, 1084–1096. [Google Scholar] [CrossRef]
- Trapnell, C.; Pachter, L.; Salzberg, S.L. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 2009, 25, 1105–1111. [Google Scholar] [CrossRef]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef]
- Chitwood, D.H.; Maloof, J.N.; Sinha, N.R. Dynamic transcriptomic profiles between tomato and a wild relative reflect distinct developmental architectures. Plant Physiol. 2013, 162, 537–552. [Google Scholar] [CrossRef] [PubMed]
- Wehrens, R.; Buydens, L.M. Self-and super-organizing maps in R: The Kohonen package. J. Stat. Softw. 2007, 21, 1–19. [Google Scholar] [CrossRef]
- Klopfenstein, D.V.; Zhang, L.; Pedersen, B.S.; Ramírez, F.; Vesztrocy, A.W.; Naldi, A.; Mungall, C.J.; Yunes, J.M.; Botvinnik, O.; Weigel, M.; et al. GOATOOLS: A Python library for gene ontology analyses. Sci. Rep. 2018, 8, 10872. [Google Scholar] [CrossRef] [PubMed]
- Harel, Y.M.; Mehari, Z.H.; Rav-David, D.; Elad, Y. Systemic resistance to gray mold induced in tomato by benzothiadiazole and Trichoderma harzianum T39. Phytopathology 2014, 104, 150–157. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.; Schmittgen, T. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Choudhary, D.K.; Prakash, A.; Johri, B.N. Induced systemic resistance (ISR) in plants: Mechanism of action. Indian J. Microbiol. 2007, 47, 289–297. [Google Scholar] [CrossRef]
- Kamle, M.; Borah, R.; Bora, H.; Jaiswal, A.K.; Singh, R.K.; Kumar, P. Systemic Acquired Resistance (SAR) and Induced Systemic Resistance (ISR): Role and mechanism of action against phytopathogens. In Fungal Biotechnology and Bioengineering; Springer: Cham, Switzerland, 2020; pp. 457–470. [Google Scholar] [CrossRef]
- Dubey, N.; Singh, K. Role of NBS-LRR proteins in plant defense. In Molecular Aspects of Plant-Pathogen Interaction; Springer: Singapore, 2018; pp. 115–138. [Google Scholar] [CrossRef]
- Coleman, A.D.; Maroschek, J.; Raasch, L.; Takken, F.L.; Ranf, S.; Hückelhoven, R. The Arabidopsis leucine-rich repeat receptor-like kinase MIK2 is a crucial component of early immune responses to a fungal-derived elicitor. New Phytol. 2021, 229, 3453–3466. [Google Scholar] [CrossRef] [PubMed]
- Limpens, E.; Franken, C.; Smit, P.; Willemse, J.; Bisseling, T.; Geurts, R. LysM domain receptor kinases regulating rhizobial Nod factor-induced infection. Science 2003, 302, 630–633. [Google Scholar] [CrossRef]
- Madsen, E.B.; Madsen, L.H.; Radutoiu, S.; Olbryt, M.; Rakwalska, M.; Szczyglowski, K.; Sato, S.; Kaneko, T.; Tabata, S.; Sandal, N.; et al. A receptor kinase gene of the LysM type is involved in legumeperception of rhizobial signals. Nature 2003, 425, 637–640. [Google Scholar] [CrossRef]
- Mulder, L.; Lefebvre, B.; Cullimore, J.; Imberty, A. LysM domains of Medicago truncatula NFP protein involved in Nod factor perception. Glycosylation state, molecular modeling and docking of chitooligosaccharides and Nod factors. Glycobiology 2006, 16, 801–809. [Google Scholar] [CrossRef] [PubMed]
- Radutoiu, S.; Madsen, L.H.; Madsen, E.B.; Jurkiewicz, A.; Fukai, E.; Quistgaard, E.M.; Albrektsen, A.S.; James, E.K.; Thirup, S.; Stougaard, J. LysM domains mediate lipochitin–oligosaccharide recognition and Nfr genes extend the symbiotic host range. EMBO J. 2007, 26, 3923–3935. [Google Scholar] [CrossRef] [PubMed]
- Smit, P.; Limpens, E.; Geurts, R.; Fedorova, E.; Dolgikh, E.; Gough, C.; Bisseling, T. Medicago LYK3, an entry receptor in rhizobial nodulation factor signaling. Plant Physiol. 2007, 145, 183–191. [Google Scholar] [CrossRef] [PubMed]
- Wan, J.; Tanaka, K.; Zhang, X.C.; Son, G.H.; Brechenmacher, L.; Nguyen, T.H.N.; Stacey, G. LYK4, a lysin motif receptor-like kinase, is important for chitin signaling and plant innate immunity in Arabidopsis. Plant Physiol. 2012, 160, 396–406. [Google Scholar] [CrossRef] [PubMed]
- Albert, I.; Böhm, H.; Albert, M.; Feiler, C.E.; Imkampe, J.; Wallmeroth, N.; Brancato, C.; Raaymakers, T.M.; Oome, S.; Zhang, H.; et al. An RLP23–SOBIR1–BAK1 complex mediates NLP-triggered immunity. Nat. Plants 2015, 1, 15140. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Fraiture, M.; Kolb, D.; Löffelhardt, B.; Desaki, Y.; Boutrot, F.F.; Tör, M.; Zipfel, C.; Gust, A.A.; Brunner, F. Arabidopsis receptor-like protein30 and receptor-like kinase suppressor of BIR1-1/EVERSHED mediate innate immunity to necrotrophic fungi. Plant Cell 2013, 25, 4227–4241. [Google Scholar] [CrossRef] [PubMed]
- Brutus, A.; Sicilia, F.; Macone, A.; Cervone, F.; De Lorenzo, G. A domain swap approach reveals a role of the plant wall-associated kinase 1 (WAK1) as a receptor of oligogalacturonides. Proc. Natl. Acad. Sci. USA 2010, 107, 9452–9457. [Google Scholar] [CrossRef] [PubMed]
- Pieterse, C.M.; Van Wees, S.C.; Hoffland, E.; Van Pelt, J.A.; Van Loon, L.C. Systemic resistance in Arabidopsis induced by biocontrol bacteria is independent of salicylic acid accumulation and pathogenesis-related gene expression. Plant Cell 1996, 8, 1225–1237. [Google Scholar] [CrossRef]
- Park, K.S.; Kloepper, J.W. Activation of PR-1a promoter by rhizobacteria that induce systemic resistance in tobacco against Pseudomonas syringae pv. tabaci. Biol. Control 2000, 18, 2–9. [Google Scholar] [CrossRef]
- Jung, H.W.; Hwang, B.K. Isolation, partial sequencing, and expression of pathogenesis-related cDNA genes from pepper leaves infected by Xanthomonas campestris pv. vesicatoria. Mol. Plant-Microbe Interact. 2000, 13, 136–142. [Google Scholar] [CrossRef] [PubMed]
- Jung, H.W.; Tschaplinski, T.J.; Wang, L.; Glazebrook, J.; Greenberg, J.T. Priming in systemic plant immunity. Science 2009, 324, 89–91. [Google Scholar] [CrossRef]
- Bertini, L.; Proietti, S.; Focaracci, F.; Sabatini, B.; Caruso, C. Epigenetic control of defense genes following MeJA-induced priming in rice (O. sativa). J. Plant Physiol. 2018, 228, 166–177. [Google Scholar] [CrossRef]
- Sun, X.C.; Gao, Y.F.; Li, H.R.; Yang, S.Z.; Liu, Y.S. Over-expression of SlWRKY39 leads to enhanced resistance to multiple stress factors in tomato. J. Plant Biol. 2015, 58, 52–60. [Google Scholar] [CrossRef]
- Wani, S.H.; Anand, S.; Singh, B.; Bohra, A.; Joshi, R. WRKY transcription factors and plant defense responses: Latest discoveries and future prospects. Plant Cell Rep. 2021, 40, 1071–1085. [Google Scholar] [CrossRef] [PubMed]
- Levine, A.; Tenhaken, R.; Dixon, R.; Lamb, C. H2O2 from the oxidative burst orchestrates the plant hypersensitive disease resistance response. Cell 1994, 79, 583–593. [Google Scholar] [CrossRef]
- Rajasekhar, V.K.; Lamb, C.; Dixon, R.A. Early events in the signal pathway for the oxidative burst in soybean cells exposed to avirulent Pseudomonas syringae pv glycinea. Plant Physiol. 1999, 120, 1137–1146. [Google Scholar] [CrossRef]
- Dutton, M.V.; Evans, C.S. Oxalate production by fungi: Its role in pathogenicity and ecology in the soil environment. Can. J. Microbiol. 1996, 42, 881–895. [Google Scholar] [CrossRef]
- Chen, W.; Chao, G.; Singh, K.B. The promoter of a H2O2-inducible, Arabidopsis glutathione S-transferase gene contains closely linked OBF-and OBP1-binding sites. Plant J. 1996, 10, 955–966. [Google Scholar] [CrossRef] [PubMed]
- Kachroo, A.; Robin, G.P. Systemic signaling during plant defense. Curr. Opin. Plant Biol. 2013, 16, 527–533. [Google Scholar] [CrossRef]
- Chanda, B.; Xia, Y.; Mandal, M.K.; Yu, K.; Sekine, K.T.; Gao, Q.M.; Selote, D.; Hu, Y.; Stromberg, A.; Navarre, D.; et al. Glycerol-3-phosphate is a critical mobile inducer of systemic immunity in plants. Nat. Genet. 2011, 43, 421–427. [Google Scholar] [CrossRef]
- Kachroo, A.; Kachroo, P. Mobile signals in systemic acquired resistance. Curr. Opin. Plant Biol. 2020, 58, 41–47. [Google Scholar] [CrossRef]
- Kachroo, A.; Kachroo, P. Fatty acid–derived signals in plant defense. Annu. Rev. Phytopathol. 2009, 47, 153–176. [Google Scholar] [CrossRef] [PubMed]
- Yu, K.; Soares, J.M.; Mandal, M.K.; Wang, C.; Chanda, B.; Gifford, A.N.; Fowler, J.S.; Navarre, D.; Kachroo, A.; Kachroo, P. A feedback regulatory loop between G3P and lipid transfer proteins DIR1 and AZI1 mediates azelaic-acid-induced systemic immunity. Cell Rep. 2013, 3, 1266–1278. [Google Scholar] [CrossRef] [PubMed]
- Munnik, T. Phosphatidic acid: An emerging plant lipid second messenger. Trends Plant Sci. 2001, 6, 227–233. [Google Scholar] [CrossRef]
- Rodas-Junco, B.A.; Nic-Can, G.I.; Muñoz-Sánchez, A.; Hernández-Sotomayor, S.M. Phospholipid signaling is a component of the salicylic acid response in plant cell suspension cultures. Int. J. Mol. Sci. 2020, 21, 5285. [Google Scholar] [CrossRef] [PubMed]
- Gallego-Giraldo, L.; Posé, S.; Pattathil, S.; Peralta, A.G.; Hahn, M.G.; Ayre, B.G.; Sunuwar, J.; Hernandez, J.; Patel, M.; Shah, J.; et al. Elicitors and defense gene induction in plants with altered lignin compositions. N. Phytol. 2018, 219, 1235–1251. [Google Scholar] [CrossRef] [PubMed]
- Zuluaga, D.L.; Gonzali, S.; Loreti, E.; Pucciariello, C.; Degl’Innocenti, E.; Guidi, L.; Alpi, A.; Perata, P. Arabidopsis thaliana MYB75/PAP1 transcription factor induces anthocyanin production in transgenic tomato plants. Funct. Plant Biol. 2008, 35, 606–618. [Google Scholar] [CrossRef] [PubMed]
- Bhargava, A.; Mansfield, S.D.; Hall, H.C.; Douglas, C.J.; Ellis, B.E. MYB75 functions in regulation of secondary cell wall formation in the Arabidopsis inflorescence stem. Plant Physiol. 2010, 154, 1428–1438. [Google Scholar] [CrossRef]
- Zhou, J.; Lee, C.; Zhong, R.; Ye, Z.H. MYB58 and MYB63 are transcriptional activators of the lignin biosynthetic pathway during secondary cell wall formation in Arabidopsis. Plant Cell 2009, 21, 248–266. [Google Scholar] [CrossRef]
- Gallego-Giraldo, L.; Escamilla-Trevino, L.; Jackson, L.A.; Dixon, R.A. Salicylic acid mediates the reduced growth of lignin down-regulated plants. Proc. Natl. Acad. Sci. USA 2011, 108, 20814–20819. [Google Scholar] [CrossRef]
- Gallego-Giraldo, L.; Jikumaru, Y.; Kamiya, Y.; Tang, Y.; Dixon, R.A. Selective lignin downregulation leads to constitutive defense response expression in alfalfa (Medicago sativa L.). N. Phytol. 2011, 190, 627–639. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Calvo, P.; Chini, A.; Fernández-Barbero, G.; Chico, J.M.; Gimenez-Ibanez, S.; Geerinck, J.; Eeckhout, D.; Schweizer, F.; Godoy, M.; Franco-Zorrilla, J.M.; et al. The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 2011, 23, 701–715. [Google Scholar] [CrossRef] [PubMed]
- Kanzaki, H.; Saitoh, H.; Ito, A.; Fujisawa, S.; Kamoun, S.; Katou, S.; Yoshioka, H.; Terauchi, R. Cytosolic HSP90 and HSP70 are essential components of INF1-mediated hypersensitive response and non-host resistance to Pseudomonas cichorii in Nicotiana benthamiana. Mol. Plant Pathol. 2003, 4, 383–391. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Lee, S.E.; Oh, S.; Seo, E.; Choi, D. HSP70s enhance a Phytophthora infestans effector-induced cell death via an MAPK cascade in Nicotiana benthamiana. Mol. Plant-Microbe Interact. 2018, 31, 356–362. [Google Scholar] [CrossRef]
- Govrin, E.M.; Levine, A. The hypersensitive response facilitates plant infection by the necrotrophic pathogen Botrytis cinerea. Curr. Biol. 2000, 10, 751–757. [Google Scholar] [CrossRef]
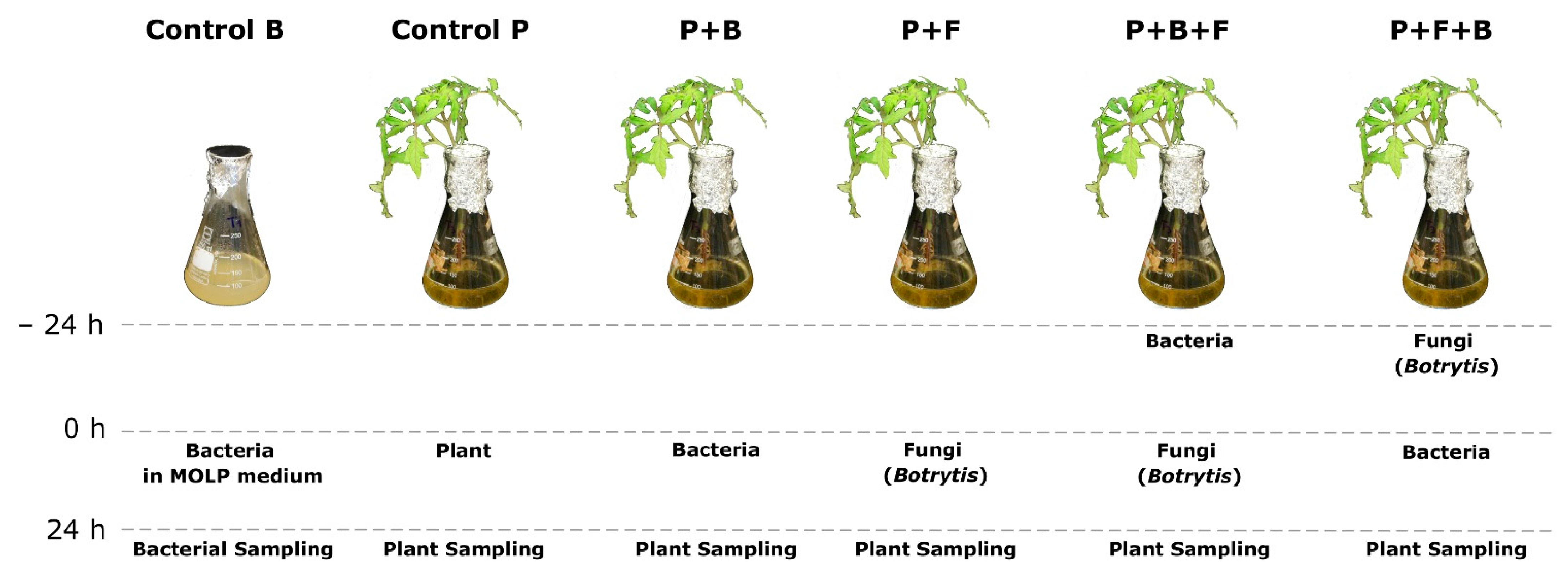



| Treatment | Surfactin C Concentration (µg/mL) | SD n = 3 | Bacterial Cell Count (cfu/mL) | Surfactin C Concentration (µg/mL) Normalized by Bacterial Cell Count |
|---|---|---|---|---|
| Control (CB) | 45.8 | 1.8 | 42,520,000 | 45.8 ± 1.8 |
| P + B | 123.4 | 4.1 | 9,242,500 | 567.6 ± 19.0 |
| P + B + F | 9.2 | 4.5 | 1,275,000 | 308.1 ± 149.6 |
| P + F + B | 15.5 | 1.2 | 1,835,000 | 358.0 ± 26.8 |
| ID | Description | Function |
|---|---|---|
| Solyc09g007010.1.1 | Pathogenesis-related protein 1, PR1 | Pathogenesis-related protein 1 |
| Solyc10g079860.2.1 | Pathogenesis-related protein 2, PR2 | Beta(1,3)glucanase |
| Solyc10g055810.2.1 | Pathogenesis-related protein 3, PR3 | Chitinase |
| Solyc01g097270.3.1 | Pathogenesis-related protein 5, PR5 | Pathogen-induced protein (pi1) |
| Solyc08g080640.2.1 | Pathogenesis-related protein 5, PR5 | Osmotin-like protein (fragment) IPR017949 Thaumatin, conserved site IPR001938 |
| Solyc08g080650.3.1 | Pathogenesis-related protein 5, PR5 | Osmotin-like protein (Fragment) IPR001938 Thaumatin, pathogenesis-related |
| Solyc00g145170.2.1 | Pathogenesis-related protein 6, PR6 | Proteinase inhibitor II |
| Solyc01g067295.1.1 | Pathogenesis-related protein 6, PR6 | Metallocarboxypeptidase inhibitor |
| Solyc03g098710.1.1 | Pathogenesis-related protein 6, PR6 | Kunitz trypsin inhibitor |
| Solyc03g098720.3.1 | Pathogenesis-related protein 6, PR6 | Kunitz trypsin inhibitor |
| Solyc03g098780.2.1 | Pathogenesis-related protein 6, PR6 | Cathepsin D Inhibitor |
| Solyc03g098790.3.1 | Pathogenesis-related protein 6, PR6 | Cathepsin D Inhibitor |
| Solyc07g007250.3.1 | Pathogenesis-related protein 6, PR6 | Metallocarboxypeptidase inhibitor |
| Solyc07g007260.3.1 | Pathogenesis-related protein 6, PR6 | Metallocarboxypeptidase inhibitor |
| Solyc09g083440.3.1 | Pathogenesis-related protein 6, PR6 | PIN-I protein |
| Solyc09g084450.3.1 | Pathogenesis-related protein 6, PR6 | Proteinase inhibitor I |
| Solyc09g084460.3.1 | Pathogenesis-related protein 6, PR6 | Proteinase inhibitor I |
| Solyc09g084465.1.1 | Pathogenesis-related protein 6, PR6 | Wound-induced proteinase inhibitor 1 |
| Solyc09g089500.3.1 | Pathogenesis-related protein 6, PR6 | Proteinase inhibitor I |
| Solyc09g089510.3.1 | Pathogenesis-related protein 6, PR6 | Proteinase inhibitor I |
| Solyc09g089530.3.1 | Pathogenesis-related protein 6, PR6 | PIN-I protein |
| Solyc09g089540.3.1 | Pathogenesis-related protein 6, PR6 | Proteinase inhibitor I |
| Solyc11g021060.2.1 | Pathogenesis-related protein 6, PR6 | TOMARPIX proteinase inhibitor |
| Solyc11g022590.1.1 | Pathogenesis-related protein 6, PR6 | Trypsin inhibitor-like protein precursor |
| Solyc00g071180.3.1 | Pathogenesis-related protein 6, PR6 | Multicystatin-cysteine protease inhibitor |
| Solyc09g089500.3.1 | Pathogenesis-related protein 6, PR6 | Proteinase inhibitor I (AHRD V3.3 K7WNW8_SOLTU) |
| Solyc09g089510.3.1 | Pathogenesis-related protein 6, PR6 | Proteinase inhibitor I (AHRD V3.3 K7WNW8_SOLTU) |
| Solyc01g067295.1.1 | Pathogenesis-related protein 6, PR6 | Metallocarboxypeptidase inhibitor (AHRD V3.3 O24373_SOLTU) |
| Solyc01g006290.3.1 | Pathogenesis-related protein 9, PR9 | Peroxidase |
| Solyc07g006560.3.1 | Pathogenesis-related protein 10, PR10 | Hypersensitive response assisting protein |
| Solyc07g006570.3.1 | Pathogenesis-related protein 10, PR10 | Ribonuclease 3 |
| ID | Description | Domain | Location |
|---|---|---|---|
| Leucine-rich receptor-like serine/threonine protein kinase | |||
| Solyc02g072470.3.1 | GSO1 | Cytoplasmic serine/threonine kinase domain Extracellular leucine-rich repeat domain | Plasma membrane |
| Solyc03g111793.1.1 | Suppressor of BIR1 1, SOBIR1 | Cytoplasmic serine/threonine kinase domain Extracellular leucine-rich repeat domain | Plasma membrane |
| Solyc04g012100.2.1 | Lipase of Fusarium solani 2, FSL2 | Cytoplasmic serine/threonine kinase domain Extracellular leucine-rich repeat domain | Plasma membrane |
| Receptor-like kinases containing leucine-rich repeats (LRRs) | |||
| Solyc04g074000.3.1 | MDIS1-interacting receptor like kinase 2, MIK2 | Extracellular leucine-rich repeat domain Cytoplasmic kinase domain | Plasma membrane |
| Solyc04g074030.3.1 | MDIS1-interacting receptor like kinase 2, MIK2 | Extracellular leucine-rich repeat domain Cytoplasmic kinase domain | Plasma membrane |
| Solyc04g074050.3.1 | MDIS1-interacting receptor like kinase 2, MIK2 | Extracellular leucine-rich repeat domain Cytoplasmic kinase domain | Plasma membrane |
| Solyc05g008950.3.1 | MDIS1-interacting receptor like kinase 2, MIK2 | Extracellular leucine-rich repeat domain Cytoplasmic kinase domain | Plasma membrane |
| Solyc05g008960.3.1 | Leaf rust 10 disease-resistance locus receptor-like protein kinase 4, LRK10L4 | Cytoplasmic serine/threonine kinase domain Extracellular leucine-rich repeat domain | Plasma membrane |
| Solyc06g048740.2.1 | Probable LRR receptor-like kinase | Cytoplasmic aerine/threonine kinase domain Extracellular leucine-rich repeat domain | Plasma membrane |
| Solyc07g055810.3.1 | Probable LRR receptor-like kinase | Cytoplasmic serine/threonine kinase domain Extracellular leucine-rich repeat domain | Plasma membrane |
| Receptor-like protein kinase | |||
| Solyc05g009040.3.1 | Suppressor of npr1-1—constitutive 4, SNC4 | Receptor-like kinase with two extracellular glycerophosphoryl diester phosphodiesterase domains | Plasma membrane |
| Solyc10g076550.1.1 | Receptor-like protein kinase. WAK1 | EGF-like domain | Plasma membrane |
| Solyc09g083200.3.1 | Nod factor receptor protein (LYK4) | Lys motif | Plasma membrane |
| Disease-resistance protein, NBS-LRR class family | |||
| Solyc04g007070.3.1 | R gene—RPP 13 | NBS-LRR class family | Cytoplasm |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Morales, P.; González, M.; Salvatierra-Martínez, R.; Araya, M.; Ostria-Gallardo, E.; Stoll, A. New Insights into Bacillus-Primed Plant Responses to a Necrotrophic Pathogen Derived from the Tomato-Botrytis Pathosystem. Microorganisms 2022, 10, 1547. https://doi.org/10.3390/microorganisms10081547
Morales P, González M, Salvatierra-Martínez R, Araya M, Ostria-Gallardo E, Stoll A. New Insights into Bacillus-Primed Plant Responses to a Necrotrophic Pathogen Derived from the Tomato-Botrytis Pathosystem. Microorganisms. 2022; 10(8):1547. https://doi.org/10.3390/microorganisms10081547
Chicago/Turabian StyleMorales, Paloma, Máximo González, Ricardo Salvatierra-Martínez, Michael Araya, Enrique Ostria-Gallardo, and Alexandra Stoll. 2022. "New Insights into Bacillus-Primed Plant Responses to a Necrotrophic Pathogen Derived from the Tomato-Botrytis Pathosystem" Microorganisms 10, no. 8: 1547. https://doi.org/10.3390/microorganisms10081547






