The Loss of focA Gene Increases the Ability of Salmonella Enteritidis to Exit from Macrophages and Boosts Early Extraintestinal Spread for Systemic Infection in a Mouse Model
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Plasmids and Cells
2.2. Mice and Animal Ethics
2.3. Construction of Tn5 Mutant Library and Mutant Screen
2.4. Identification of Sequence Flanking Tn5 Inserted in Bacterial Genome
2.5. Cosntruction of SEΔfocA and SEΔfocA::focA
2.6. In Vitro Exiting Ability of Intramacrophage Salmonella focA Mutants
2.7. Virulence Analysis of Salmonella focA Mutant in Mice
2.7.1. Extraintestinal Spread
2.7.2. Lethal Ability
2.7.3. mRNA Level of Cytokines in Murine Spleen
2.8. Biological Feature Analysis for Possible Mechanisms
2.8.1. Formate Level in the Bacteria
2.8.2. Formate Level in Salmonella-Infected Macrophages
2.8.3. LDH Assay for Cytotoxicity
2.8.4. Pyroptosis and Apoptosis Assessment for Cell Death
2.8.5. Motility Analysis for Flagella
2.9. Statistical Analysis
3. Results
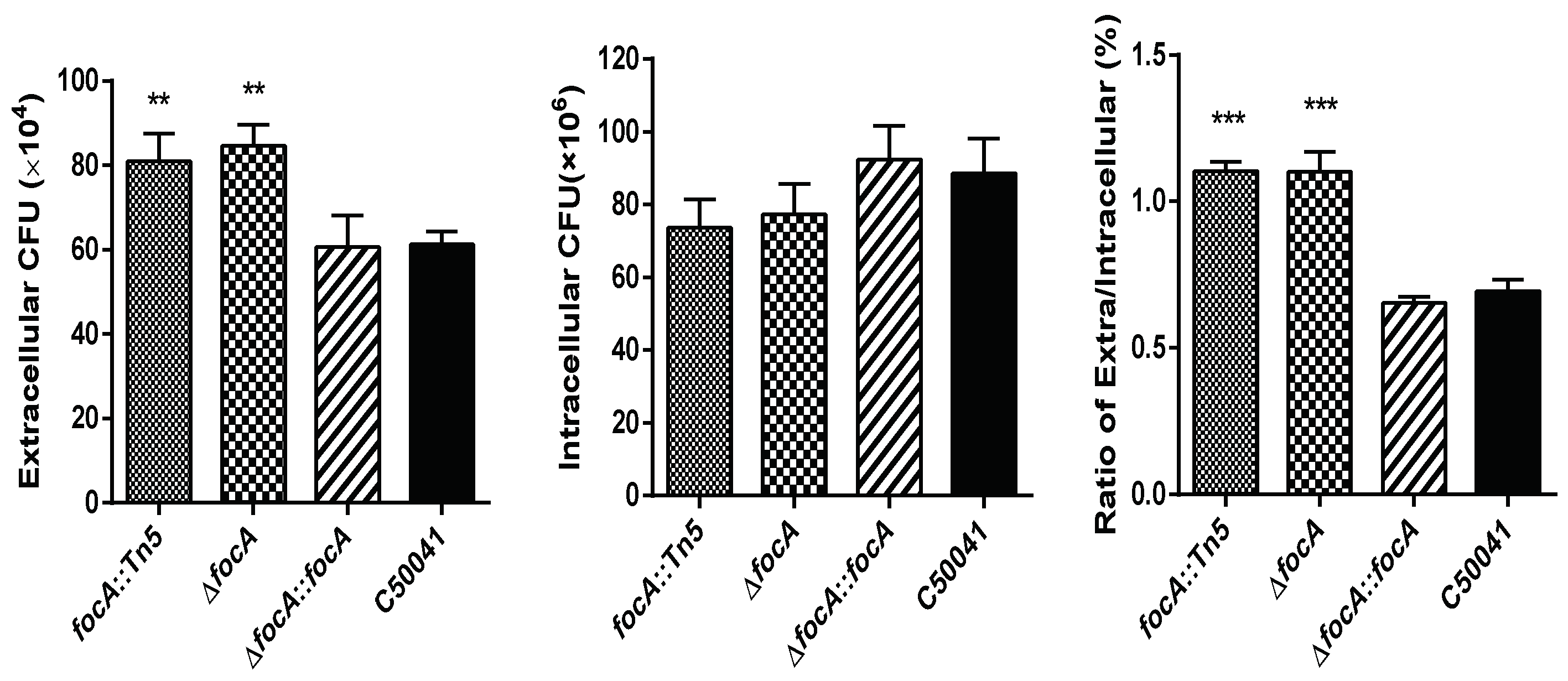
3.1. focA::Tn5 Mutant with Imrpoved Exiting Ability from Macrophages
3.2. SEΔfocA Mutant Reconfirmed
3.3. Virulence Analysis of Salmonella focA Mutants in Mice
3.3.1. Improved Early Extraintestinal Spreading Ability
3.3.2. No Obvious Change in Lethal Ability in Mice
3.3.3. Increased Ability to Promote Murine Proinflammatory Cytokines
3.4. Biological Phenotype Analysis of focA Mutant for Possible Mechanisms
3.4.1. Less Formate Produced by focA Mutant
3.4.2. Less Formate in focA Mutant-Infected RAW264.7
3.4.3. Increased Cytotoxicity by focA Mutants
3.4.4. No Obvious Change in Cellular Pyroptosis Based on Caspase-1 Protein Measurement by focA Mutants
3.4.5. No Obvious Change in Cellular Apoptosis Based on Annexin V-FITC/PI Staining by focA Mutants
3.4.6. No Obvious Change of Flagella Based on Bacterial Motility by focA Mutants
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Strains or Plasmids | Characteristic | Reference |
|---|---|---|
| Strains | ||
| SE C50041 | Wild-type Salmonella Enteritidis C50041 | Lab collection |
| SE C50041focA::Tn5 | C50041 with Tn5 inserted in focA gene | This study |
| SE C50041ΔfocA | C50041 with a defined deletion of the focA gene | This study |
| SE C50041ΔfocA::focA | C50041∆focA with pBR322 expressing the focA gene | This study |
| E.coli χ7213 | Its growth for pGMB152 with DAP, as conjugal donor | [27] |
| Plasmids | ||
| pUT mini-Tn5Km2(Cm) | Transposon delivery vector, Cmr, Kmr | [28] |
| pGMB152 | pGMB151 derivative, suicide vector, Ampr, Smr, LacZYA | [27] |
| pBR322 | For construction of C50041ΔfocA::focA, Cmr | [29] |
| pBR322-focA | pPR322 derivative containing focA, Ampr and Tetr | This study |
| Cell | ||
| RAW264.7 J774A.1 | Murine macrophages Murine macrophages | This study This study |
Appendix B
| Primer Name | Primer Sequences (5′-3′) | Target |
|---|---|---|
| Y-linker | CTGCTCGAATTCAAGCTTCT | PCR of sequence-flanking Tn5 in bacterial genome |
| P6U | CGAGCTCGAATTCGGCCTAG | |
| Tn5-P | GGCCAGATCTGACAAGAGA | |
| Adapter | TTTCTGCTCGAATTCAAGCTTC TAACGATGTACGGGGACACATG | |
| TGTCCCCGTACATCGTTAGAACTACTCGTACCATCCACAT | ||
| focA-up-F | CCCCCCCTGCAGGTCGACGTGCGTCTTGCTCGGTGAT | Construction of SEΔfocA |
| focA-up-R | CAGCCTACACAATCGCTCAA GATGCCCATTACACGCAGTAA | |
| focA-Cm-F | TTACTGCGTGTAATGGGCATC TTGAGCGATTGTGTAGGCTG | |
| focA-Cm-R | CTTTGTTAGTATCTCGTCGCCG ATGGGAATTAGCCATGGTCC | |
| focA-down-F | GGACCATGGCTAATTCCCAT CGGCGACGAGATACTAACAAAG | |
| focA-down-R | CTTATCGATACCGTCGACTGCGTGAACTGTTGGGTCTG | |
| focA-in-F | ACGCAGGTAAATGACCCAGT | |
| focA-in-R | TTTTCGTGTTACTGATGTGGC | |
| pGMB152-F | CGTGGAGGCCATCAAACCAC | |
| pGMB152-R | CGCGAAATAAACGACCGGGA | |
| R-focA-F | TTATCATCGATAAGCTTTTGTTAGTATCTCGTCGCCGACT | Construction of SEΔfocA::focA |
| R-focA-R | TCCGGCGTAGAGGATCCTGCGTGTAATGGGCATCAAC |
Appendix C
| Primer Name | Primer Sequence (5′-3′) | Size (bp) |
|---|---|---|
| IL-1β-F | TGGCCTTCAAAGGAAAGAATCTATACCTGTCC | 167 |
| IL-1β-R | GTTGGGGAACTCTGCAGACTCAAACTCCAC | |
| IL-12-F | TGCCCCCACAGAAGACGTCTTTGATGAT | 138 |
| IL-12-R | GATGGCCACCAGCATGCCCTTGTC | |
| TNF-α-F | CAGGCCTTCCTACCTTCAGACCTTTCCAGAT | 122 |
| TNF-α-R | ACACCCCGCCCTTCCAAATAAATACATTCAT | |
| IFN-γ-F | GCCAAGACTGTGATTGCGGGGTTGTATCT | 198 |
| IFN-γ-R | TAAAGCGCTGGCCCGGAGTGTAGACA | |
| GAPDH-F | CAGCCTCGTCCCGTAGACAA | 156 |
| GAPDH-R | ACCCCGTCTCCGGAGTCCATCACAAT |
References
- WHO. Critically Important Antimicrobials for Human Medicine; World Health Organization: Geneva, Switzerland, 2017.
- Ao, T.T.; Feasey, N.A.; Gordon, M.A.; Keddy, K.H.; Angulo, F.; Crump, J.A. Global burden of invasive nontyphoidal Salmonella disease. Emerg. Infect. Dis. 2010, 21, 941–949. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pullinger, G.D.; Paulin, S.M.; Charleston, B.; Watson, P.R.; Bowen, A.J.; Dziva, F.; Morgan, E.; Villarreal-Ramos, B.; Wallis, T.S.; Stevens, M.P. Systemic translocation of Salmonella enterica serovar Dublin in cattle occurs predominantly via efferent lymphatics in a cell-free niche and requires type III secretion system 1 (T3SS-1) but not T3SS-2. Infect. Immun. 2007, 75, 5191–5199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mastroeni, P.; Grant, A.J. Spread of Salmonella enterica in the body during systemic infection: Unravelling host and pathogen determinants. Expert. Rev. Mol. Med. 2011, 13, e12. [Google Scholar] [CrossRef]
- García-del Portillo, F. Salmonella intracellular proliferation: Where, when and how? Microbes. Infect. 2001, 3, 1305–1311. [Google Scholar] [CrossRef]
- Lathrop, S.K.; Binder, K.A.; Starr, T.; Cooper, K.G.; Chong, A.; Carmody, A.B.; Steele-Mortimer, O. Replication of Salmonella enterica Serovar Typhimurium in Human Monocyte-Derived Macrophages. Infect. Immun. 2015, 83, 2661–2671. [Google Scholar] [CrossRef] [Green Version]
- Pucciarelli, M.G.; García-Del Portillo, F. Salmonella intracellular lifestyles and their impact on host-to-host transmission. Microbiol. Spectr. 2017, 5. [Google Scholar] [CrossRef]
- Gog, J.R.; Murcia, A.; Osterman, N.; Restif, O.; McKinley, T.J.; Sheppard, M.; Achouri, S.; Wei, B.; Mastroeni, P.; Wood, J.L. Dynamics of Salmonella infection of macrophages at the single cell level. J. R. Soc. Interface 2012, 9, 2696–2707. [Google Scholar] [CrossRef] [Green Version]
- Hybiske, K.; Stephens, R.S. Exit strategies of intracellular pathogens. Nat. Rev. Microbiol. 2008, 6, 99–110. [Google Scholar] [CrossRef]
- Jennison, A.V.; Verma, N.K. Shigella flexneri infection: Pathogenesis and vaccine development. FEMS Microbiol. Rev. 2003, 28, 43–58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mounier, J.; Ryter, A.; Coquis-Rondon, M.; Sansonetti, P.J. Intracellular and cell-to-cell spread of Listeria monocytogenes involves interaction with F-actin in the enterocytelike cell line Caco-2. Infect. Immun. 1990, 58, 1048–1058. [Google Scholar] [CrossRef] [Green Version]
- Hybiske, K.; Stephens, R.S. Mechanisms of host cell exit by the intracellular bacterium Chlamydia. Proc. Natl. Acad. Sci. USA 2007, 104, 11430–11435. [Google Scholar] [CrossRef] [Green Version]
- Alli, O.A.; Gao, L.Y.; Pedersen, L.L.; Zink, S.; Radulic, M.; Doric, M.; Abu Kwaik, Y. Temporal pore formation-mediated egress from macrophages and alveolar epithelial cells by Legionella pneumophila. Infect. Immun. 2000, 68, 6431–6440. [Google Scholar] [CrossRef] [PubMed]
- Galluzzi, L.; Vitale, I.; Aaronson, S.A.; Abrams, J.M.; Adam, D.; Agostinis, P.; Alnemri, E.S.; Altucci, L.; Amelio, I.; Andrews, D.W.; et al. Molecular mechanisms of cell death: Recommendations of the Nomenclature Committee on Cell Death. Cell Death Differ. 2018, 25, 486–541. [Google Scholar] [CrossRef] [PubMed]
- Guiney, D.G. The role of host cell death in Salmonella infections. Curr. Top. Microbiol. Immunol. 2005, 289, 131–150. [Google Scholar]
- Hueffer, K.; Galan, J.E. Salmonella-induced macrophage death: Multiple mechanisms, different outcomes. Cell. Microbiol. 2004, 6, 1019–1025. [Google Scholar] [CrossRef]
- Chen, L.M.; Kaniga, K.; Galan, J.E. Salmonella spp. are cytotoxic for cultured macrophages. Mol. Microbiol. 1996, 21, 1101–1115. [Google Scholar] [CrossRef] [PubMed]
- Monack, D.M.; Raupach, B.; Hromockyj, A.E.; Falkow, S. Salmonella typhimurium invasion induces apoptosis in infected macrophages. Proc. Natl. Acad. Sci. USA 1996, 93, 9833–9838. [Google Scholar] [CrossRef] [Green Version]
- Hersh, D.; Monack, D.M.; Smith, M.R.; Ghori, N.; Falkow, S.; Zychlinsky, A. The Salmonella invasin SipB induces macrophage apoptosis by binding to caspase-1. Proc. Natl. Acad. Sci. USA 1999, 96, 2396–2401. [Google Scholar] [CrossRef] [Green Version]
- Brennan, M.A.; Cookson, B.T. Salmonella induces macrophage death by caspase-1-dependent necrosis. Mol. Microbiol. 2000, 38, 31–40. [Google Scholar] [CrossRef] [Green Version]
- Sano, G.-I.; Yasunari, T.; Sinichi, G.; Matuyama, K.; Shindo, Y.; Oka, K.; Matsui, H.; Matsuo, K. Flagella facilitate escape of Salmonella from oncotic macrophages. J. Bacteriol. 2007, 189, 8224–8232. [Google Scholar] [CrossRef] [Green Version]
- Beuzón, C.R.; Méresse, S.; Unsworth, K.E.; Ruíz-Albert, J.; Garvis, S.; Waterman, S.R.; Ryder, T.A.; Boucrot, E.; Holden, D.W. Salmonella maintains the integrity of its intracellular vacuole through the action of SifA. EMBO J. 2000, 19, 3235–3249. [Google Scholar] [CrossRef] [PubMed]
- Sukhan, A.; Kubori, K.; Galan, J.E. Synthesis and Localization of the Salmonella SPI-1 Type III Secretion Needle Complex Proteins PrgI and PrgJ. J. Bacteriol. 2003, 185, 3480–3483. [Google Scholar] [CrossRef] [Green Version]
- Martins, R.P.; Aguilar, C.; Graham, J.E.; Carvajal, A.; Bautista, R.; Claros, M.G.; Garrido, J.J. Pyroptosis and adaptive immunity mechanisms are promptly engendered in mesenteric lymph-nodes during pig infections with Salmonella enterica serovar Typhimurium. Vet. Res. 2013, 44, 120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.; Watarai, M.; Kondo, Y.; Erdenebaatar, J.; Makino, S.; Shirahata, T. Isolation and characterization of mini-Tn5Km2 insertion mutants of Brucella abortus deficient in internalization and intracellular growth in HeLa cells. Infect. Immun. 2003, 71, 3020–3027. [Google Scholar] [CrossRef] [Green Version]
- Lü, W.; Du, J.; Schwarzer, N.J.; Gerbig-Smentek, E.; Einsle, O.; Andrade, S.L. The formate channel FocA exports the products of mixed-acid fermentation. Proc. Natl. Acad. Sci. USA 2012, 109, 13254–13259. [Google Scholar] [CrossRef] [Green Version]
- Geng, S.Z.; Tian, Q.; An, S.M.; Pan, Z.M.; Chen, X.; Jiao, X.A. High-efficiency, two-step scarless-markerless genome genetic modification in Salmonella enterica. Curr. Microbiol. 2016, 72, 700–706. [Google Scholar] [CrossRef] [Green Version]
- Geng, S.Z.; Jiao, X.A.; Barrow, P.; Pan, Z.M.; Chen, X. Virulence determinants of Salmonella Gallinarum biovar Pullorum identified by PCR signature-tagged mutagenesis and the spiC mutant as a candidate live attenuated vaccine. Vet. Microbiol. 2014, 168, 388–394. [Google Scholar] [CrossRef]
- Geng, S.Z.; Wang, Y.N.; Xue, Y.; Wang, H.Q.; Cai, Y.; Zhang, J.; Barrow, P.; Pan, Z.M.; Jiao, X.A. The SseL protein inhibits the intracellular NF-κB pathway to enhance the virulence of Salmonella Pullorum in a chicken model. Microb. Pathog. 2019, 129, 1–6. [Google Scholar] [CrossRef]
- Wang, J.; Wang, Y.; Wang, Z.Y.; Wu, H.; Mei, C.Y.; Shen, P.C.; Pan, Z.M.; Jiao, X.A. Chromosomally located fosA7 in Salmonella isolates from China. Front. Microbiol. 2021, 12, 781306. [Google Scholar] [CrossRef]
- Oizel, K.; Tait-Mulder, J.; Fernandez-de-Cossio-Diaz, J.; Pietzke, M.; Brunton, H.; Lilla, S.; Dhayade, S.; Athineos, D.; Blanco, G.R.; Sumpton, D. Formate induces a metabolic switch in nucleotide and energy metabolism. Cell. Death. Dis. 2020, 11, 310. [Google Scholar] [CrossRef]
- Lin, H.H.; Chen, H.L.; Weng, C.C.; Janapatla, R.P.; Chen, C.L.; Chiu, C.H. Activation of apoptosis by Salmonella pathogenicity island-1 effectors through both intrinsic and extrinsic pathways in Salmonella-infected macrophages. J. Microbiol. Immunol. Infect. 2021, 54, 616–626. [Google Scholar] [CrossRef] [PubMed]
- He, W.T.; Wan, H.; Hu, L.; Chen, P.; Wang, X.; Huang, Z.; Yang, Z.H.; Zhong, C.Q.; Han, J. Gasdermin D is an executor of pyroptosis and required for interleukin-1β secretion. Cell. Res. 2015, 12, 1285–1298. [Google Scholar] [CrossRef] [PubMed]
- Kaiser, P.; Rothwell, L.; Galyov, E.E.; Barrow, P.A.; Burnside, J.; Wigley, P. Differential cytokine expression in avian cells in response to invasion by Salmonella typhimurium, Salmonella enteritidis and Salmonella gallinarum. Microbiology 2000, 146, 3217–3226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Winter, S.E.; Rafatellu, M.; Wilson, R.P.; Russmann, H.; Baumler, A.J. The Salmonella enterica serotype Typhi regulator TviA reduces interleukin-8 production in intestinal epithelial cells by repressing flagellin secretion. Cell. Microbiol. 2008, 10, 247–261. [Google Scholar] [CrossRef]
- Doberenz, C.; Zorn, M.; Falke, D.; Nannemann, D.; Hunger, D.; Beyer, L.; Ihling, C.H.; Meiler, J.; Sinz, A.; Sawers, R.G. Pyruvate formate-lyase interacts directly with the formate channel FocA to regulate formate translocation. J. Mol. Biol. 2014, 426, 2827–2839. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barker, H.C.; Kinsella, N.; Jaspe, N.A.; Friedrich, A.T.; O’Connor, C.D. Formate protects stationary-phase Escherichia coli and Salmonella cells from killing by a cationic antimicrobial peptide. Mol. Microbiol. 2000, 35, 1518–1529. [Google Scholar] [CrossRef] [Green Version]
- Koestler, B.; Fisher, C.R.; Payne, S.M. Formate promotes Shigella intercellular spread and virulence gene expression. mBio 2018, 9, e01777-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fink, S.L.; Cookson, B.T. Pyroptosis and host cell death responses during Salmonella infection. Cell. Microbiol. 2007, 9, 2562–2570. [Google Scholar] [CrossRef]
- Brokatzky, D.; Mostowy, S. Pyroptosis in host defence against bacterial infection. Dis. Model. Mech. 2022, 7, dmm049414. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Abu Kwaik, Y. Hijacking of apoptotic pathways by bacterial pathogens. Microbes. Infect. 2000, 14, 1705–1719. [Google Scholar] [CrossRef]









Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, R.; Zhang, J.; Geng, H.; Wang, Y.; Kang, X.; Geng, S.; Jiao, X.; Barrow, P. The Loss of focA Gene Increases the Ability of Salmonella Enteritidis to Exit from Macrophages and Boosts Early Extraintestinal Spread for Systemic Infection in a Mouse Model. Microorganisms 2022, 10, 1557. https://doi.org/10.3390/microorganisms10081557
Gao R, Zhang J, Geng H, Wang Y, Kang X, Geng S, Jiao X, Barrow P. The Loss of focA Gene Increases the Ability of Salmonella Enteritidis to Exit from Macrophages and Boosts Early Extraintestinal Spread for Systemic Infection in a Mouse Model. Microorganisms. 2022; 10(8):1557. https://doi.org/10.3390/microorganisms10081557
Chicago/Turabian StyleGao, Ran, Jian Zhang, Haoyu Geng, Yaonan Wang, Xilong Kang, Shizhong Geng, Xin’an Jiao, and Paul Barrow. 2022. "The Loss of focA Gene Increases the Ability of Salmonella Enteritidis to Exit from Macrophages and Boosts Early Extraintestinal Spread for Systemic Infection in a Mouse Model" Microorganisms 10, no. 8: 1557. https://doi.org/10.3390/microorganisms10081557
APA StyleGao, R., Zhang, J., Geng, H., Wang, Y., Kang, X., Geng, S., Jiao, X., & Barrow, P. (2022). The Loss of focA Gene Increases the Ability of Salmonella Enteritidis to Exit from Macrophages and Boosts Early Extraintestinal Spread for Systemic Infection in a Mouse Model. Microorganisms, 10(8), 1557. https://doi.org/10.3390/microorganisms10081557






