Synthetic Peptides against Plant Pathogenic Bacteria
Abstract
:1. Introduction
2. Peptides with Distinct Activity: Design and Identification of Leads
2.1. Antimicrobial Peptides (AMPs)
2.1.1. Linear Peptides
2.1.2. Cyclic Peptides

| Peptide | Sequence a | MIC Intervals (μM) | Hemolysis (%) c | ||
|---|---|---|---|---|---|
| Ea b | Pss b | Xav b | |||
| Linear | |||||
| BP76 | KKLFKKILKFL-NH2 | 2.5–5.0 | 2.5–5.0 | 2.5–5.0 | 34 |
| BP100 | KKLFKKILKYL-NH2 | 2.5–5.0 | 2.5–5.0 | 5.0–7.5 | 22 |
| BP143 | KKLfKKILKYL-NH2 | 2.5–5.0 | 2.5–5.0 | 5.0–7.5 | 4 |
| BP250 | KKLA(Tr-Bn)KKILKYL-NH2 | 3.1–6.2 | 1.6–3.1 | 6.2–12.5 | 0 |
| BP275 | Ts-FKLHKKILKVL-NH2 | 12.5–25 | 3.1–6.2 | 3.1–6.2 | 4 |
| BP279 | Ac-FKLH(5-Ph)KKILKVL-NH2 | 12.5–25 | 12.5–25 | 3.1–6.2 | 25 |
| BP387 | Ac-KKLFKKIK(COC3H7)KYL-NH2 | 3.1–6.2 | 3.1–6.2 | 1.6–3.1 | 11 |
| BP389 | Ac-KKLFKKILKK(COC3H7)L-NH2 | 3.1–6.2 | 6.2–12.5 | 0.8–1.6 | 16 |
| BP475 | Ac-KKLfKKILKK(COC3H7)L-NH2 | 3.1–6.2 | 3.1–6.2 | 1.6–3.1 | 0 |
| BP209 | G-KKLFKKILKYL-AGPA-GIGKFLHSAK-OH | 1.2–2.5 | 2.5–5 | <1.2 | 13 |
| BP210 | S-KKLFKKILKYL-AGPA-GIGKFLHSAK-OH | 1.2–2.5 | 2.5–5 | <1.2 | 17 |
| BP211 | G-KKLFKKILKYL-AGPA-KFLHSAK-OH | 1.2–2.5 | 2.5–5 | <1.2 | 1 |
| BP178 | KKLFKKILKYL-AGPA-GIGKFLHSAK-KDEL-OH | 2.5–5.0 | 2.5–5.0 | 2.5–5.0 | 3 |
| Cyclic | |||||
| BPC16 | c(KLKLKFKLKQ) | >100 | 12.5–25 | 6.2–12.5 | 17 |
| BPC194 | c(KKLKKFKKLQ) | 6.2–12.5 | 3.1–6.2 | 3.1–6.2 | 13 |
| BPC198 | c(KLKKKFKKLQ) | 12.5–25 | 3.1–6.2 | 3.1–6.2 | 10 |
| BPC086W | c(LKKKLWKKLQ) | 6.2–12.5 | 3.1–6.2 | 0.8–1.6 | 8 |
| BPC108W | c(LKKKKWLLKQ) | 6.2–12.5 | 1.6–3.1 | 1.6–3.1 | 4 |
| BPC548 | c(KK-Nle(Tr-Ph-Me)-KKFKKLQ) | 12.5–25 | 3.1–6.2 | 3.1–6.2 | 12 |
| BPC550 | c(KK-Nle(Tr-Ph-OMe)-KKFKKLQ) | 12.5–25 | 3.1–6.2 | 3.1–6.2 | 7 |
| BPC702 | c(KKLKk(COC3H7)FKKLQ) | 25–50 | 6.2–12.5 | 6.2–12.5 | 1 |
2.2. Plant Defense Elicitor Peptides
2.3. Bifunctional Peptides
3. Synthesis of Peptides
4. Activity of Peptides against Plant Pathogenic Bacteria
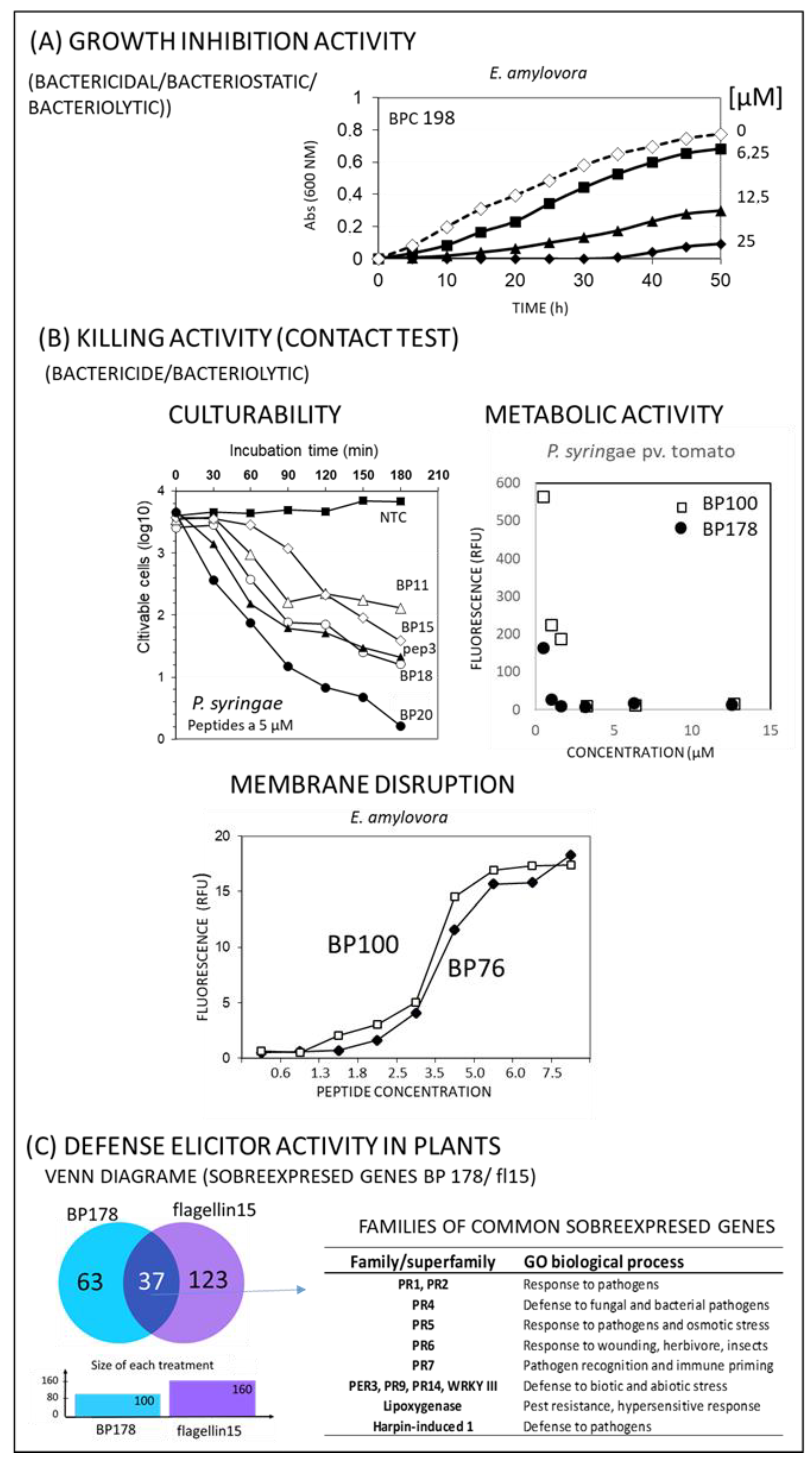
4.1. In Vitro Activity
4.2. Defense Elicitor Activity of Peptides in Plants
4.3. Other Activities

5. Control of Infections in Plant Material Caused by Phytopathogenic Bacteria in Controlled Environment Conditions
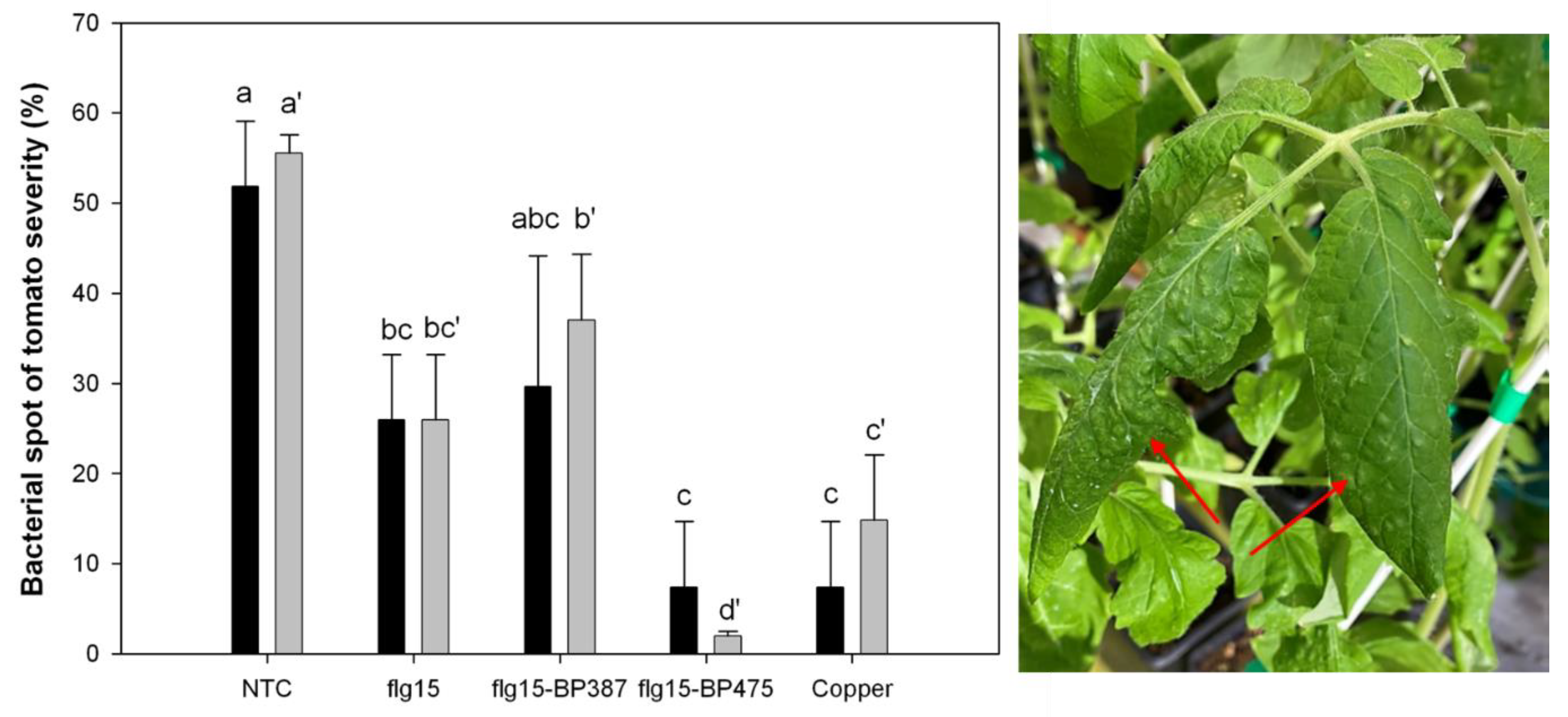
| Peptide | Sequence a | Pathosystem | Gene Overexpression b |
|---|---|---|---|
| BP13 | FKLFKKILKVL-NH2 | Ea/pear | 3/11 |
| BP16 | KKLFKKILKKL-NH2 | Ea/pear | 1/11 |
| BP100 | KKLFKKILKYL-NH2 | Xav/pepper, Pss/pear, Ea/pear | 2/11 |
| BP143 | KKLfKKILKYL-NH2 | Xav/pepper, Pss/pear, Ea/pear | 2/11 |
| BP378 | Ac-KKLFKKILKYK(COC5H11)-NH2 | - | 3/11 |
| BPC200W | c(LLLLKWKKLQ) | Ea/pear | 4/11 |
| BP178 | KKLFKKILKYL-AGPA-GIGKFLHSAK-KDEL-OH | Pst/Xav/tomato | 8/11 |
| flg15-BP16 | RINSAKDDAAGLQIA-KKLFKKILKKL-NH2 | Ea/pear | 6/11 |
| flg15-BP475 | Ac-RINSAKDDAAGLQIA-KKLfKKILKK(COC3H7)L-NH2 | Xav/tomato | 7/11 |
6. Concluding Remarks
7. Ongoing Research
Author Contributions
Funding
Conflicts of Interest
References
- Oerke, E.C. Crop losses to pests. J. Agric. Sci. 2006, 144, 31–43. [Google Scholar] [CrossRef]
- Mansfield, J.; Genin, S.; Magori, S.; Citovsky, V.; Sriariyanum, M.; Ronald, P.; Dow, M.; Verdier, V.; Beer, S.V.; Machado, M.A.; et al. Top 10 plant pathogenic bacteria in molecular plant pathology. Mol. Plant Pathol. 2012, 13, 614–629. [Google Scholar] [CrossRef] [PubMed]
- López, M.M.; Jesús, M.; Montesinos, E.; Palacio-Bielsa, A. Enfermedades de Plantas Causadas por Bacterias; Sociedad Española de Fitopatología (SEF) y Bubok Publishing S.L.: Madrid, Spain, 2013. [Google Scholar] [CrossRef]
- La Torre, A.; Jovino, V.; Caradonia, F. Copper in plant protection: Current situation and prospects. Phytopathol. Mediterrr. 2018, 57, 201–236. [Google Scholar]
- Taylor, P.; Reeder, R. Antibiotic use on crops in low and middle-income countries based on recommendations made by agricultural advisors. CABI Agric. Biosci. 2020, 1, 1. [Google Scholar] [CrossRef]
- Manyi-Loh, C.; Mamphweli, S.; Meyer, E.; Okoh, A. Antibiotic Use in Agriculture and Its Consequential Resistance in Environmental Sources: Potential Public Health Implications. Molecules 2018, 23, 795. [Google Scholar] [CrossRef]
- European Food Safety Authority; Parnell, S.; Camilleri, M.; Diakaki, M.; Schrader Gand Vos, S. Pest survey card on huanglongbing and its vectors. EFSA Supporting Publ. 2019, 16, 1574E. [Google Scholar] [CrossRef]
- Montesinos, E. Antimicrobial peptides and plant disease control. FEMS Microbial. Lett. 2007, 270, 1–11. [Google Scholar] [CrossRef]
- Huan, Y.; Kong, Q.; Mou, H.; Yi, H. Antimicrobial Peptides: Classification, Design, Application and Research Progress in Multiple Fields. Front. Microbiol. 2020, 11, 582779. [Google Scholar] [CrossRef] [PubMed]
- Montesinos, L.; Gascón, B.; Ruz, L.; Badosa, E.; Planas, M.; Feliu, L.; Montesinos, E. A Bifunctional Synthetic Peptide With Antimicrobial and Plant Elicitation Properties That Protect Tomato Plants From Bacterial and Fungal Infections. Appl. Environ. Microbiol. 2020, 12, 756357. [Google Scholar] [CrossRef]
- Benfield, A.H.; Henriques, S.T. Mode-of-Action of Antimicrobial Peptides: Membrane Disruption vs. Intracellular Mechanisms. Front. Med. Technol. 2020, 2, 25–28. [Google Scholar] [CrossRef]
- Schäfer, A.B.; Wenzel, M. A How-To Guide for Mode of Action Analysis of Antimicrobial Peptides. Front. Cell. Infect. Microbiol. 2020, 10, 540898. [Google Scholar] [CrossRef] [PubMed]
- Czékus, Z.; Kukri, A.; Hamow, K.Á.; Szalai, G.; Tari, I.; Ördög, A.; Poór, P. Activation of local and systemic defence responses by flg22 is dependent on daytime and ethylene in intact tomato plants. Int. J. Mol. Sci. 2021, 22, 8354. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, C.; Nadal, A.; Montesinos, E.; Pla, M. Novel Rosaceae plant elicitor peptides as sustainable tools to control Xanthomonas arboricola pv. pruni in Prunus spp. Mol. Plant Pathol. 2018, 19, 418–431, Erratum in Mol. Plant Pathol. 2021, 22, 896. [Google Scholar] [CrossRef] [PubMed]
- Foix, L.; Nadal, A.; Zagorščak, M.; Ramšak, Ž.; Esteve-Codina, A.; Gruden, K.; Pla, M. Prunus persica plant endogenous peptides PpPep1 and PpPep2 cause PTI-like transcriptome reprogramming in peach and enhance resistance to Xanthomonas arboricola pv. pruni. BMC Genom. 2021, 22, 360. [Google Scholar] [CrossRef]
- Santos-Silva, C.A.D.; Zupin, L.; Oliveira-Lima, M.; Vilela, L.M.B.; Bezerra-Neto, J.P.; Ferreira-Neto, J.R.; Ferreira, J.D.C.; Oliveira-Silva, R.L.D.; Pires, C.D.J.; Aburjaile, F.F.; et al. Plant Antimicrobial Peptides: State of the Art, In Silico Prediction and Perspectives in the Omics Era. Bioinform. Biol. Insights 2020, 14, 1177932220952739. [Google Scholar] [CrossRef]
- Iqbal, A.; Khan, R.S.; Shehryar, K.; Imran, A.; Ali, F.; Attia, S.; Shah, S.; Mii, M. Antimicrobial Peptides as Effective Tools for Enhanced Disease Resistance in Plants. Plant Cell. Tissue Organ Cult. 2019, 139, 1–15. [Google Scholar] [CrossRef]
- Phazang, P.; Negi, N.P.; Raina, M.; Kumar, D. Plant Antimicrobial Peptides: Next-Generation Bioactive Molecules for Plant Protection. In Phyto-Microbiome in Stress Regulation; Springer: Singapore, 2020; pp. 281–293. [Google Scholar] [CrossRef]
- Li, J.; Hu, S.; Jian, W.; Xie, C.; Yang, X. Plant Antimicrobial Peptides: Structures, Functions, and Applications. Bot. Stud. 2021, 62, 5. [Google Scholar] [CrossRef]
- Wu, Q.; Patočka, J.; Kuča, K. Insect Antimicrobial Peptides, a Mini Review. Toxins 2018, 10, 461. [Google Scholar] [CrossRef]
- Józefiak, A.; Engberg, R.M. Insect Proteins as a Potential Source of Antimicrobial Peptides in Livestock Production. A Review. J. Anim. Feed Sci. 2017, 26, 87–99. [Google Scholar] [CrossRef]
- Lee, H.; Lee, D.G. Mode of Action of Antimicrobial Peptides Identified from Insects. J. Life Sci. 2015, 25, 715–723. [Google Scholar] [CrossRef]
- Kumar, R.; Ali, S.A.; Singh, S.K.; Bhushan, V.; Mathur, M.; Jamwal, S.; Mohanty, A.K.; Kaushik, J.K.; Kumar, S. Antimicrobial Peptides in Farm Animals: An Updated Review on Its Diversity, Function, Modes of Action and Therapeutic Prospects. Vet. Sci. 2020, 7, 206. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.W.; Kim, B.S. Antimicrobial Cyclic Peptides for Plant Disease Control. Plant Pathol. J. 2015, 31, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Kundu, R. Cationic Amphiphilic Peptides: Synthetic Antimicrobial Agents Inspired by Nature. ChemMedChem 2020, 15, 1887–1896. [Google Scholar] [CrossRef] [PubMed]
- Kishi, R.N.I.; Stach-Machado, D.; de Lacorte Singulani, J.; Santos, C.T.D.; Fusco-Almeida, A.M.; Cilli, E.M.; Freitas-Astúa, J.; Picchi, S.C.; Machado, M.A. Evaluation of Cytotoxicity Features of Antimicrobial Peptides with Potential to Control Bacterial Diseases of Citrus. PLoS ONE 2018, 13, e0203451. [Google Scholar] [CrossRef]
- Kumar, P.; Kizhakkedathu, J.N.; Straus, S.K. Antimicrobial Peptides: Diversity, Mechanism of Action and Strategies to Improve the Activity and Biocompatibility in Vivo. Biomolecules 2018, 8, 4. [Google Scholar] [CrossRef]
- Manniello, M.D.; Moretta, A.; Salvia, R.; Scieuzo, C.; Lucchetti, D.; Vogel, H.; Sgambato, A.; Falabella, P. Insect Antimicrobial Peptides: Potential Weapons to Counteract the Antibiotic Resistance. Cell. Mol. Life Sci. 2021, 78, 4259–4282. [Google Scholar] [CrossRef]
- Zhang, C.; Yang, M. Antimicrobial Peptides: From Design to Clinical Application. Antibiotics 2022, 11, 349. [Google Scholar] [CrossRef]
- EFSA. Update of the Xylella spp. host plant database—Systematic literature search up to 31 December 2020. EFSA J. 2021, 19, 6674. [Google Scholar]
- Baldi, P.; La Porta, N. Xylella fastidiosa: Host range and advance in molecular identification techniques. Front. Plant Sci. 2017, 8, 944. [Google Scholar] [CrossRef]
- Gottwald, T.R.; Bassanezi, R.B. Citrus Huanglongbing: The Pathogen and Its Impact. Plant Health Prog. 2007, 8, 31. [Google Scholar] [CrossRef] [Green Version]
- Blaustein, R.A.; Lorca, G.L.; Teplitski, M. Challenges for Managing Candidatus Liberibacter spp. (Huanglongbing Disease Pathogen): Current Control Measures and Future Directions. Phytopathology 2019, 108, 424–435. [Google Scholar] [CrossRef] [PubMed]
- Cavallarin, L.; Andreu, D.; San Segundo, B. Cecropin A-derived peptides are potent inhibitors of fungal plant pathogens. Mol. Plant Microbe Interact 1998, 11, 218–227. [Google Scholar] [PubMed]
- Ali, G.S.; Reddy, A.S.N. Inhibition of fungal and bacterial plant pathogens by synthetic peptides: In vitro growth inhibition, interaction between peptides, and inhibition of disease progression. Mol. Plant Microbe Interact 2000, 13, 847–859. [Google Scholar] [CrossRef] [PubMed]
- Ferre, R.; Badosa, E.; Feliu, L.; Planas, M.; Montesino, E.; Bardají, E. Inhibition of plant-pathogenic bacteria by short synthetic cecropin A-melittin hybrid peptides. Appl. Environ. Microbiol. 2006, 72, 3302–3308. [Google Scholar] [CrossRef] [PubMed]
- Badosa, E.; Ferre, R.; Planas, M.; Feliu, L.; Besalú, E.; Cabrefiga, J.; Bardají, E.; Montesinos, E. A library of lineal undecapeptides with bactericidal activity against phytopathogenic bacteria. Peptides 2007, 28, 2276–2285. [Google Scholar] [CrossRef]
- Montesinos, E.; Bardají, E. Synthetic antimicrobial peptides as agricultural pesticides for plant-disease control. Chem. Biodivers. 2008, 5, 1225–1237. [Google Scholar] [CrossRef]
- Ng-Choi, I.; Soler, M.; Güell, I.; Badosa, E.; Cabrefiga, J.; Bardají, E.; Montesinos, E.; Planas, M.; Feliu, L. Antimicrobial peptides incorporating non-natural amino acids as agents for plant protection. Protein Pept. Lett. 2014, 21, 357–367. [Google Scholar] [CrossRef]
- Güell, I.; Cabrefiga, J.; Badosa, E.; Ferre, R.; Talleda, M.; Bardají, E.; Planas, M.; Feliu, L.; Montesinos, E. Improvement of the efficacy of linear undecapeptides against plant-pathogenic bacteria by incorporation of D-amino acids. Appl. Environ. Microbiol. 2011, 77, 2667–2675. [Google Scholar] [CrossRef]
- Güell, I.; Micaló, L.; Cano, L.; Badosa, E.; Ferre, R.; Montesinos, E.; Bardají, E.; Feliu, L.; Planas, M. Peptidotriazoles with antimicrobial activity against bacterial and fungal plant pathogens. Peptides 2012, 33, 9–17. [Google Scholar] [CrossRef]
- Ng-Choi, I.; Soler, M.; Cerezo, V.; Badosa, E.; Montesinos, E.; Planas, M.; Feliu, L. Solid-phase synthesis of 5-arylhistidine-containing peptides with antimicrobial activity through a microwave-assisted Suzuki-Miyaura cross-coupling. Eur. J. Org. Chem. 2012, 2012, 4321–4332. [Google Scholar] [CrossRef]
- Oliveras, À.; Baró, A.; Montesinos, L.; Badosa, E.; Montesinos, E.; Feliu, L.; Planas, M. Antimicrobial activity of linear lipopeptides derived from BP100 towards plant pathogens. PLoS ONE. 2018, 13, e0201571. [Google Scholar] [CrossRef] [PubMed]
- Oliveras, À.; Moll, L.; Riesco-Llach, G.; Tolosa-Canudas, A.; Gil-Caballero, S.; Badosa, E.; Bonaterra, A.; Montesinos, E.; Planas, M.; Feliu, L. D-amino acid-containing lipopeptides derived from the lead peptide bp100 with activity against plant pathogens. Inter. J. Mol. Sci. 2021, 22, 6631. [Google Scholar] [CrossRef] [PubMed]
- Zamora-Carreras, H.; Strandberg, E.; Muhlhauser, P.; Burch, J.; Wadhwani, P.; Jimenez, M.A.; Bruix, M.; Ulrich, A.S. Alanine scan and 2H NMR analysis of the membrane-active peptide BP100 point to a distinct carpet mechanism of action. Biochim. Biophys. Acta 2016, 1858, 1328–1338. [Google Scholar] [CrossRef] [PubMed]
- Badosa, E.; Moiset, M.; Montesinos, L.; Talleda, M.; Bardají, E.; Feliu, L.; Planas, M.; Montesinos, E. Derivatives of the antimicrobial peptide BP100 for expression in plant systems. PLoS ONE 2013, 8, e85515. [Google Scholar] [CrossRef]
- Montesinos, L.; Bundó, M.; Badosa, E.; San Segundo, B.; Coca, M.; Montesinos, E. Production of BP178, a derivative of the synthetic antibacterial peptide BP100, in the rice seed endosperm. BMC Plant Biol. 2017, 17, 63. [Google Scholar] [CrossRef]
- Monroc, S.; Badosa, E.; Feliu, L.; Planas, M.; Montesinos, E.; Bardají, E. De novo designed cyclic cationic peptides as inhibitors of plant pathogenic bacteria. Peptides 2006, 27, 2567–2574. [Google Scholar] [CrossRef]
- Monroc, S.; Badosa, E.; Besalú, E.; Planas, M.; Bardají, E.; Montesinos, E.; Feliu, L. Improvement of cyclic decapeptides against plant pathogenic bacteria using a combinatorial chemistry approach. Peptides 2006, 27, 2575–2584. [Google Scholar] [CrossRef]
- Camo, C.; Torné, M.; Besalú, E.; Rosés, C.; Cirac, A.D.; Moiset, G.; Badosa, E.; Bardaji, E.; Montesinos, E.; Planas, M.; et al. Tryptophan-containing cyclic decapeptides with activity against plant pathogenic bacteria. Molecules 2017, 22, 1817. [Google Scholar] [CrossRef]
- Cirac, A.D.; Torné, M.; Badosa, E.; Montesinos, E.; Salvador, P.; Feliu, L.; Planas, M. Rational design of cyclic antimicrobial peptides based on BPC194 and BPC198. Molecules 2017, 22, 1054. [Google Scholar] [CrossRef]
- Güell, I.; Vilà, S.; Micaló, L.; Badosa, E.; Montesinos, E.; Planas, M.; Feliu, L. Synthesis of cyclic peptidotriazoles with activity against phytopathogenic bacteria. Eur. J. Org. Chem. 2013, 22, 4933–4943. [Google Scholar] [CrossRef]
- Güell, I.; Vilà, S.; Badosa, E.; Montesinos, E.; Feliu, L.; Planas, M. Design, synthesis, and biological evaluation of cyclic peptidotriazoles derived from BPC194 as novel agents for plant protection. Pept. Sci. 2017, 108, e23012. [Google Scholar] [CrossRef] [PubMed]
- Vilà, S.; Badosa, E.; Montesinos, E.; Feliu, L.; Planas, M. A convenient solid-phase strategy for the synthesis of antimicrobial cyclic lipopeptides. Org. Biomol. Chem. 2013, 11, 3365–3374. [Google Scholar] [CrossRef] [PubMed]
- Vilà, S.; Badosa, E.; Montesinos, E.; Planas, M.; Feliu, L. Synthetic Cyclolipopeptides Selective against Microbial, Plant and Animal Cell Targets by Incorporation of D-Amino Acids or Histidine. PLoS ONE 2016, 11, e0151639. [Google Scholar] [CrossRef] [PubMed]
- Malik, N.A.A.; Kumar, I.S.; Nadarajah, K. Elicitor and receptor molecules: Orchestrators of plant defense and immunity. Int. J. Mol. Sci. 2020, 21, 963. [Google Scholar] [CrossRef]
- Brunner, Â.; Rosahl, S.; Lee, J.; Rudd, J.J.; Geiler, C.; Scheel, D.; Nu, T. Pep-13, a plant defense-inducing pathogen associated pattern from Phytophthora transglutaminases. EMBO J. 2002, 21, 6681–6688. [Google Scholar] [CrossRef]
- Miyashita, M.; Oda, M.; Ono, Y.; Komoda, E.; Miyagawa, H. Discovery of a Small Peptide from Combinatorial Libraries That Can Activate the Plant Immune System by a Jasmonic Acid Signaling Pathway. Chem. Biol. Chem. 2011, 8502, 1323–1329. [Google Scholar] [CrossRef]
- Pieterse, C.M.J.; Van der Does, D.; Zamioudis, C.; Leon-Reyes, A.; Van Wees, S.C.M. Hormonal modulation of plant immunity. Ann. Rev. Cell Dev. Biol. 2012, 28, 489–521. [Google Scholar] [CrossRef]
- Hajam, I.; Dar, P.; Shahnawaz, I.; Jaume, J.C.; Hwa Lee, J. Bacterial flagellin—A potent immunomodulatory agent. Exp. Mol. Med. 2017, 49, e373. [Google Scholar] [CrossRef]
- Badosa, E.; Montesinos, L.; Camó, C.; Ruz, L.; Cabrefiga, J.; Francés, J.; Gascón, B.; Planas, M.; Feliu, L.; Montesinos, E. Control of fire blight infections with synthetic peptides that elicit plant defense responses. J. Plant Pathol. 2017, 99, 65–73. [Google Scholar] [CrossRef]
- Caravaca-Fuentes, P.; Camó, C.; Oliveras, À.; Baró, A.; Francés, J.; Badosa, E.; Planas, M.; Feliu, L.; Montesinos, E.; Bonaterra, A. A bifunctional peptide conjugate that controls infections of erwinia amylovora in pear plants. Molecules 2021, 26, 3426. [Google Scholar] [CrossRef]
- Oliveras, À.; Camó, C.; Caravaca-fuentes, P.; Moll, L.; Riesco-llach, G.; Badosa, E.; Bonaterra, A.; Montesinos, E.; Feliu, L. Peptide conjugates derived from flg15, Pep13 and PIP1 that are active against plant pathogenic bacteria and trigger plant defense responses. Appl. Environ. Microbiol. 2022, 88, e00574-22. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.; Schmittgen, T. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Camó, C.; Bonaterra, A.; Badosa, E.; Baró, A.; Montesinos, L.; Montesinos, E.; Planas, M.; Feliu, L. Antimicrobial peptide KSL-W and analogues: Promising agents to control plant diseases. Peptides 2019, 112, 85–95. [Google Scholar] [CrossRef] [PubMed]
- Montesinos, E.; Badosa, E.; Cabrefiga, J.; Planas, M.; Feliu, L.; Bardajı, E. Antimicrobial Peptides for Plant Disease Control. From Discovery to Application. In Small Wonders: Peptides for Disease Control; ACS Symposium Series; American Chemical Society: Washington, DC, USA, 2012; pp. 235–261. [Google Scholar] [CrossRef]
- Baró, A.; Mora, I.; Montesinos, L.; Montesinos, E. Differential susceptibility of Xylella fastidiosa strains to synthetic bactericidal peptides. Phytopathology 2020, 110, 1018–1026. [Google Scholar] [CrossRef] [PubMed]
- Baró, A.; Badosa, E.; Montesinos, L.; Feliu, L.; Planas, M.; Montesinos, E.; Bonaterra, A. Screening and identification of BP100 peptide conjugates active against Xylella fastidiosa using a viability-qPCR method. BMC Microbiol. 2020, 20, 229. [Google Scholar] [CrossRef] [PubMed]
- Zaini, P.A.; Fuente, L.D.L.; Hoch, H.C.; Burr, T.J. Grapevine xylem sap enhances biofilm development by Xylella fastidiosa. FEMS Microb. Lett. 2009, 295, 129–134. [Google Scholar] [CrossRef] [PubMed]
- Moll, L.; Badosa, E.; Planas, M.; Feliu, L.; Montesinos, E.; Bonaterra, A. Antimicrobial Peptides With Antibiofilm Activity Against Xylella fastidiosa. Front. Microbiol. 2021, 12, 3243. [Google Scholar] [CrossRef]
- Moll, L.; Baró, A.; Montesinos, L.; Badosa, E.; Bonaterra, A.; Montesinos, E. Induction of defense responses and protection of almond plants against Xylella fastidiosa by endotherapy with a bifunctional peptide. Phytopathology 2022, 112, 1907–1916. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Badosa, E.; Planas, M.; Feliu, L.; Montesinos, L.; Bonaterra, A.; Montesinos, E. Synthetic Peptides against Plant Pathogenic Bacteria. Microorganisms 2022, 10, 1784. https://doi.org/10.3390/microorganisms10091784
Badosa E, Planas M, Feliu L, Montesinos L, Bonaterra A, Montesinos E. Synthetic Peptides against Plant Pathogenic Bacteria. Microorganisms. 2022; 10(9):1784. https://doi.org/10.3390/microorganisms10091784
Chicago/Turabian StyleBadosa, Esther, Marta Planas, Lidia Feliu, Laura Montesinos, Anna Bonaterra, and Emilio Montesinos. 2022. "Synthetic Peptides against Plant Pathogenic Bacteria" Microorganisms 10, no. 9: 1784. https://doi.org/10.3390/microorganisms10091784
APA StyleBadosa, E., Planas, M., Feliu, L., Montesinos, L., Bonaterra, A., & Montesinos, E. (2022). Synthetic Peptides against Plant Pathogenic Bacteria. Microorganisms, 10(9), 1784. https://doi.org/10.3390/microorganisms10091784









