Abstract
Carnobacterium maltaromaticum is a non-starter lactic acid bacterium (LAB) of interest in the dairy industry for biopreservation. This study investigated the interference competition network and the specialized metabolites biosynthetic gene clusters (BGCs) content in this LAB in order to explore the relationship between the antimicrobial properties and the genome content. Network analysis revealed that the potency of inhibition tended to increase when the inhibition spectrum broadened, but also that several strains exhibited a high potency and narrow spectrum of inhibition. The C. maltaromaticum strains with potent anti-L. monocytogenes were characterized by high potency and a wide intraspecific spectrum. Genome mining of 29 strains revealed the presence of 12 bacteriocin BGCs: four of class I and eight of class II, among which seven belong to class IIa and one to class IIc. Overall, eight bacteriocins and one nonribosomal peptide synthetase and polyketide synthase (NRPS-PKS) BGCs were newly described. The comparison of the antimicrobial properties resulting from the analysis of the network and the BGC genome content allowed us to delineate candidate BGCs responsible for anti-L. monocytogenes and anti-C. maltaromaticum activity. However, it also highlighted that genome analysis is not suitable in the current state of the databases for the prediction of genes involved in the antimicrobial activity of strains with a narrow anti-C. maltaromaticum activity.
Keywords:
biopreservation; bioprotection; bacteriocin; NRPS-PKS; lactic acid bacteria; network; genome mining 1. Introduction
Lactic acid bacteria (LAB) are widely industrially used as bioprotective cultures due to their capacity to synthesize antimicrobial compounds, such as primary metabolites resulting from primary metabolism and bacteriocins [1]. Bacteriocins are ribosomally synthesized bacterial peptides or proteins produced by bacteria that may favor niche occupancy over competitors by (i) conquering or replacing a bacterial competitor in an ecological niche; or (ii) maintaining an already colonized niche [2,3]. Bacteriocins can be classified into three classes [4]: class I comprises peptides with less than 50 amino acids that are extensively post-translationally modified; class II refers to small (<10 kDa) and heat-stable, non-modified peptides; and class III brings together large (>10 kDa) and heat-labile proteins.
Although LAB are reported to produce mainly specialized metabolites of the bacteriocin type, the genome of several strains has been described to harbor nonribosomal peptide synthetase (NRPS) and polyketide synthase (PKS) biosynthesis gene clusters (BGCs) [5,6,7]. Nonribosomal peptides (NRPs) comprise a diverse range of natural products of bacterial or fungal origin [8]. Polyketides (PKs) form a large family of molecules synthesized by bacteria, fungi, and plants. Both NRPs and PKs exhibit a wide range of biological functions, including antimicrobial activity [9]. NRPS and PKS systems are typically composed of several enzymatic domains organized in subunits [10]. The basic unit incorporated in the polymers consists of amino acids for NRPs and acyl-coenzyme A for PKs [11].
Carnobacterium maltaromaticum is a non-starter LAB [12,13] of technological interest in cheese production as non-starter culture [14,15,16], as bioprotective cultures in Ricotta fresca cheese [16], meat [17] and seafood [18], or as probiotics in fish [19,20] and rabbits [21]. The biopreservative potential of the C. maltaromaticum species is partly due to the production of bacteriocins of wide inhibition spectra [16,18]. Carnobacterium spp. are reported to produce the class I Carnocin UI49 [22], carnolysin A1-A2 [23] and carnocyclin A [24]; the class IIa carnobacteriocins BM1 and B2 [25], piscicolin 126 [26], piscicocins CS526 [27], V1a and V1b [28]; and the class IIc piscicolin 61 [29] and carnobacteriocin A [30].
Several strains of C. maltaromaticum are commercially available for biopreservation purposes. The strain C. maltaromaticum CB1, which produces the three bacteriocins carnocyclin A, piscicolin 126, and carnobacteriocin BM1, is commercially used to inhibit spoilage and the pathogenic bacterium Listeria monocytogenes during the storage of vacuum packaged meat products [21,31]. Lately, the two strains of C. maltaromaticum F88 and B10 (CNCM I-5242 and CNCM I-5243, respectively) were selected for potent anti-L. monocytogenes in soft cheeses and one of these strains is commercially available for biopreservation applications [32]. In packed Ricotta fresca cheese, a commercial biopreservative Lyofast CNBAL containing Carnobacterium spp. showed a significant reduction of Pseudomonas spp. of 0.83 log without changing the intrinsic parameters and the physicochemical composition of the product [16].
Besides their interest in biopreservation, the impact of the production of inhibitory compounds between non-pathogenic food microorganisms has been studied lately [33]. High-throughput competition assays allowed for investigating 5329 pairwise interactions within a collection of 73 C. maltaromaticum strains that revealed that competition is major within this species. Approximately 56% of the tested strains showed antagonistic activity toward at least one other strain. A high diversity of the inhibitory and sensitivity spectra was found with a majority of narrow spectra, suggesting that interference competition plays a major role in this species. In addition, the network of interference competition in this species is not random but rather structured according to a nested pattern. However, the compounds involved in the inhibitory activities recorded in this study are not well known [33].
The aim of this study was to investigate the relationship between the antimicrobial properties described in a competition network and the genomic features of 29 C. maltaromaticum strains, in order to gain insight into the potential specialized antimicrobial metabolites produced by these strains.
2. Materials and Methods
2.1. Bacterial Strains and Culture Conditions
The strains of C. maltaromaticum (Table S1) and L. monocytogenes EGDe were cultivated in Trypticase Soy Broth medium (bioMérieux, Marcy-l’Etoile, France) supplemented with 6 g·L−1 of bacto-Yeast Extract (TSBYE) at 30 °C without agitation. The cultures in broth were either performed in standing tubes containing 10 or 50 mL of TSBYE medium or in 96-well sterile microplates filled with 195 µL of TSBYE per well. Cultures on a solid medium were performed in TSBYE supplemented with 15 g·L−1 agar and incubated at 30 °C in aerobic conditions.
2.2. Competition Assays and Data Processing
The competition assays and the data processing were performed as previously described [33]. When the strains of C. maltaromaticum were used as the target, the inhibition potency was expressed as the growth inhibition indicator (GII), which was calculated as t2-t1, where t2 is the time (in min) required for the receiver strain culture to reach an optical density value of 0.2 at 595 nm (OD595 nm) when it is cultivated in a fresh medium supplemented with cell-free supernatant, and t1 the time (in min) required for the receiver strain to reach an OD595 nm value of 0.2 when cultivated in fresh medium only. As previously described, GII values higher than the threshold of 300 min were considered inhibitions [33]. When the strain L. monocytogenes EGDe was used as the target, the classification of C. maltaromaticum sender strains as inhibitory or non-inhibitory was performed by non-supervised clustering. To do so, the software R (version 4.1.0, sourced at CRAN) [34] and the package factoextra (version 1.0.7) [35] were used to define the optimal number of groups with the silhouette method and to perform kmeans clustering.
2.3. Genome Sequencing and Analysis
The genome of the C. maltaromaticum strains was sequenced using an Illumina MiSeq sequencer (Eurofins genomics, Les Ulis, France) in order to generate paired-end reads. De novo assembly was performed using QIAGEN CLC Genomics Workbench 10. Genome annotations were carried out using the platform MicroScope (Evry-Courcouronnes, France) [36,37,38,39].
2.4. Data Analysis
Secondary metabolite gene clusters were predicted using BAGEL [40,41,42,43] and antiSMASH [44,45,46,47,48]. Genome clustering was conducted on the MicroScope platform which uses an approach that originates from network science called community detection. The resulting pairwise inter-genomics distances, which are approximately equal to 1-ANI (Average Nucleotide Identity), are used to construct a tree using the neighbour-joining method [39]. The heatmaps were constructed with the R package ComplexHeatmap (version 2.12.1, sourced at Bioconductor) [49]. Hierarchical clustering of the inhibitory profiles was performed by using Euclidean distances and the average method with the R package ComplexHeatmap [49]. Simple linear regression modelling, descriptive statistics, ANOVA, and Tukey’s test were performed in base R [34]. The data manipulation and the graphical representations were performed using the R packages tidyverse (version 1.3.1, sourced at CRAN) [50], ggprism (version 1.0.3, sourced at CRAN) [51], ggrepel (version 0.9.1, sourced at CRAN) [52], and viridis (version 0.6.2, sourced at CRAN) [53].
2.5. Accession Numbers
The accession numbers of enterocin F4-9, carnobacteriocin BM1, carnobacteriocin B2, piscicolin 126, hiracin JM79, lactococcin 972, are BAR87969, AAA23014.1, AAB81310.1, AAX21354.1, Q0Z8B6.1, WP_000726502.1, respectively. The genome sequences and annotations are publicly available on the MicroScope MaGe web portal (Table S1, https://mage.genoscope.cns.fr, last accession on 21 July 2022).
3. Results
3.1. Adjacency Matrix of Inhibition
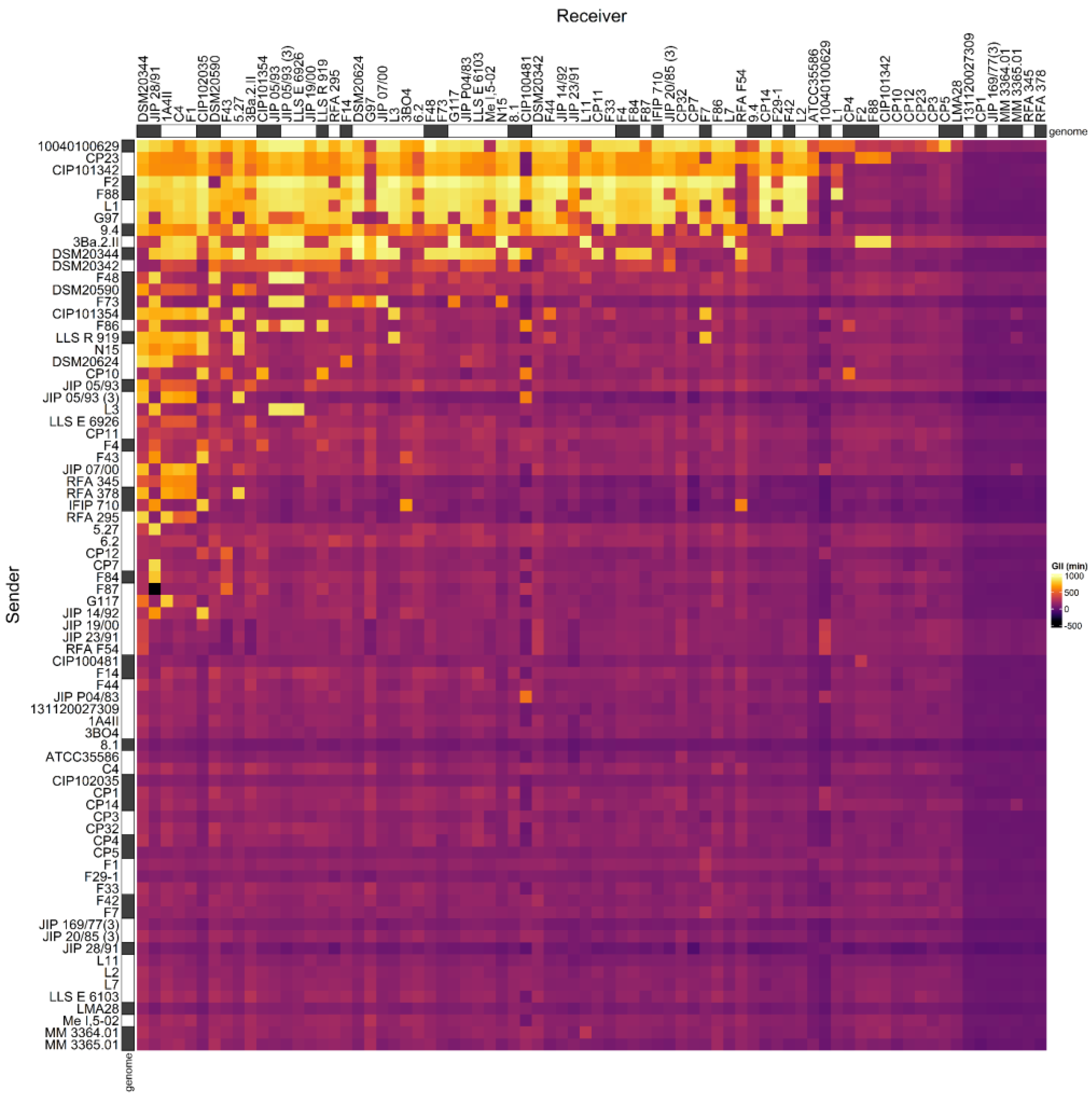
We previously described the antagonistic properties of a set of C. maltaromaticum strains by considering the data in a qualitative binary approach, without considering the intensity of inhibitions [33]. Here, we reconsidered these data by taking into account the inhibition intensities. To do so, the data produced by [33], as well as the data obtained for three additional strains (C. maltaromaticum 10040100629, 3Ba.2.II, and DSM20590) were used to construct an adjacency matrix describing the intensity of inhibition between each pair of strains that can be formed from the set of 76 selected C. maltaromaticum strains (Figure 1).
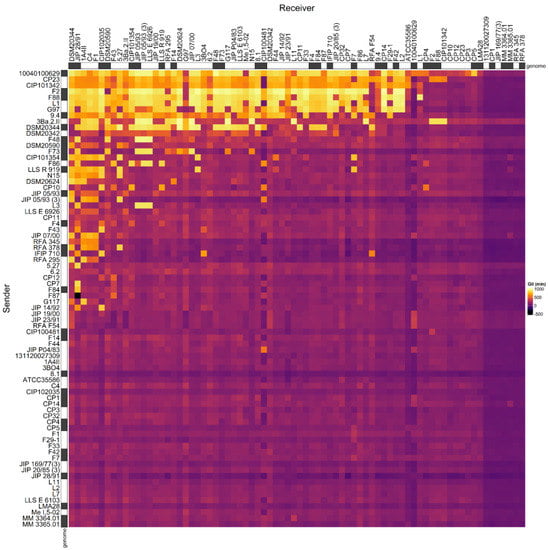
Figure 1.
Adjacency matrix representing the GII inhibition values resulting from pairwise competition assays in C. maltaromaticum. The matrix was constructed with the data produced for 73 strains by [33] and the three additional strains C. maltaromaticum 10040100629, 3Ba.2.II, and DSM20590. The supernatant of sender strains (in rows) was added to cultures of the receiver strains (in columns) in order to measure their impact on the growth of the receivers. The color scale from purple to yellow represents increasing GII values. On the X-axis the strains are sorted in the descending order of sender degree from the left to the right. On the Y-axis, the strains are sorted in descending order of receiver degree, from the top to the bottom. The strains for which the genome was analyzed in this study are indicated with a grey box next to the name of the strains.
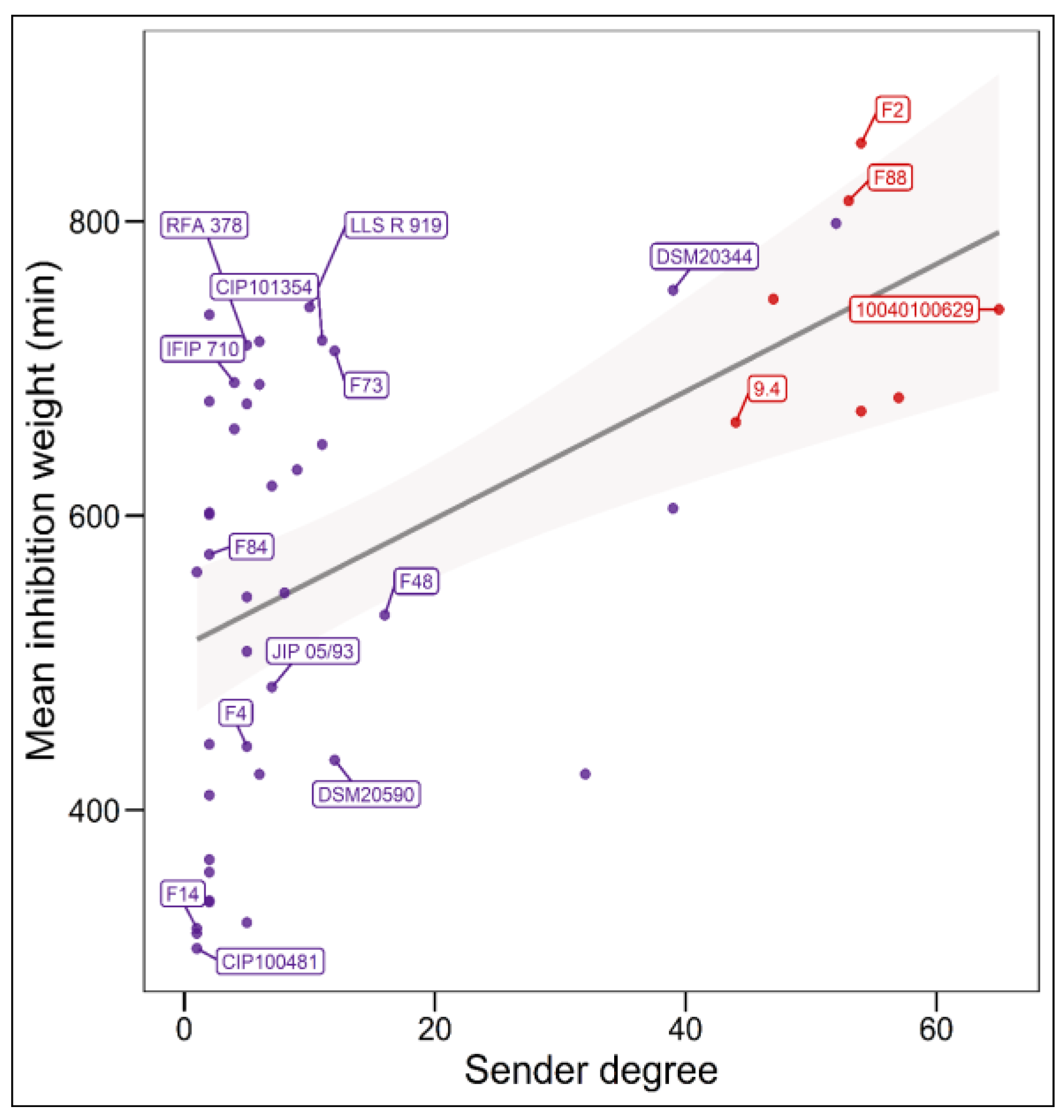
The analysis of the relationship between the mean intensity of inhibition of C. maltaromaticum receiver strains and the sender degree (the number of strains inhibited by a given strain, i.e., the width of the inhibitory spectrum) revealed that the intraspecific inhibition spectra are related to the inhibition potency. Indeed, the broader the spectrum of inhibition of C. maltaromaticum strains, the higher the potency of inhibition (p-value < 8.8 × 10−5) (Figure 2). In addition, the strains that were able to strongly inhibit L. monocytogenes EGDe, including the strains F2, F88, 10040100629, and 9.4, show a wide spectrum of inhibition of conspecific strains. However, the linear model only explains approximately 29% of the variability (R2 = 0.29). This is due to strains with low sender degrees and high inhibition potency, namely strains RFA 378, LLS R 919, IFIP 710, CIP 101354, and F73 (Figure 2). This indicates that strains with narrow intraspecific inhibitory spectra can also exhibit potent antimicrobial properties.
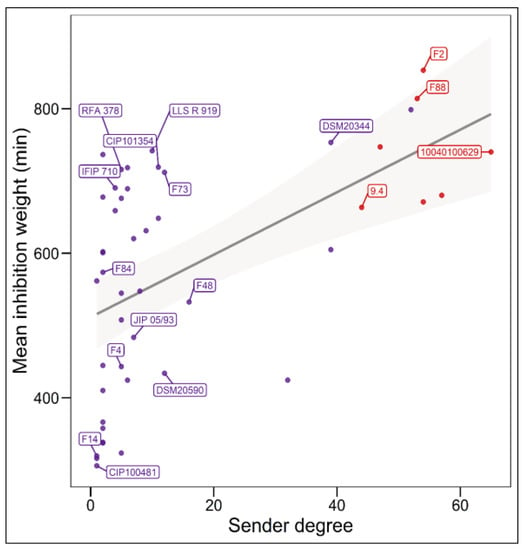
Figure 2.
Relationship between the sender degree, the inhibitory potency, and the ability to inhibit L. monocytogenes EGDe. The sender degree refers to the number of receiver C. maltaromaticum strains inhibited by a given sender C. maltaromaticum strain. The mean inhibition weight is the mean of GII values corresponding to inhibitions and recorded for each sender strain. Only strains for which the sender degree is different from 0 and for which the genome was analysed in this study are labelled. Simple linear regression modelling was performed (p-value = 8.8 × 10−5, R2 = 0.29), the grey shaded area denotes the 95% confidence intervals (p-value < 0.05).
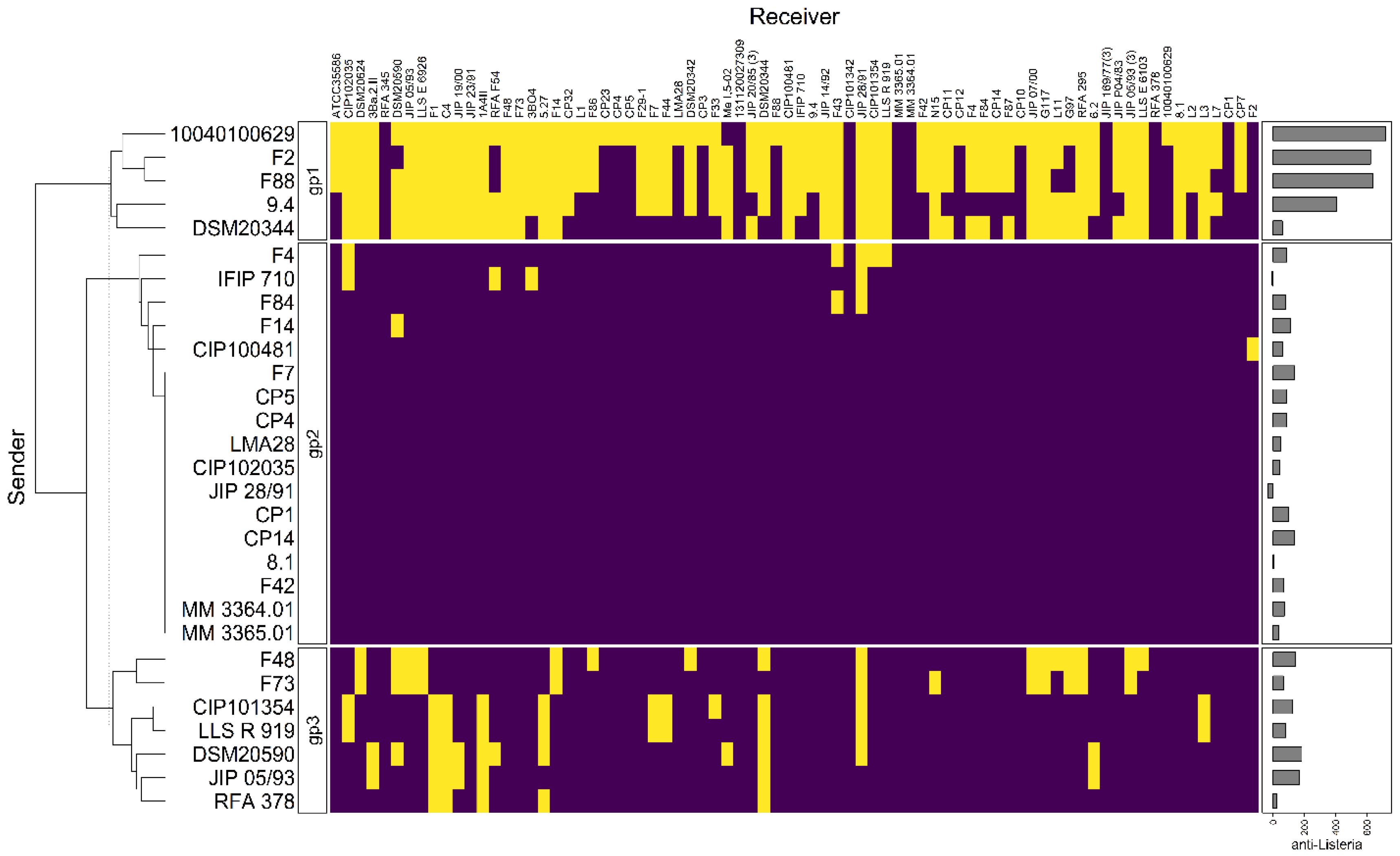
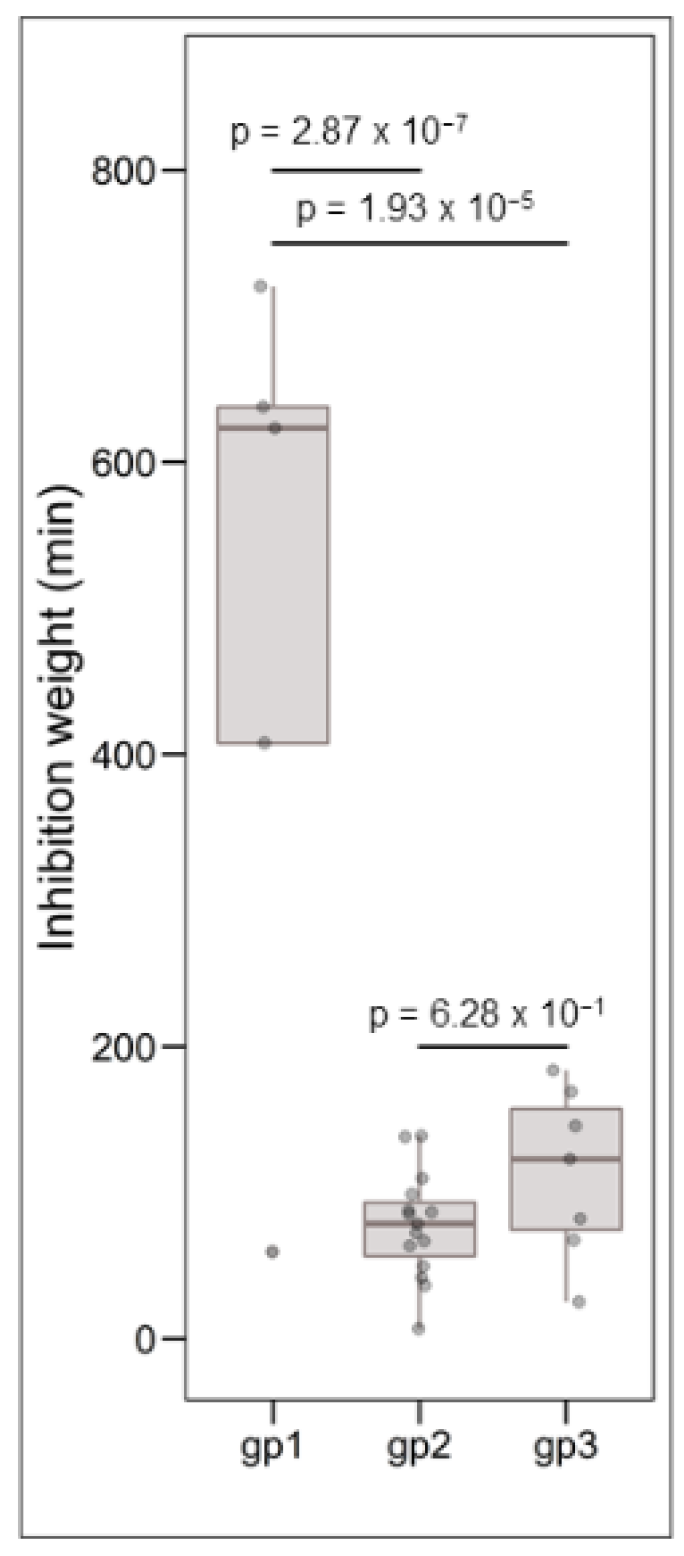
Clustering analysis was performed on the basis of intraspecific inhibition spectra of the 29 strains that were subsequently selected for genome mining. The strains were clustered into three groups: gp1, gp2, and gp3 encompassing 5, 17, and 7 strains, respectively (Figure 3). The strains from gp2 and gp3 have a narrow inhibitory spectrum of C. maltaromaticum strains and weakly inhibit L. monocytogenes EGDe. Conversely, the strains that belong to gp1 have a wide spectrum of inhibition of C. maltaromaticum as well as a high potency of inhibition of L. monocytogenes EGDe (p-values < 0.001, Figure 4). However, not all strains of gp1 are characterized by high anti-L. monocytogenes EGDe potency since among the five strains, the strain DSM20344 is a weak inhibitor of the pathogen (Figure 3).
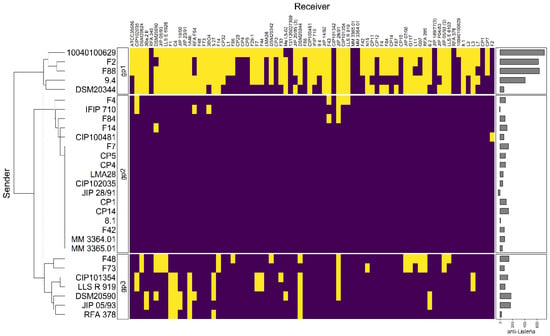
Figure 3.
Hierarchical clustering analysis of the sender strains. The sender strains subjected to genome analysis in this study (in rows) were clustered based on their inhibition profiles of the C. maltaromaticum receiver strains (in columns). Yellow cells represent inhibition, purple cells non-inhibition. The clustering divides the senders into three groups, noted “gp1”, “gp2”, and “gp3”. The barplot annotation on the right represents the quantitative inhibition of L monocytogenes EGDe (in min) by the sender strains.
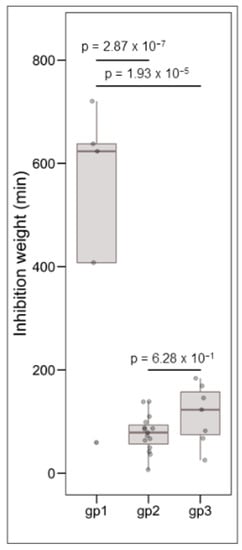
Figure 4.
Boxplots representing the distribution of L. monocytogenes EGDe inhibition, the grouping variable being the group defined by hierarchical clustering (see Figure 3). The data were analysed by one-way ANOVA followed by a Tukey’s test. The resulting p-values are indicated above the compared groups.
3.2. Bacteriocin Gene Clusters
A subset of 29 strains with varying sender and receiver profiles were selected (Figure 1) for genome mining in order to identify potential bacteriocin BGCs. Overall, 13 bacteriocin structural genes encoding 12 bacteriocins were predicted in the genome of these strains (Figure 5, Table 1, Tables S2 and S3).
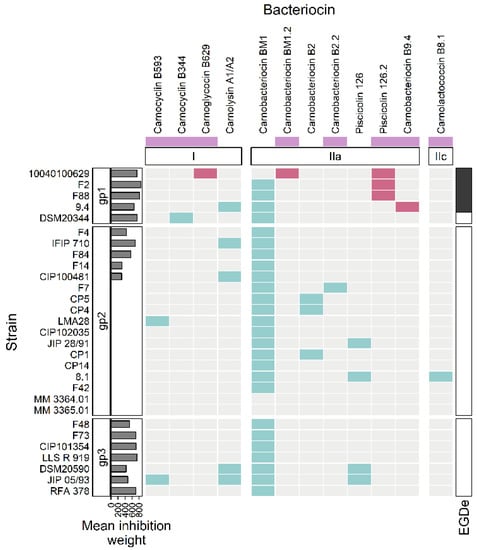
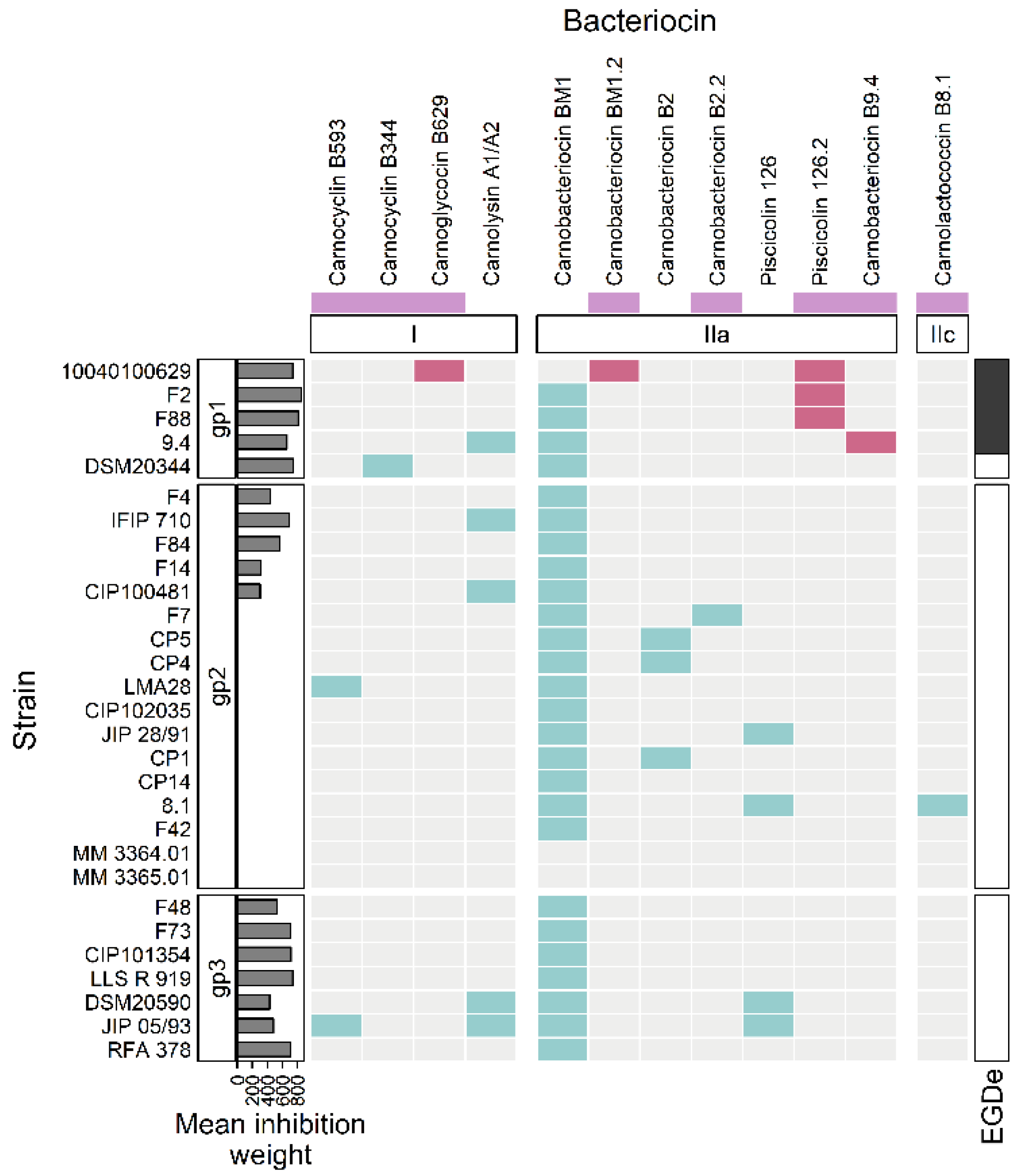
Figure 5.
Heatmap representing the occurrence of bacteriocin BGCs in the genomes. The strains of C. maltaromaticum (in rows) are grouped according to the groups “gp1”, “gp2”, and “gp3”. The considered bacteriocin BGCs (in columns) are grouped according to the class of bacteriocin to which they belong (class I, IIa, IIc). The mauve boxes, below the bacteriocin names, denotes newly described bacteriocins in this study. The barplot annotation on the left represents the mean inhibition weight (in min) of C. maltaromaticum receiver strains by the sender strains. In the annotation block on the right, black boxes indicate inhibition of L. monocytogenes EGDe by the C. maltaromaticum strains. The presence of a bacteriocin BGC is indicated by turquoise or red cells. The red cells denote bacteriocin BGCs specifically found in C. maltaromaticum strains with anti-L. monocytogenes activity, the other BGC are denoted in turquoise.

Table 1.
Bacteriocins predicted from the genome of 29 strains of C. maltaromaticum.
More precisely and according to the classification proposed by Alvares-Siero et al. [4], five structural genes would encode four class I bacteriocins: two head-to-tail cyclic peptides (carnocyclin B593 and carnocyclin B344), one glycocin (carnoglycocin B629) and one two-component lantibiotic (carnolysin A1-A2, encoded by two Coding DNA Sequences, CDSs). Eight clusters were associated with class II bacteriocins, among which seven structural genes of class IIa (carnobacteriocin BM1 and BM1.2, carnobacteriocin B2 and B2.2, piscicolin 126, and 126.2, and carnobacteriocin B9.4), and one structural gene related to class IIc (carnolactococcin B8.1).
Among the 12 possible bacteriocin homologs found in this study, four have 100% identity with previously described bacteriocins: carnolysin A1-A2, carnobacteriocin BM1, carnobacteriocin B2, and piscicolin 126. The other eight bacteriocins have newly identified sequences either because they are variants with slight changes in the amino acid sequences compared to known bacteriocins (carnobacteriocin BM1.2, carnobacteriocin B2.2, and piscicoline 126.2), or because they are distantly related to other bacteriocins (carnocyclin B593, carnocyclin B344, carnoglycocin B629, carnobacteriocin B9.4, and carnolactococcin B8.1).
3.2.1. Class I
The putative carnocyclin B593 (CMALT398_v1_10035, BN424_2342) and carnocyclin B344 (CMDSM20344_v1_130013) are both annotated as belonging to the uberolysin/carnocyclin family of circular bacteriocins (Table 2). Carnocyclin B593 has been predicted in the genome of C. maltaromaticum LMA28 and JIP 05/93, while carnocyclin B344 has been predicted in the genome of the strain DSM20344 (Figure 5). Circular bacteriocins have N-to-C- terminal covalent linkages forming a circular peptide backbone [54]. As expected for group “i” of circular bacteriocins (please refer to [54] for group definition), the predicted isoelectric point is high (8.9 and 10.1 for carnocyclin B593 and B344, respectively). As for other circular bacteriocin BGCs, most of the CDSs encode proteins with membrane-associated function (seven out of nine or eleven CDSs for carnocyclin B593, depending on the strain, and six out of nine CDSs carnocyclin B344), of which several would be involved in transport across the membrane (Figure 6, Table 2).

Table 2.
BGCs potentially encoding the synthesis of specialized metabolites.
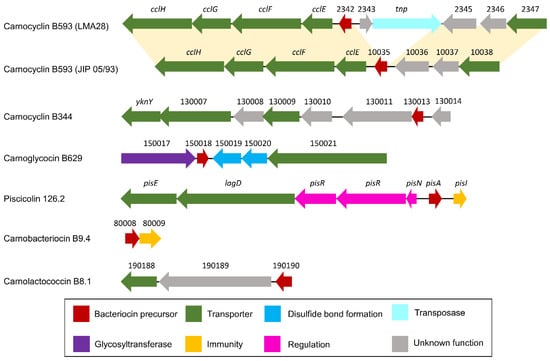
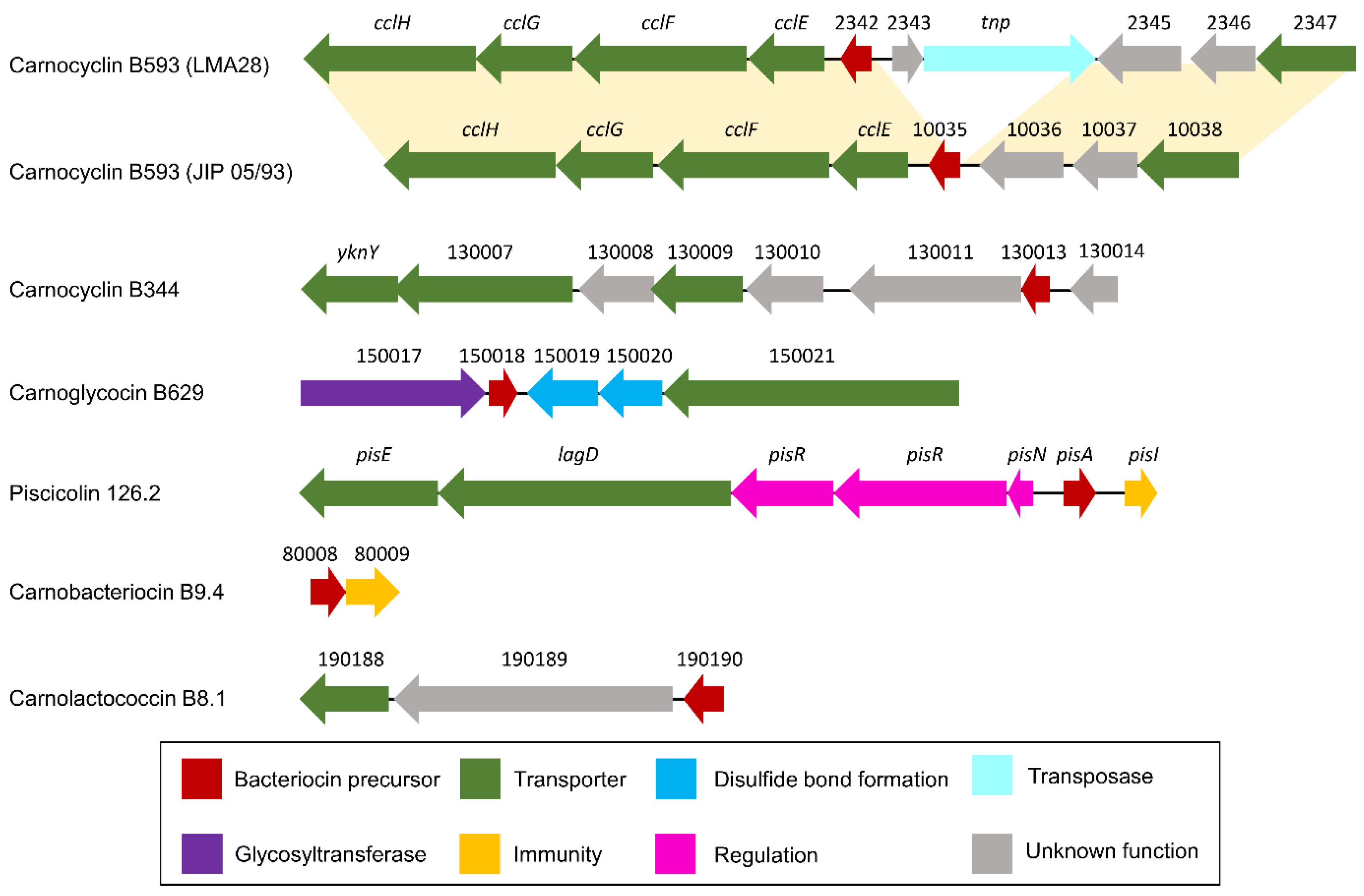
Figure 6.
Schematic representation of bacteriocin BGCs. Below the arrows which represent CDSs, the short names of the locus tags are indicated (the prefix have been omitted: “BN424_”, “CMALT398_v1_”, “CMDSM20344_v1_”, “CMALT430_v1_”, “CMALT94_v1_”, “CMALT81_v1_” for LMA28, JIP 05/93, DSM20344, 10040100629, 9.4, and 8.1, respectively). The yellow shading connects homologous regions between BGCs.
Genes homologous to cclF and cclE of the carnoclycin A BGC were found in the carnocyclin B593 BGC and would encode transporters [55] (Figure 6, Table 2). No CDS clearly associated with an immunity function has been identified. In addition, and surprisingly, no homolog of the DUF95 family was found, neither in the vicinity of the bacteriocin structural genes nor elsewhere in the genome of these two strains. Yet, the presence of such a gene is conserved in clusters of described circular bacteriocins. In addition, this protein is suspected to be directly involved in the cyclization of these circular bacteriocins even if experimental data are missing in the literature [54]. This suggests that these bacteriocins could be linear instead of circular.
The putative carnoglycocin B629 (CMALT430_v1_150018) from strain C. maltaromaticum 10040100629 is homologous to the glycocin Enterocin F4-9 (query coverage 83%, identity 52%, similarity 64%, Figure 7). Glycocins are glycosylated peptides where the carbohydrate group is covalently linked to a serine/threonine residue (O-linked) or a cysteine residue (S-linked). In addition, glycocins have four cysteine residues that are engaged in two disulfide bonds, thus stabilizing the helix-loop-helix structures of the polypeptide [56]. The sequence of carnoglycocin B629 contains four Cys residues (Cys26, Cys50, Cys56, and Cys64, Table S2) that could be involved in two disulfide bonds. Consistently, two CDSs potentially encoding thiol-disulfide isomerases (CMALT430_v1_150019 and CMALT430_v1_150020, Figure 6, Table 2) were predicted in the BGC. These enzymes could be involved in the formation of two disulfide bonds in carnoglycocin B629. Upstream of the carnoglycocin B629 CDS, a CDS encoding a glycosyltransferase (CMALT430_v1_150017) was predicted and could be involved in the glycosylation of the bacteriocin (Figure 6, Table 2). Twelve possible sites for O-glycosylation are present in the sequence of carnoglycocin B629 (Ser12, Ser17, Ser39, Ser40, Ser51, Ser52, Ser54, Ser55, Ser57, Thr59, Thr60, and Thr63, Table S2). The carnoglycocin B629 BGC also encompasses a CDS (CMALT430_v1_10021) encoding a peptidase domain-containing ABC transporter that may be involved in the export and signal peptide removal of the bacteriocin (Figure 6, Table 2).
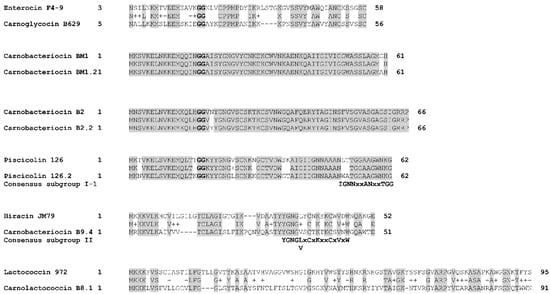
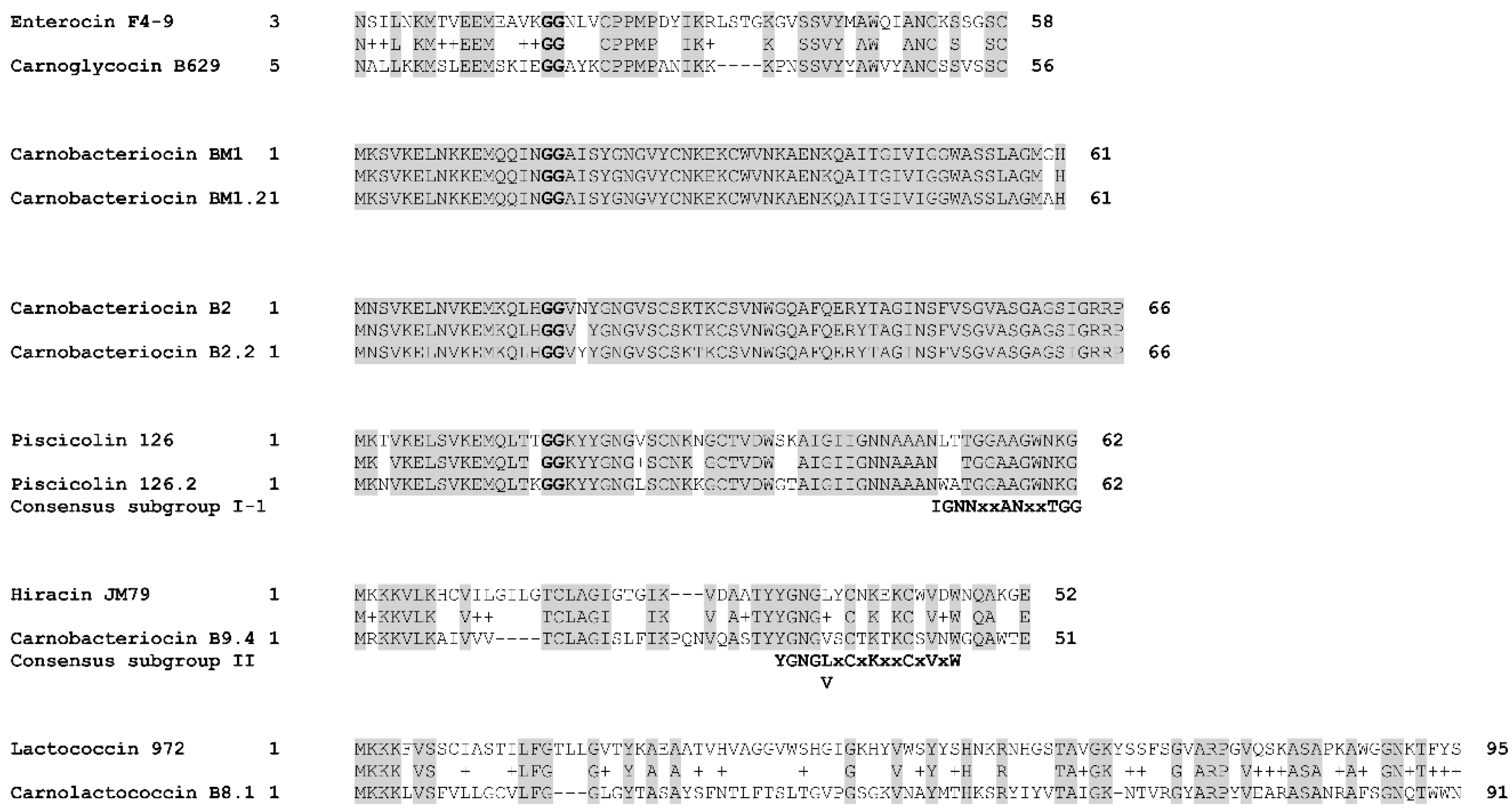
Figure 7.
Alignment of bacteriocins homologs. The alignment was performed with BLASTp by using the BLOSUM62 matrix. The double glycines of the leader peptides are indicated in bold. The consensus sequences described for class IIa bacteriocins by [57] are indicated below the alignments. Identical amino acids are shaded in grey. Chemically similar amino acids are connected by a “+” symbol.
The carnolysin A1-A2 BGC was identified in the genome of five strains, namely C. maltaromaticum 9.4, IFIP 710, CIP100481, DSM20590, and JIP 05/93 (Figure 5). The primary structure of the bacteriocins shares 100% identity with the carnolysin A1-A2 described in the databases (query coverage of 100%). Carnolysins A1 and A2 combine to form a two-peptide lantibiotic synthesized by C. maltaromaticum C2. It is an extensively modified bacteriocin containing D-alanine and D-aminobutyrate derived from serine and threonine, respectively. This bacteriocin shows antimicrobial activity against other LAB, such as Lactilactobacillus sakei, as well as Listeria innocua and L. monocytogenes [58].
3.2.2. Class IIa
Seven class IIa bacteriocins were predicted: carnobacteriocin BM1 and BM1.2, carnobacteriocin B2 and B2.2, piscicolin 126 and 126.2, and carnobacteriocin B9.4 (Figure 5). Class IIa bacteriocins are heat stable, non-modified, cationic, hydrophilic, and hydrophobic peptides that are exported thanks to a double–glycine-containing leader peptide. They classically have a molecular weight of less than 10 kDa [59,60]. The pediocin-like bacteriocins are particularly active against Listeria spp. The N-terminal region contains two or four cysteine residues joined by one or two disulfide bridges, and a conserved YGNGV-C motif (less frequently the valine is replaced by leucine) while the C-terminus is less conserved [57].
The variant carnobacteriocin BM1.2 was predicted in the genome of the strain C. maltaromaticum 10040100629 and exhibits an alanine residue instead of glycine at position 60 (Figure 7). Carnobacteriocin B2.2 was found in the genome of C. maltaromaticum F7 (Figure 5) and presents a tyrosine residue instead of asparagine at position 20 (Figure 7). The variant piscicolin 126.2 was found in the genome of the three strains C. maltaromaticum 10040100629, F2, and F88. It shows eight substitutions of which two are localized in the leader peptide (T3N, T16K), and could influence the export efficiency. The remaining six other substitutions are localized in the mature bacteriocin, including a V25L substitution in the YGNGV motif. Although piscicolin 126.2 exhibits five substitutions compared to piscicolin 126, it contains the consensus sequence IGNNxxANxxTGG which is typically found in the subgroup I-1 of class IIa bacteriocins [57]. It can be noticed that the nucleotide sequence of the piscicolin 126.2 BGC, and more specifically of the intergenic region between pisN and pisA (Figure 6), exhibit 94% and 88% identity, respectively, with the piscicolin 126 BGC.
The genome of the strain C. maltaromaticum 9.4 is predicted to encode the class IIa carnobacteriocin B9.4 (CMALT94_v1_80008, Figure 5, Table 1) which is a homolog of hiracin JM79 (query coverage 70%, 56% identity, Figure 7). Hiracin JM79 is a bacteriocin with potent anti-L. monocytogenes activity [61]. The CDS CMALT94_v1_80008 is localized next to a homolog of the leucocin A immunity encoding gene (CMALT94_v1_80009, Figure 6). The sequence of carnobacteriocin B9.4 matches with the consensus sequence YGNGL(V)xCxKxxCxVxW described for the group II of class IIa bacteriocins to which hiracin JM79 belongs [57]. Similar to hiracin JM79 and other bacteriocins of group II, carnobacteriocin B9.4 is predicted to contain a sec signal peptide of 29 amino acid residues suggesting that this bacteriocin is secreted thanks to the sec translocase [57]. The two CDSs encoding carnobacteriocin B9.4 and the immunity factor belong to a genomic island of 27 kbp which encompasses 40 CDSs and is localized downstream of a Leu-tRNA gene. This genomic island contains CDSs potentially encoding an integrase, an excisionase, a conjugation protein (FtsK-like DNA-translocation protein), and other functions typically found in mobile genetic elements (CDSs encoding a LPXTG-surface protein and site-specific DNA-methyltransferase), suggesting that the CDS encoding carnobacteriocin B9.4 is localized within an Integrative and Conjugative Element (ICE) and can be subjected to horizontal transfer [62].
3.2.3. Class IIc
The CDS CMALT81_v1_190190, from the genome of C. maltaromaticum 8.1, is predicted to encode a class IIc bacteriocin, called carnolactococcin B8.1 (Table 1). Class IIc bacteriocins are small, heat-stable, leaderless peptides that do not undergo any post-translational modifications [54]. Carnolactococcin B8.1 shows a weak resemblance with lactococcin 972 (query coverage 100%, identity 36%, similarity 56%, Figure 7). The BGC predicted in the genome of C. maltaromaticum 8.1 comprises one putative ABC-type transporter that could be involved in bacteriocin export and a membrane protein of unknown function (Figure 6, Table 2).
3.3. NRPS-BGC
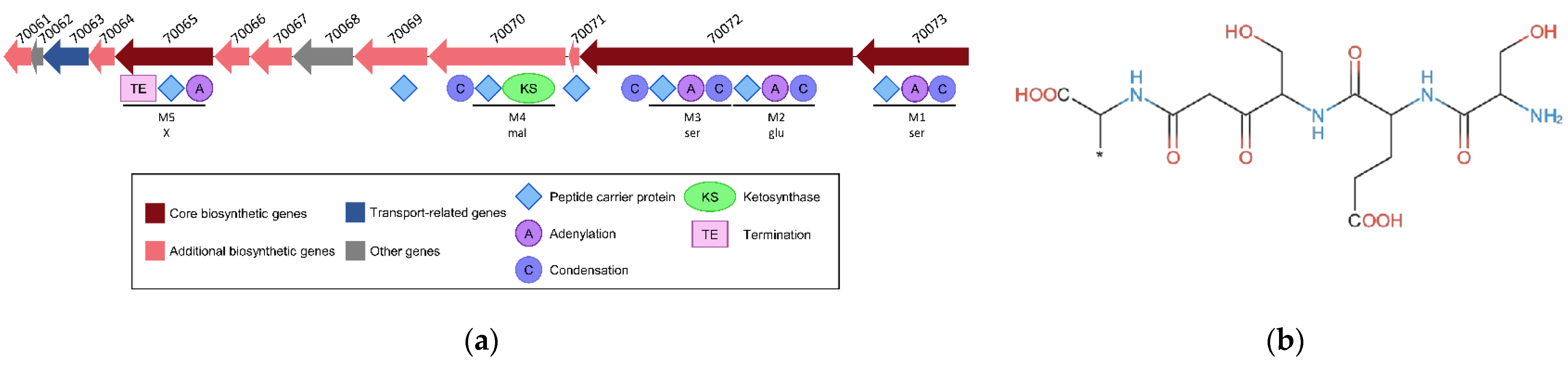
An NRPS-PKS BGC of 26.3 kbp comprising 13 CDSs was predicted in the genome of C. maltaromaticum 10040100629 (Figure 8, Table 2). Altogether, the CDSs encode seven peptide carrier protein domains, four adenylation domains, five condensation domains, one ketosynthase domain, and one termination domain (Figure 8a). Five modules (Figure 8a) were predicted to synthesize a metabolite with the following structure: (ser) + (glu − ser) + (mal) + (X) (Figure 8b). Additional CDSs whose function is compatible with the synthesis of specialized metabolites are also present and encode one 4’-phosphopantetheinyl transferase, two thioesterases, two Acyl-CoA dehydrogenases, and one aminopeptidase (Table 2).
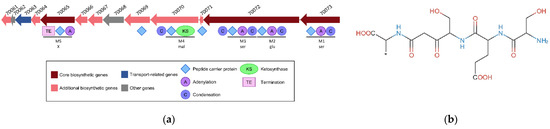
Figure 8.
NRPS-PKS BGC was predicted in the genome of the strain C. maltaromaticum 10040100629. (a) Representation of the genetic organization where the CDSs are represented by arrows. The domains predicted in the encoded proteins are represented below the CDSs. The locus tag is indicated above the CDSs, the prefix “CMALT430_v1_” of the locus tag was omitted; (b) core structure of the predicted metabolite.
3.4. Genome Clustering
Genome clustering revealed that the carnobacteriocin B2 BGC is distributed among the closely related strains CP1, CP4, and CP5 (Figure 9). By contrast, the carnocyclin B593, the carnolysin A1/A2, and the piscicolin 126 BGCs are distributed in several clades. Indeed, the carnocyclin B593 BGC is present in two distant strains: JIP 05/93 and LM128 (Figure 9). Similarly, the carnolysin A1/A2 BGC is present in two clades: one formed by the strains JIP 05/93, and DSM20590, and the other by CIP100481, 9.4, and IFIP 710; and the piscicolin 126.2 BGC in other two clades: one formed by the strain 10040100629 and the other encompassing the strains F2 and F88. Furthermore, the piscicolin 126 BGC is present in three lineages: one formed by JIP 28/91, the second by JIP 05/93 and DSM20590, and the third by 8.1 (Figure 9). The distribution of these BGCs in multiple lineages suggests that they evolved by horizontal transfer.
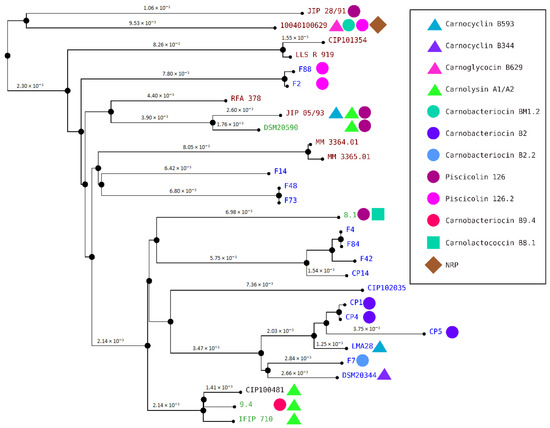
Figure 9.
C. maltaromaticum genome clustering. The names of the C. maltaromaticum strains are indicated, in blue, green, dark red, and black when the strains were isolated from a dairy product, other food products, diseased fish, and human blood, respectively. Next to strain names, the symbols indicate the bacteriocin and the NRP predicted from the genome of the strains. The bacteriocin BM1 is not represented.
3.5. Comparisons of Antimicrobial Phenotypes and Genome Predictions
The three strains C. maltaromaticum 10040100629, F2, and F88, which belong to gp1, are characterized by a potent anti-L. monocytogenes EGDe activity and a high sender degree (Figure 2 and Figure 3). As these three strains are the only ones to potentially produce piscicolin 126.2 (Figure 5), it is likely that their remarkable antimicrobial properties, and in particular their anti-L. monocytogenes activity is at least in part due to the production of piscicolin 126.2.
The strain C. maltaromaticum 10040100629 is characterized by a higher inhibition potency of L. monocytogenes EGDe compared to strains F2 and F88 (Figure 2). Interestingly, this strain would produce two bacteriocins in addition to piscicolin 126.2: carnoglycocin B629, and carnobacteriocin BM1.2, as well as an NRP-PK compound. All or part of these compounds could be responsible for the higher potency of the strain 10040100629.
According to the hierarchical clustering analysis, the inhibition profile of strain 9.4 is relatively distant from the other three 10040100629, F2, and F88, even though all these strains belong to gp1 and exhibit anti-L. monocytogenes activity (Figure 3). Interestingly, strain 9.4 is the only one that potentially produces carnobacteriocin B9.4 which could therefore be responsible for the potent anti-L. monocytogenes activity of this strain.
Several strains are predicted to produce the single bacteriocin BM1. However, it is described in the literature that the production of this bacteriocin is highly dependent on the presence of carnobacteriocin B2 BGC, which contains the CDSs required for the synthesis of the two-component regulatory system CbnRK which induces the expression of both carnobacteriocin B2 and BM1 [63]. It is therefore unlikely that the anti-C. maltaromaticum activity of the strains for which the carnobacteriocin BM1 is the only bacteriocin-encoding CDS found in the genome, namely the strains F4, F84, F14, F48, F73, CIP101354, and LLS R 919 (Figure 5), results from the production of this bacteriocin. In addition, all the strains carrying the CDS encoding carnobacteriocin BM1 (26 strains out of 29) also have the associated CDS encoding the immunity protein, thus reinforcing this hypothesis. Furthermore, the strains CIP102035, CP14, and F42 have no detected antimicrobial activity under our experimental conditions even though they possess the CDS encoding carnobacteriocin BM1. All these observations suggest that the anti-C. maltaromaticum activity of the strains F4, F84, F14, F48, F73, CIP101354, F73, CIP101354, and LLS R 919 is not due to BM1 but rather to one or several compounds produced thanks to BGCs that were not detected by genome mining.
4. Discussion
In this study, the antimicrobial properties of C. maltaromaticum were investigated by combining interference competition network analysis and genome mining. From the inhibition data obtained with 76 C. maltaromaticum strains, the relationship between the potency of inhibition, the sender degree (i.e., inhibitory spectrum), and the anti-L. monocytogenes EGDe properties were studied. The genome of a selection of 29 strains, representative of the diversity of the inhibition profiles, was analyzed in order to predict the genes with functions associated with antimicrobial activity.
The results have shown that the potency of inhibition tended to increase when the inhibition spectrum broadened. In addition, the strains that were able to effectively inhibit L. monocytogenes EGDe also exhibited a broad inhibition spectrum of C. maltaromaticum strains. This suggests that the use of C. maltaromaticum strains with anti-L. monocytogenes for biopreservation purposes may reduce the diversity of autochthonous C. maltaromaticum populations in food microbiota. This would be in agreement with a previous report showing that bioprotective Lactilactobacillus curvatus markedly decrease the diversity of the metabolically active microbial community in fermented sausages [64].
Although the potency of inhibition tended to increase when the sender degree increased, several strains were characterized by high potency of inhibition and narrow anti-C. maltaromaticum spectrum. This suggests that the antimicrobial compounds involved in these inhibitions exhibit high molecular specificity and target receptors characterized by inter-strain variability. Little is known about the bacteriocins involved in conspecific inhibitions in LAB since the majority of studies are conducted with the perspective of engineering biopreservation systems targeting unwanted microorganisms. However, it is known that bacteriocins produced by LAB can specifically target bacteria that are closely related to the producers. For instance, the bacteriocin mesenterocin 52B is produced by Leuconostoc mesenteroides subsp. mesenteroides FR52 only inhibits bacteria from the genus Leuconostoc [65].
The mining of 29 genomes of C. maltaromaticum allowed us to predict 12 bacteriocins and one NRPS-PKS BGCs. This is consistent with previous genome mining analysis in the species C. maltaromaticum and C. divergens [66]. This suggests that bacteriocins are the main antimicrobial specialized metabolites produced by C. maltaromaticum. Similarly, LAB are mainly reported to produce specialized metabolites of the type bacteriocin [67]. By contrast, few BGCs potentially encoding NRPS/PKS are described in the literature for LAB [5,6,7]. The tremendous amount of available bacterial genomic and metagenomic data provided opportunities to investigate the distribution of BGCs in bacterial taxa. Genome mining of the human microbiome project’s reference genome database revealed that bacteriocin BGCs are mainly found in the phyla Firmicutes and Proteobacteria and less frequently encountered in other phyla including Actinobacteria [68]. Conversely, NRPS/PKS BGCs are widely encountered in Actinobacteria, Proteobacteria, Verrumicrobia, and Bacteroidetes [69]. In the phylum Firmicutes, PKS BGC is preferably encountered in the order Bacillales of the phylum Firmicutes [70]. The reason why these genes are heterogeneously distributed is not known.
The genome of C. maltaromaticum 10040100629 was predicted to contain an NRPS-PKS BGC allowing the potential biosynthesis of a compound with the structure (ser) + (glu − ser) + (mal) + (X). The genome of Carnobacterium divergens V41 also contains a genomic island of 39 kbp encompassing three core synthetic CDSs which encodes seven modules that would synthesize a compound with the following structure: (mal) + (leu − D-ser) + (pro − X − cys − gly) [5]. Nonribosomal compounds vary from about 3 to 15 amino acids in length and can exhibit a linear, cyclic, or branched-cyclic structure [71,72]. It has been previously described that NRPs as short as tetrapeptides can exhibit antimicrobial activity [73], opening the possibility that the specialized metabolites potentially produced by C. maltaromaticum 10040100629 and C. divergens V41 have antimicrobial activity. However, the biological properties of NRP/PK compounds are not exclusively restricted to antimicrobial activities. In L. lactis KF147, an NRPS-PKS system improves tolerance to reactive oxygen species [6]. Further studies are required to better understand the role of these compounds in LAB.
Among the bacteriocins predicted in this study, piscicolin B126.2 is likely involved in the remarkable antimicrobial activity of the strains C. maltaromaticum 10040100629, F2, and F88. The sequence of piscicolin 126.2 precursor shows eight amino acid substitutions. It has been shown that amino acid substitutions can change the stability, potency, and spectrum of activity of class IIa bacteriocins [74]. Amino acid changes can either reduce or increase the potency of the bacteriocins. For instance, the addition of lysine at the C-terminus and T20K mutants of sakacin P shows increased cell binding and potency [74,75]. The increase in bacteriocin activity thanks to amino acid substitutions was also observed for nisin [76]. Alternatively, the high activity of the strains C. maltaromaticum 10040100629, F2, and F88 could result from high expression of piscicolin 126.2 since multiple point mutations are present in the piscicolin 126.2 BGC compared to the already described piscicolin 126 BGC, including in the intergenic region which likely contains the promoters the bacteriocin structural gene (pisA) and the export, regulation, and maturation system (pisNKR, lagD, and pisE, Figure 6).
Several strains were predicted to produce known bacteriocins, yet these strains have no antimicrobial activity under our experimental conditions. It has been described that the regulation of piscicolin 126 BGC is variable, depending on the strains considered. More precisely, the strain C. maltaromaticum JG126, produces piscicolin 126 regardless of the cultivation temperature, while the strain C. maltaromaticum UAL26 produces the bacteriocin only at temperatures below 19 °C. When this latter strain is cultivated at 25 °C, it does not produce piscicolin 126 [77]. Apart from the strains containing the piscicolin 126 BGC, other strains such as C. maltaromaticum CP5 carry the carnobacteriocin B2 BGC in their genome and yet were not found as inhibitory. The strain C. maltaromaticum CP5 was studied in our laboratory 30 years ago and revealed potent anti-L. monocytogenes activity [78]. However, our latest attempts to show any antimicrobial activity with this strain were not successful. The historical frozen culture stocks of this strain are no longer available in our laboratory. The instability of bacteriocin production was also observed for Lactobacillus sake L45, for which the propagation in a liquid medium results in the emergence of non-bacteriocin-producing variants at high frequency [79].
Several strains were found to exhibit potent anti-C. maltaromaticum with a narrow spectrum. This activity could be attributable to the production of several bacteriocins (carnobacteriocin B593, carnolysin A1/A2, and piscicolin 126), alone or in combination for the strains IFIP 710, and CIP 100481, DSM20590, and JIP 05/93 (Figure 5). However, no BGC candidates were predicted for the other anti-C. maltaromaticum strains F4, F84, F14, F48, F73, CIP101354, LLS R 919, and RFA 378. It can be noticed that among these strains, RFA 378, LLS R 919, CIP101354, IFIP 710, and F73 have a potent anti-C. maltaromaticum activity (Figure 2). This suggests that genome mining would not be able to identify BGCs allowing the production of antimicrobial substances acting on conspecific strains. The basis of functional prediction in genome mining is the availability of experimental data providing information on the function of genes. In the field of antimicrobials produced by LAB, the literature describing data obtained from wet experiments is tremendously dedicated to the characterization of compounds inhibiting unwanted microorganisms, such as L. monocytogenes. This documentary bias may have led to missing genes encoding the synthesis of antimicrobial substances whose sequence properties are unrelated to those already described in the databases. However, these substances could play an important role in ecosystems by contributing to the structuring of microbial food communities. Recent advances in the identification techniques, in particular those involving artificial intelligence such as deep learning approaches [80], could be a real game changer in the area.
5. Conclusions
C. maltaromaticum is a species of high interest for dairy products, especially for biopreservation purposes, that has been extensively studied for the production of bacteriocins. However, it has been shown here using a combined approach of inhibition network analysis and genomics that this species harbors an unexplored potential that could be of direct interest specifically for biopreservation but also more globally for the understanding and engineering of dairy ecosystems. The further use of these high throughput approaches should lead to new technological innovations for better control of the quality of dairy products.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/microorganisms10091794/s1, Table S1: Carnobacterium maltaromaticum strains used in this study; Table S2: Sequences of the bacteriocins predicted from the genome of 29 strains of C. maltaromaticum; Table S3: CDSs encoding structural bacteriocins in C. maltaromaticum strains. References in Supplementary Materials can be found in [18,33,81,82,83,84,85,86,87,88,89,90,91].
Author Contributions
Conceptualization, F.B., A.-M.R.-J., C.M.; methodology, M.T.P.G., N.E.R., A.D. and A.E.; formal analysis, M.T.P.G., N.E.R. and F.B.; investigation, M.T.P.G., N.E.R., A.D. and F.B.; resources, A.E.; data curation, M.T.P.G., N.E.R. and F.B.; writing—original draft preparation, M.T.P.G. and N.E.R.; writing—review and editing, F.B.; supervision, A.-M.R.-J., F.B. and S.T.; project administration, A.-M.R.-J. and F.B.; funding acquisition, A.-M.R.-J., F.B. and S.T. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Société d’Accélération du Transfert de Technologie (SATT, Technology Transfer Acceleration Company) SAYENS grant number VALICARNO.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data and the code used in R are available on GitHub: https://github.com/fredericborges/network_genome_antimic (accessed on 21 July 2022).
Acknowledgments
We thank Myriam Michel and Sylvie Wolff for their technical assistance during this work.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Borges, F.; Briandet, R.; Callon, C.; Champomier-Vergès, M.-C.; Christieans, S.; Chuzeville, S.; Denis, C.; Desmasures, N.; Desmonts, M.-H.; Feurer, C.; et al. Contribution of Omics to Biopreservation: Toward Food Microbiome Engineering. Front. Microbiol. 2022, 13, 951182. [Google Scholar] [CrossRef] [PubMed]
- Kommineni, S.; Bretl, D.J.; Lam, V.; Chakraborty, R.; Hayward, M.; Simpson, P.; Cao, Y.; Bousounis, P.; Kristich, C.J.; Salzman, N.H. Bacteriocin Production Augments Niche Competition by Enterococci in the Mammalian Gastrointestinal Tract. Nature 2015, 526, 719–722. [Google Scholar] [CrossRef] [PubMed]
- Riley, M.A.; Wertz, J.E. Bacteriocin Diversity: Ecological and Evolutionary Perspectives. Biochimie 2002, 84, 357–364. [Google Scholar] [CrossRef]
- Alvarez-Sieiro, P.; Montalbán-López, M.; Mu, D.; Kuipers, O.P. Bacteriocins of Lactic Acid Bacteria: Extending the Family. Appl. Microbiol. Biotechnol. 2016, 100, 2939–2951. [Google Scholar] [CrossRef]
- Remenant, B.; Borges, F.; Cailliez-Grimal, C.; Revol-Junelles, A.-M.; Marché, L.; Lajus, A.; Médigue, C.; Pilet, M.-F.; Prévost, H.; Zagorec, M. Draft Genome Sequence of Carnobacterium divergens V41, a Bacteriocin-Producing Strain. Genome Announc. 2016, 4, e01109-16. [Google Scholar] [CrossRef]
- Golomb, B.L.; Yu, A.O.; Coates, L.C.; Marco, M.L. The Lactococcus Lactis KF147 Nonribosomal Peptide Synthetase/Polyketide Synthase System Confers Resistance to Oxidative Stress during Growth on Plant Leaf Tissue Lysate. MicrobiologyOpen 2018, 7, e00531. [Google Scholar] [CrossRef]
- Khayatt, B.I.; van Noort, V.; Siezen, R.J. The Genome of the Plant-Associated Lactic Acid Bacterium Lactococcus Lactis KF147 Harbors a Hybrid NRPS-PKS System Conserved in Strains of the Dental Cariogenic Streptococcus mutans. Curr. Microbiol. 2020, 77, 136–145. [Google Scholar] [CrossRef]
- Süssmuth, R.D.; Mainz, A. Nonribosomal Peptide Synthesis-Principles and Prospects. Angew. Chem. Int. Ed. 2017, 56, 3770–3821. [Google Scholar] [CrossRef]
- Miyanaga, A. Structure and Function of Polyketide Biosynthetic Enzymes: Various Strategies for Production of Structurally Diverse Polyketides. Biosci. Biotechnol. Biochem. 2017, 81, 2227–2236. [Google Scholar] [CrossRef]
- Skiba, M.A.; Maloney, F.P.; Dan, Q.; Fraley, A.E.; Aldrich, C.C.; Smith, J.L.; Brown, W.C. PKS-NRPS Enzymology and Structural Biology: Considerations in Protein Production. Methods Enzymol. 2018, 604, 45–88. [Google Scholar] [CrossRef]
- Fisch, K.M. Biosynthesis of Natural Products by Microbial Iterative Hybrid PKS–NRPS. RSC Adv. 2013, 3, 18228–18247. [Google Scholar] [CrossRef]
- Afzal, M.I.; Jacquet, T.; Delaunay, S.; Borges, F.; Milliere, J.B.; Revol-Junelles, A.M.; Cailliez-Grimal, C. Carnobacterium maltaromaticum: Identification, Isolation Tools, Ecology and Technological Aspects in Dairy Products. Food Microbiol. 2010, 27, 573–579. [Google Scholar] [CrossRef] [PubMed]
- Leisner, J.J.; Laursen, B.G.; Prevost, H.; Drider, D.; Dalgaard, P. Carnobacterium: Positive and Negative Effects in the Environment and in Foods. FEMS Microbiol. Rev. 2007, 31, 592–613. [Google Scholar] [CrossRef]
- Afzal, M.I.; Gonzalez Ariceaga, C.C.; Lhomme, E.; Kamel Ali, N.; Payot, S.; Burgain, J.; Gaiani, C.; Borges, F.; Revol-Junelles, A.M.; Delaunay, S.; et al. Characterization of Carnobacterium maltaromaticum LMA 28 for Its Positive Technological Role in Soft Cheese Making. Food Microbiol. 2013, 36, 223–230. [Google Scholar] [CrossRef] [PubMed]
- Hammi, I.; Delalande, F.; Belkhou, R.; Marchioni, E.; Cianferani, S.; Ennahar, S. Maltaricin CPN, a New Class IIa Bacteriocin Produced by Carnobacterium maltaromaticum CPN Isolated from Mould-Ripened Cheese. J. Appl. Microbiol. 2016, 121, 1268–1274. [Google Scholar] [CrossRef]
- Spanu, C.; Piras, F.; Mocci, A.M.; Nieddu, G.; De Santis, E.P.L.; Scarano, C. Use of Carnobacterium spp. Protective Culture in MAP Packed Ricotta Fresca Cheese to Control Pseudomonas spp. Food Microbiol. 2018, 74, 50–56. [Google Scholar] [CrossRef]
- Danielski, G.M.; Imazaki, P.H.; de Andrade Cavalari, C.M.; Daube, G.; Clinquart, A.; de Macedo, R.E.F. Carnobacterium maltaromaticum as Bioprotective Culture in Vitro and in Cooked Ham. Meat Sci. 2020, 162, 108035. [Google Scholar] [CrossRef] [PubMed]
- Laursen, B.G.; Bay, L.; Cleenwerck, I.; Vancanneyt, M.; Swings, J.; Dalgaard, P.; Leisner, J.J. Carnobacterium Divergens and Carnobacterium maltaromaticum as Spoilers or Protective Cultures in Meat and Seafood: Phenotypic and Genotypic Characterization. Syst. Appl. Microbiol. 2005, 28, 151–164. [Google Scholar] [CrossRef]
- Dos Reis, F.B.; de Souza, V.M.; Thomaz, M.R.S.; Fernandes, L.P.; de Oliveira, W.P.; De Martinis, E.C.P. Use of Carnobacterium maltaromaticum Cultures and Hydroalcoholic Extract of Lippia Sidoides Cham. against Listeria monocytogenes in Fish Model Systems. Int. J. Food Microbiol. 2011, 146, 228–234. [Google Scholar] [CrossRef]
- Kim, D.H.; Austin, B. Innate Immune Responses in Rainbow Trout (Oncorhynchus mykiss, Walbaum) Induced by Probiotics. Fish Shellfish Immunol. 2006, 21, 513–524. [Google Scholar] [CrossRef]
- Koné, A.P.; Zea, J.M.V.; Gagné, D.; Cinq-Mars, D.; Guay, F.; Saucier, L. Application of Carnobacterium maltaromaticum as a Feed Additive for Weaned Rabbits to Improve Meat Microbial Quality and Safety. Meat Sci. 2018, 135, 174–188. [Google Scholar] [CrossRef] [PubMed]
- Stoffels, G.; Nes, I.F.; Guthmundsdottir, A. Isolation and Properties of a Bacteriocin-Producing Carnobacterium piscicola Isolated from Fish. J. Appl. Bacteriol. 1992, 73, 309–316. [Google Scholar] [CrossRef] [PubMed]
- Tulini, F.L.; Lohans, C.T.; Bordon, K.C.F.; Zheng, J.; Arantes, E.C.; Vederas, J.C.; De Martinis, E.C.P. Purification and Characterization of Antimicrobial Peptides from Fish Isolate Carnobacterium maltaromaticum C2: Carnobacteriocin X and Carnolysins A1 and A2. Int. J. Food Microbiol. 2014, 173, 81–88. [Google Scholar] [CrossRef]
- Martin-Visscher, L.A.; van Belkum, M.J.; Garneau-Tsodikova, S.; Whittal, R.M.; Zheng, J.; McMullen, L.M.; Vederas, J.C. Isolation and Characterization of Carnocyclin a, a Novel Circular Bacteriocin Produced by Carnobacterium maltaromaticum UAL307. Appl. Environ. Microbiol. 2008, 74, 4756–4763. [Google Scholar] [CrossRef] [PubMed]
- Quadri, L.E.; Sailer, M.; Roy, K.L.; Vederas, J.C.; Stiles, M.E. Chemical and Genetic Characterization of Bacteriocins Produced by Carnobacterium piscicola LV17B. J. Biol. Chem. 1994, 269, 12204–12211. [Google Scholar] [CrossRef]
- Jack, R.W.; Wan, J.; Gordon, J.; Harmark, K.; Davidson, B.E.; Hillier, A.J.; Wettenhall, R.E.; Hickey, M.W.; Coventry, M.J. Characterization of the Chemical and Antimicrobial Properties of Piscicolin 126, a Bacteriocin Produced by Carnobacterium piscicola JG126. Appl. Environ. Microbiol. 1996, 62, 2897–2903. [Google Scholar] [CrossRef]
- Yamazaki, K.; Suzuki, M.; Kawai, Y.; Inoue, N.; Montville, T.J. Purification and Characterization of a Novel Class IIa Bacteriocin, Piscicocin CS526, from Surimi-Associated Carnobacterium piscicola CS526. Appl. Environ. Microbiol. 2005, 71, 554–557. [Google Scholar] [CrossRef]
- Bhugaloo-Vial, P.; Dousset, X.; Metivier, A.; Sorokine, O.; Anglade, P.; Boyaval, P.; Marion, D. Purification and Amino Acid Sequences of Piscicocins V1a and V1b, Two Class IIa Bacteriocins Secreted by Carnobacterium piscicola V1 That Display Significantly Different Levels of Specific Inhibitory Activity. Appl Environ Microbiol 1996, 62, 4410–4416. [Google Scholar] [CrossRef]
- Holck, A.L.; Axelsson, L.; Schillinger, U. Purification and Cloning of Piscicolin 61, a Bacteriocin from Carnobacterium Piscicola LV61. Curr. Microbiol. 1994, 29, 63–68. [Google Scholar] [CrossRef]
- Worobo, R.W.; Henkel, T.; Sailer, M.; Roy, K.L.; Vederas, J.C.; Stiles, M.E. Characteristics and Genetic Determinant of a Hydrophobic Peptide Bacteriocin, Carnobacteriocin A, Produced by Carnobacterium piscicola LV17A. Microbiology 1994, 140 Pt 3, 517–526. [Google Scholar] [CrossRef][Green Version]
- Stiles, M.E.; Calrson, D.; Smith, D.C. Enhanced Preservation of Processed Food. U.S. Patent 9,066,521 B2, 30 June 2015. [Google Scholar]
- Borges, F.; Revol-Junelles, A.-M. Nouvelles Souches de Carnobacterium maltaromaticum et Leurs Utilisations. International Patent WO2021078612A1, 29 April 2021. [Google Scholar]
- Ramia, N.E.; Mangavel, C.; Gaiani, C.; Muller-Gueudin, A.; Taha, S.; Revol-Junelles, A.-M.; Borges, F. Nested Structure of Intraspecific Competition Network in Carnobacterium maltaromaticum. Sci. Rep. 2020, 10, 7335. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing. Foundation for Statistical Computing: Vienna, Austria. 2021. Available online: https://www.R-project.org/ (accessed on 28 August 2022).
- Kassambara, A.; Mundt, F. Factoextra: Extract and Visualize the Results of Multivariate Data Analyses. 2020. Available online: https://CRAN.R-project.org/package=factoextra (accessed on 28 August 2022).
- Vallenet, D.; Engelen, S.; Mornico, D.; Cruveiller, S.; Fleury, L.; Lajus, A.; Rouy, Z.; Roche, D.; Salvignol, G.; Scarpelli, C.; et al. MicroScope: A Platform for Microbial Genome Annotation and Comparative Genomics. Database 2009, 2009, bap021. [Google Scholar] [CrossRef] [PubMed]
- Vallenet, D.; Belda, E.; Calteau, A.; Cruveiller, S.; Engelen, S.; Lajus, A.; Le Fevre, F.; Longin, C.; Mornico, D.; Roche, D.; et al. MicroScope—An Integrated Microbial Resource for the Curation and Comparative Analysis of Genomic and Metabolic Data. Nucleic Acids Res. 2013, 41, D636–D647. [Google Scholar] [CrossRef] [PubMed]
- Vallenet, D.; Calteau, A.; Cruveiller, S.; Gachet, M.; Lajus, A.; Josso, A.; Mercier, J.; Renaux, A.; Rollin, J.; Rouy, Z.; et al. MicroScope in 2017: An Expanding and Evolving Integrated Resource for Community Expertise of Microbial Genomes. Nucleic Acids Res. 2017, 45, D517–D528. [Google Scholar] [CrossRef]
- Vallenet, D.; Calteau, A.; Dubois, M.; Amours, P.; Bazin, A.; Beuvin, M.; Burlot, L.; Bussell, X.; Fouteau, S.; Gautreau, G.; et al. MicroScope: An Integrated Platform for the Annotation and Exploration of Microbial Gene Functions through Genomic, Pangenomic and Metabolic Comparative Analysis. Nucleic Acids Res. 2020, 48, D579–D589. [Google Scholar] [CrossRef]
- De Jong, A.; van Hijum, S.A.F.T.; Bijlsma, J.J.E.; Kok, J.; Kuipers, O.P. BAGEL: A Web-Based Bacteriocin Genome Mining Tool. Nucleic Acids Res. 2006, 34, W273–W279. [Google Scholar] [CrossRef]
- De Jong, A.; van Heel, A.J.; Kok, J.; Kuipers, O.P. BAGEL2: Mining for Bacteriocins in Genomic Data. Nucleic Acids Res. 2010, 38, W647–W651. [Google Scholar] [CrossRef]
- Van Heel, A.J.; de Jong, A.; Montalbán-López, M.; Kok, J.; Kuipers, O.P. BAGEL3: Automated Identification of Genes Encoding Bacteriocins and (Non-)Bactericidal Posttranslationally Modified Peptides. Nucleic Acids Res. 2013, 41, W448–W453. [Google Scholar] [CrossRef]
- Van Heel, A.J.; de Jong, A.; Song, C.; Viel, J.H.; Kok, J.; Kuipers, O.P. BAGEL4: A User-Friendly Web Server to Thoroughly Mine RiPPs and Bacteriocins. Nucleic Acids Res. 2018, 46, W278–W281. [Google Scholar] [CrossRef]
- Blin, K.; Wolf, T.; Chevrette, M.G.; Lu, X.; Schwalen, C.J.; Kautsar, S.A.; Suarez Duran, H.G.; de Los Santos, E.L.C.; Kim, H.U.; Nave, M.; et al. AntiSMASH 4.0—Improvements in Chemistry Prediction and Gene Cluster Boundary Identification. Nucleic Acids Res. 2017, 45, W36–W41. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, S.Y.; Medema, M.H.; Weber, T. AntiSMASH 5.0: Updates to the Secondary Metabolite Genome Mining Pipeline. Nucleic Acids Res. 2019, 47, W81–W87. [Google Scholar] [CrossRef] [PubMed]
- Weber, T.; Blin, K.; Duddela, S.; Krug, D.; Kim, H.U.; Bruccoleri, R.; Lee, S.Y.; Fischbach, M.A.; Müller, R.; Wohlleben, W.; et al. AntiSMASH 3.0—A Comprehensive Resource for the Genome Mining of Biosynthetic Gene Clusters. Nucleic Acids Res. 2015, 43, W237–W243. [Google Scholar] [CrossRef] [PubMed]
- Medema, M.H.; Blin, K.; Cimermancic, P.; de Jager, V.; Zakrzewski, P.; Fischbach, M.A.; Weber, T.; Takano, E.; Breitling, R. AntiSMASH: Rapid Identification, Annotation and Analysis of Secondary Metabolite Biosynthesis Gene Clusters in Bacterial and Fungal Genome Sequences. Nucleic Acids Res. 2011, 39, W339–W346. [Google Scholar] [CrossRef]
- Blin, K.; Medema, M.H.; Kazempour, D.; Fischbach, M.A.; Breitling, R.; Takano, E.; Weber, T. AntiSMASH 2.0—A Versatile Platform for Genome Mining of Secondary Metabolite Producers. Nucleic Acids Res. 2013, 41, W204–W212. [Google Scholar] [CrossRef] [PubMed]
- Gu, Z.; Eils, R.; Schlesner, M. Complex Heatmaps Reveal Patterns and Correlations in Multidimensional Genomic Data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef]
- Wickham, H.; Averick, M.; Bryan, J.; Chang, W.; McGowan, L.D.; François, R.; Grolemund, G.; Hayes, A.; Henry, L.; Hester, J.; et al. Welcome to the Tidyverse. J. Open Source Softw. 2019, 4, 1686. [Google Scholar] [CrossRef]
- Dawson, C. Ggprism: A “ggplot2” Extension Inspired by “GraphPad Prism”. 2021. Available online: https://cran.r-project.org/web/packages/ggprism/index.html (accessed on 28 August 2022).
- Slowikowski, K. Ggrepel: Automatically Position Non-Overlapping Text Labels with “Ggplot2”. 2021. Available online: https://cran.r-project.org/web/packages/ggrepel/ (accessed on 28 August 2022).
- Garnier, S.; Ross, N.; Rudis, B.; Sciaini, M.; Camargo, A.P.; Scherer, C. Viridis-Colorblind-Friendly Color Maps for R. 2021. Available online: https://cran.r-project.org/web/packages/viridis/ (accessed on 28 August 2022).
- Perez, R.H.; Zendo, T.; Sonomoto, K. Circular and Leaderless Bacteriocins: Biosynthesis, Mode of Action, Applications, and Prospects. Front. Microbiol. 2018, 9, 2085. [Google Scholar] [CrossRef]
- Van Belkum, M.J.; Martin-Visscher, L.A.; Vederas, J.C. Cloning and Characterization of the Gene Cluster Involved in the Production of the Circular Bacteriocin Carnocyclin A. Probiotics Antimicrob. Proteins 2010, 2, 218–225. [Google Scholar] [CrossRef]
- Kaunietis, A.; Buivydas, A.; Čitavičius, D.J.; Kuipers, O.P. Heterologous Biosynthesis and Characterization of a Glycocin from a Thermophilic Bacterium. Nat. Commun. 2019, 10, 1115. [Google Scholar] [CrossRef]
- Cui, Y.; Zhang, C.; Wang, Y.; Shi, J.; Zhang, L.; Ding, Z.; Qu, X.; Cui, H. Class IIa Bacteriocins: Diversity and New Developments. Int. J. Mol. Sci. 2012, 13, 16668. [Google Scholar] [CrossRef]
- Lohans, C.T.; Li, J.L.; Vederas, J.C. Structure and Biosynthesis of Carnolysin, a Homologue of Enterococcal Cytolysin with D-Amino Acids. J. Am. Chem. Soc. 2014, 136, 13150–13153. [Google Scholar] [CrossRef] [PubMed]
- Drider, D.; Fimland, G.; Hechard, Y.; McMullen, L.M.; Prevost, H. The Continuing Story of Class IIa Bacteriocins. Microbiol. Mol. Biol. Rev. 2006, 70, 564–582. [Google Scholar] [CrossRef] [PubMed]
- Balciunas, E.M.; Castillo Martinez, F.A.; Todorov, S.D.; de Melo Franco, B.D.G.; Converti, A.; de Souza Oliveira, R.P. Novel Biotechnological Applications of Bacteriocins: A Review. Food Control 2013, 32, 134–142. [Google Scholar] [CrossRef]
- Sánchez, J.; Diep, D.B.; Herranz, C.; Nes, I.F.; Cintas, L.M.; Hernández, P.E. Amino Acid and Nucleotide Sequence, Adjacent Genes, and Heterologous Expression of Hiracin JM79, a Sec-Dependent Bacteriocin Produced by Enterococcus Hirae DCH5, Isolated from Mallard Ducks (Anas platyrhynchos). FEMS Microbiol. Lett. 2007, 270, 227–236. [Google Scholar] [CrossRef]
- Bellanger, X.; Payot, S.; Leblond-Bourget, N.; Guédon, G. Conjugative and Mobilizable Genomic Islands in Bacteria: Evolution and Diversity. FEMS Microbiol. Rev. 2014, 38, 720–760. [Google Scholar] [CrossRef]
- Quadri, L.E.N. Regulation of Class II Bacteriocin Production by Cell-Cell Signaling. J. Microbiol. 2003, 41, 175–182. [Google Scholar]
- Giello, M.; La Storia, A.; De Filippis, F.; Ercolini, D.; Villani, F. Impact of Lactobacillus curvatus 54M16 on Microbiota Composition and Growth of Listeria Monocytogenes in Fermented Sausages. Food Microbiol. 2018, 72, 1–15. [Google Scholar] [CrossRef]
- Revol-Junelles, A.M.; Mathis, R.; Krier, F.; Fleury, Y.; Delfour, A.; Lefebvre, G. Leuconostoc Mesenteroides Subsp. Mesenteroides FR52 Synthesizes Two Distinct Bacteriocins. Lett. Appl. Microbiol. 1996, 23, 120–124. [Google Scholar]
- Begrem, S.; Ivaniuk, F.; Gigout-Chevalier, F.; Kolypczuk, L.; Bonnetot, S.; Leroi, F.; Grovel, O.; Delbarre-Ladrat, C.; Passerini, D. New Insight into Antimicrobial Compounds from Food and Marine-Sourced Carnobacterium Species through Phenotype and Genome Analyses. Microorganisms 2020, 8, 1093. [Google Scholar] [CrossRef]
- Reis, J.A.; Paula, A.T.; Casarotti, S.N.; Penna, A.L.B. Lactic Acid Bacteria Antimicrobial Compounds: Characteristics and Applications. Food Eng. Rev. 2012, 4, 124–140. [Google Scholar] [CrossRef]
- Walsh, C.J.; Guinane, C.M.; Hill, C.; Ross, R.P.; O’Toole, P.W.; Cotter, P.D. In Silico Identification of Bacteriocin Gene Clusters in the Gastrointestinal Tract, Based on the Human Microbiome Project’s Reference Genome Database. BMC Microbiol. 2015, 15, 183. [Google Scholar] [CrossRef] [PubMed]
- Borsetto, C.; Amos, G.C.A.; da Rocha, U.N.; Mitchell, A.L.; Finn, R.D.; Laidi, R.F.; Vallin, C.; Pearce, D.A.; Newsham, K.K.; Wellington, E.M.H. Microbial Community Drivers of PK/NRP Gene Diversity in Selected Global Soils. Microbiome 2019, 7, 78. [Google Scholar] [CrossRef] [PubMed]
- Aleti, G.; Sessitsch, A.; Brader, G. Genome Mining: Prediction of Lipopeptides and Polyketides from Bacillus and Related Firmicutes. Comput. Struct. Biotechnol. J. 2015, 13, 192–203. [Google Scholar] [CrossRef]
- Bloudoff, K.; Schmeing, T.M. Structural and Functional Aspects of the Nonribosomal Peptide Synthetase Condensation Domain Superfamily: Discovery, Dissection and Diversity. Biochim. Biophys. Acta Proteins Proteom. 2017, 1865, 1587–1604. [Google Scholar] [CrossRef]
- Martínez-Núñez, M.; López, V.E. Nonribosomal Peptides Synthetases and Their Applications in Industry. Sustain. Chem. Process. 2016, 4, 13. [Google Scholar] [CrossRef]
- Agrawal, S.; Acharya, D.; Adholeya, A.; Barrow, C.J.; Deshmukh, S.K. Nonribosomal Peptides from Marine Microbes and Their Antimicrobial and Anticancer Potential. Front. Pharmacol. 2017, 8, 828. [Google Scholar] [CrossRef] [PubMed]
- Lohans, C.T.; Vederas, J.C. Development of Class IIa Bacteriocins as Therapeutic Agents. Int. J. Microbiol. 2012, 2012, 386410. [Google Scholar] [CrossRef]
- Kazazic, M.; Nissen-Meyer, J.; Fimland, G. Mutational Analysis of the Role of Charged Residues in Target-Cell Binding, Potency and Specificity of the Pediocin-like Bacteriocin Sakacin P. Microbiology 2002, 148, 2019–2027. [Google Scholar] [CrossRef][Green Version]
- Zheng, Y.; Du, Y.; Qiu, Z.; Liu, Z.; Qiao, J.; Li, Y.; Caiyin, Q. Nisin Variants Generated by Protein Engineering and Their Properties. Bioengineering 2022, 9, 251. [Google Scholar] [CrossRef]
- Gursky, L.J.; Martin, N.I.; Derksen, D.J.; van Belkum, M.J.; Kaur, K.; Vederas, J.C.; Stiles, M.E.; McMullen, L.M. Production of Piscicolin 126 by Carnobacterium maltaromaticum UAL26 Is Controlled by Temperature and Induction Peptide Concentration. Arch. Microbiol. 2006, 186, 317–325. [Google Scholar] [CrossRef]
- Mathieu, F.; Michel, M.; Lebrihi, A.; Lefebvre, G. Effect of the Bacteriocin Carnocin CP5 and of the Producing Strain Carnobacterium piscicola CP5 on the Viability of Listeria monocytogenes ATCC 15313 in Salt Solution, Broth and Skimmed Milk, at Various Incubation Temperatures. Int J Food Microbiol 1994, 22, 155–172. [Google Scholar] [CrossRef]
- Mørtvedt, C.; Nes, I. Plasmid-Associated Bacteriocin Production by a Lactobacillus sake Strain. Microbiology 1990, 136, 1601–1607. [Google Scholar] [CrossRef]
- Stokes, J.M.; Yang, K.; Swanson, K.; Jin, W.; Cubillos-Ruiz, A.; Donghia, N.M.; MacNair, C.R.; French, S.; Carfrae, L.A.; Bloom-Ackermann, Z.; et al. A Deep Learning Approach to Antibiotic Discovery. Cell 2020, 180, 688–702.e13. [Google Scholar] [CrossRef] [PubMed]
- Rahman, A.; Cailliez-Grimal, C.; Bontemps, C.; Payot, S.; Chaillou, S.; Revol-Junelles, A.-M.; Borges, F. High Genetic Diversity among Strains of the Unindustrialized Lactic Acid Bacterium Carnobacterium maltaromaticum in Dairy Products as Revealed by Multilocus Sequence Typing. Appl. Environ. Microbiol. 2014, 80, 3920–3929. [Google Scholar] [CrossRef]
- Millière, J.B.; Michel, M.; Mathieu, F.; Lefebvre, G. Presence of Carnobacterium Spp. in French Surface Mould-Ripened Soft-Cheese. J. Appl. Bacteriol. 1994, 76, 264–269. [Google Scholar] [CrossRef]
- Miller, A.; Morgan, M.E.; Libbey, L.M. Lactobacillus maltaromicus, a New Species Producing a Malty Aroma. Int. J. Syst. Bacteriol. 1974, 24, 346–354. [Google Scholar] [CrossRef]
- Ramia, N.E.; El Kheir, S.M.; Taha, S.; Mangavel, C.; Revol-Junelles, A.M.; Borges, F. Multilocus Sequence Typing of Carnobacterium maltaromaticum Strains Associated with Fish Disease and Dairy Products. J. Appl. Microbiol. 2019, 126, 377–387. [Google Scholar] [CrossRef]
- Cailliez-Grimal, C.; Edima, H.C.; Revol-Junelles, A.M.; Millière, J.B. Short Communication: Carnobacterium maltaromaticum: The Only Carnobacterium Species in French Ripened Soft Cheeses as Revealed by Polymerase Chain Reaction Detection. J. Dairy Sci. 2007, 90, 1133–1138. [Google Scholar] [CrossRef]
- Hantsis-Zacharov, E.; Halpern, M. Culturable Psychrotrophic Bacterial Communities in Raw Milk and Their Proteolytic and Lipolytic Traits. Appl. Environ. Microbiol. 2007, 73, 7162–7168. [Google Scholar] [CrossRef]
- Walker, V.K.; Palmer, G.R.; Voordouw, G. Freeze-Thaw Tolerance and Clues to the Winter Survival of a Soil Community. Appl. Environ. Microbiol. 2006, 72, 1784–1792. [Google Scholar] [CrossRef]
- Leisner, J.J.; Vogensen, F.K.; Kollmann, J.; Aideh, B.; Vandamme, P.; Vancanneyt, M.; Ingmer, H. α-Chitinase Activity among Lactic Acid Bacteria. Syst. Appl. Microbiol. 2008, 31, 151–156. [Google Scholar] [CrossRef] [PubMed]
- Hiu, S.F.; Holt, R.A.; Sriranganathan, N.; Seidler, R.J.; Fryer, J.L. Lactobacillus piscicola, a New Species from Salmonid Fish. Int. J. Syst. Bacteriol. 1984, 34, 393–400. [Google Scholar] [CrossRef]
- Holzapfel, W.H.; Gerber, E.S. Lactobacillus divergens Sp. Nov., a New Heterofermentative Lactobacillus Species Producing L(+)-Lactate. Syst. Appl. Microbiol. 1983, 4, 522–534. [Google Scholar] [CrossRef]
- Feurer, C.; Ellouze, M. Utilisation Raisonnée de La Flore Naturelle de Viande de Porc Fraîche Pour Améliorer Sa Qualité Sanitaire. IFIP 2010, 34, 125. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).