An Insight into Goat Cheese: The Tales of Artisanal and Industrial Gidotyri Microbiota
Abstract
:1. Introduction
2. Materials and Methods
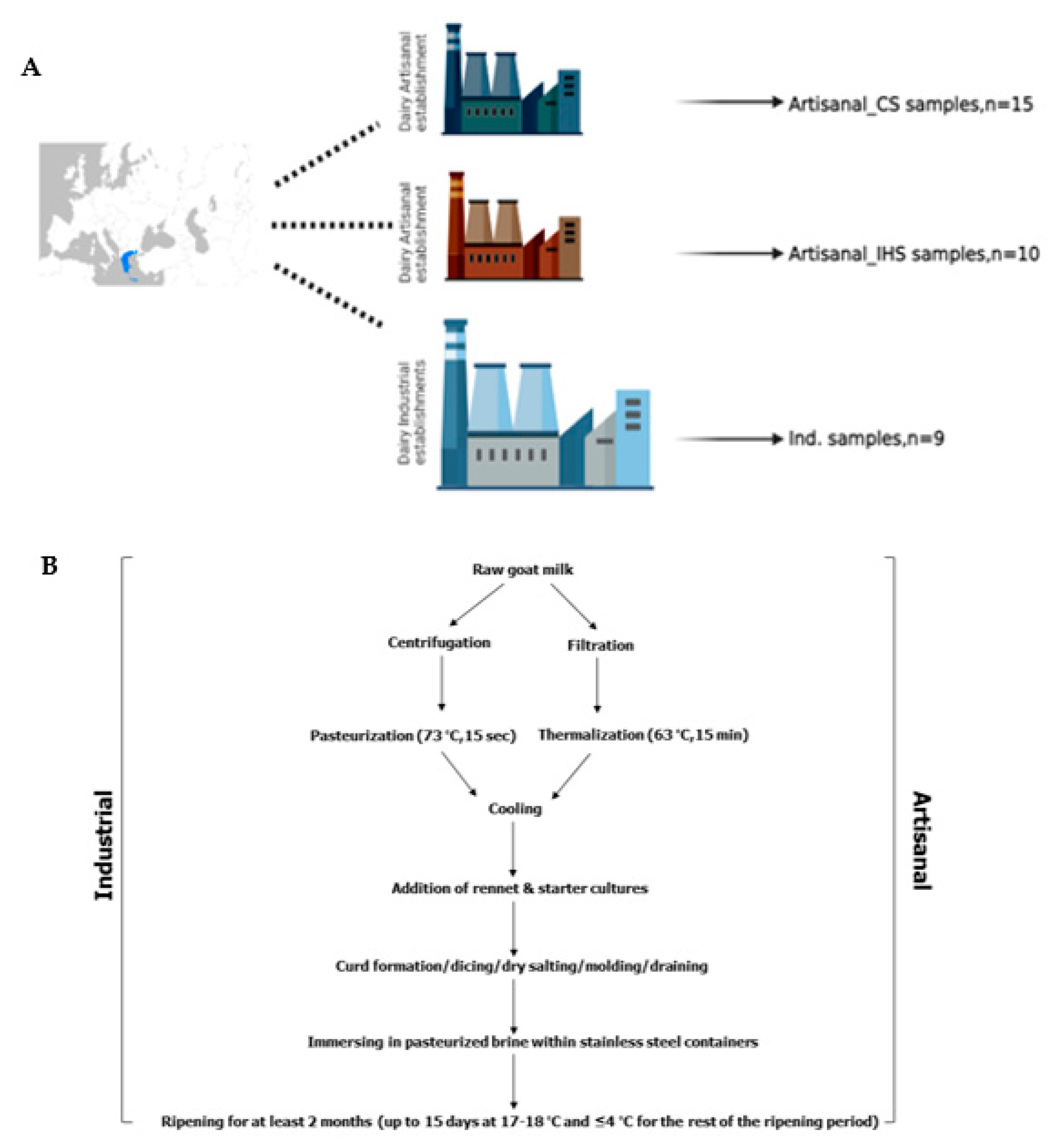
2.1. Collection of Gidotyri Cheese (Goat Cheese) Samples
2.2. DNA Extraction
2.3. High Throughput 16S rRNA Sequencing
2.4. Data Analysis and Bioinformatics
3. Results and Discussion
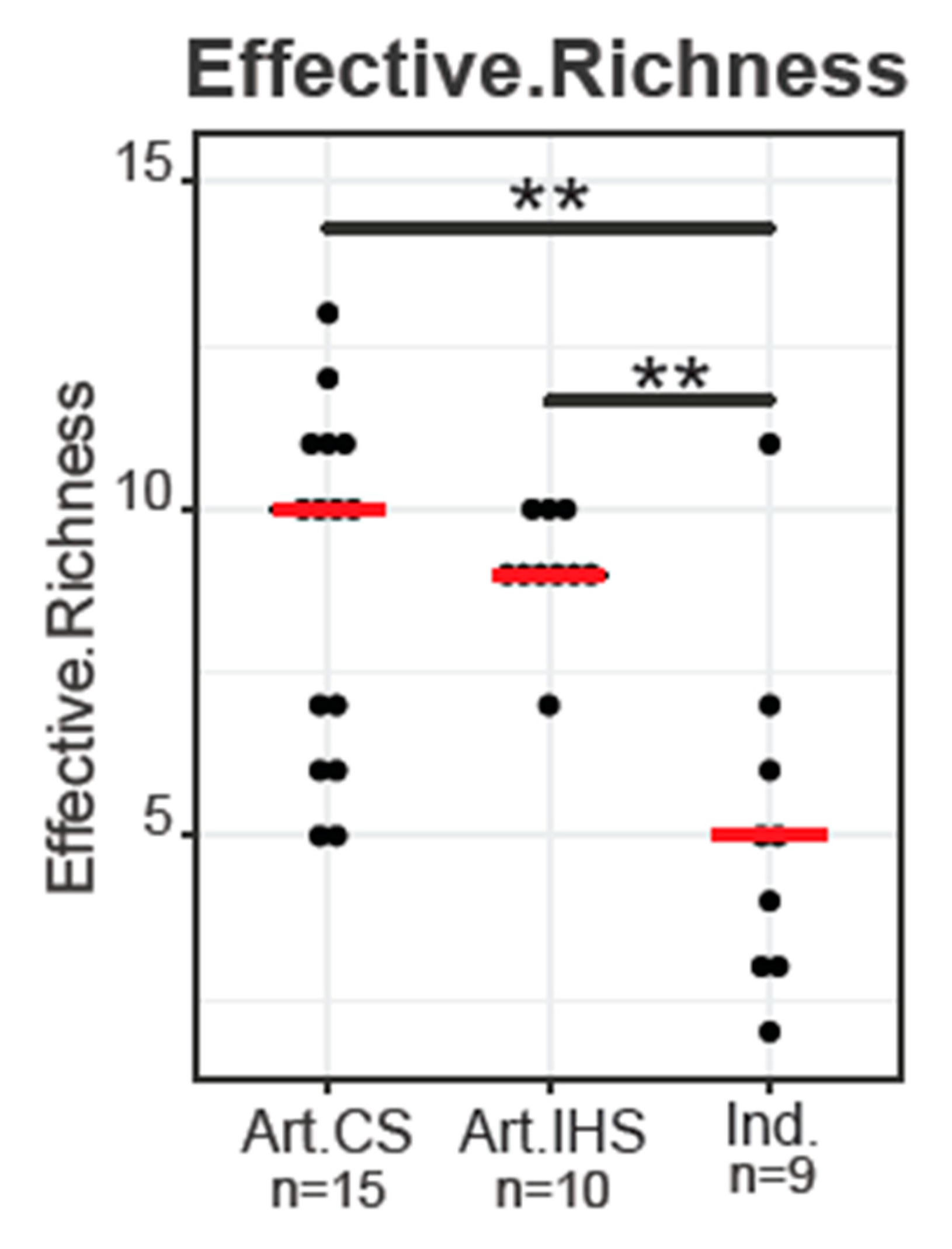
3.1. DNA Sequencing Analysis and Alpha Diversity
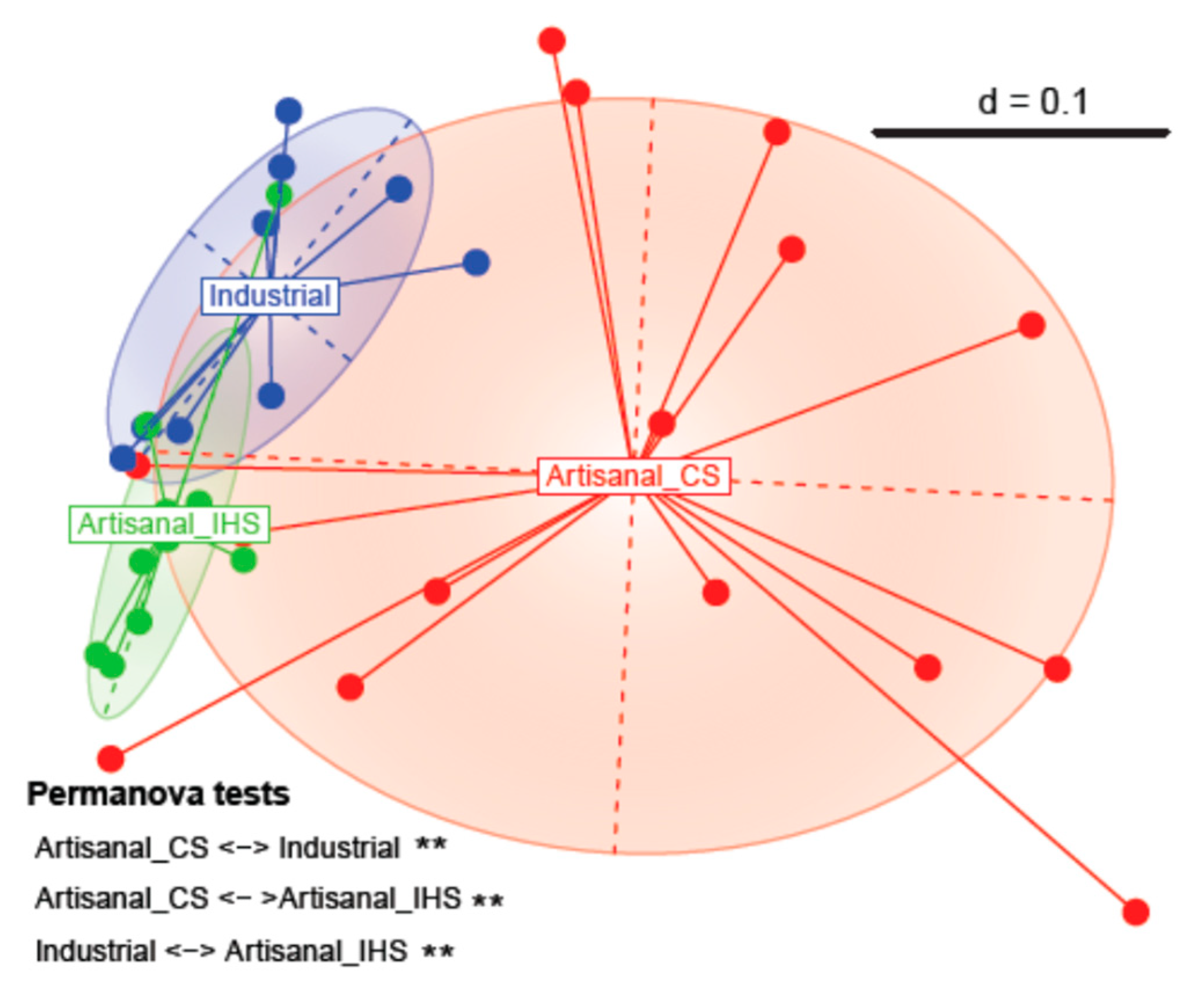
3.2. Beta Diversity
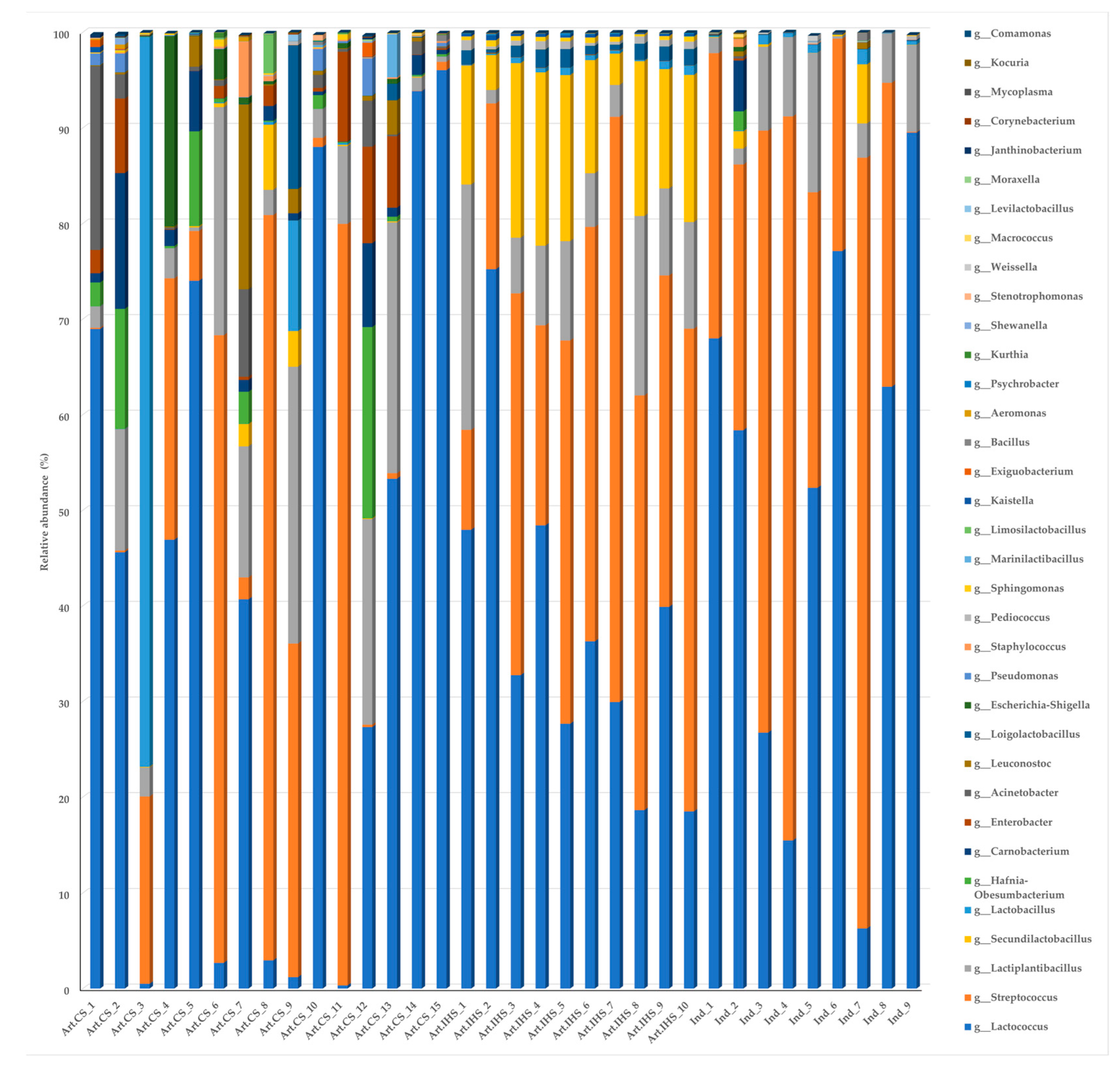
3.3. Microbiota Diversity in Gidotyri Cheese Samples
3.4. The Most Prevalent Bacterial Species in Artisanal- and Industrial-Type Gidotyri Cheese
| Bacterial Species | Identification Similarity (%) * | Relative Abundance (%) | Spore Forming | Role | Ref. | ||
|---|---|---|---|---|---|---|---|
| Art.CS | Art. IHS | Ind. | |||||
| Lactococcus lactis subsp. hordniae/subsp. lactis/Lactococcus cremoris subsp. tructae | 100 | 42.816 | 37.530 | 50.749 | no | Starter culture in dairy industry, food safety (production of bacteriocins, nisin) | [74] |
| Streptococcus thermophilus | 99.78 | 20.214 | 34.533 | 39.863 | no | Traditional starter culture, acidifying activity, food safety, organoleptic properties | [71] |
| Lactobacillus delbrueckii subsp. bulgaricus | 100 | 5.588 | 0.465 | 0.173 | no | Typically found in artisanal cheese, involved in cheese fermentation, production of folates | [75,76] |
| Lactiplantibacillus paraplantarum/pentosus/argentoratensis/pingfangensis | 100 | 10.100 | 9.966 | 5.887 | no | Lb. paraplantarum: found in artisanal cheeses, improving texture (viscosity) by exopolysaccharide production, food safety (production of bacteriocins, paraplantaricin) | [77] |
| Lb. pentosus: involved in the production of beneficial metabolites (indolepyruvate and pantothenic acid), improves intestinal barrier function (probiotic potential) | [78] | ||||||
| Lb. argentoratensis: capacity to metabolize different carbohydrates, involved in cheese fermentation, riboflavin and folate biosynthesis | [79] | ||||||
| Lb. pingfangensis: isolated from traditional Chinese pickle | [80] | ||||||
| Hafnia alvei | 100 | 3.434 | 0.011 | 0.228 | no | Development of favourable organoleptic properties in cheese, anti-obesity properties in mice | [69] |
| Escherichia fergusonii/Shigella sonnei/Shigella flexneri | 100 | 1.684 | 0.022 | 0.055 | no | Poor hygiene indicator of cheese production | [81,82] |
| Leuconostoc mesenteroides subsp. mesenteroides/subsp. cremoris/subsp. dextranicum/subsp. Jonggajibkimchii/Leuconostoc suionicum | 100 | 2.047 | 0.000 | 0.132 | no | Flavor-producing starter or adjunct cultures in dairy products | [83] |
| Acinetobacter colistiniresistens | 98.88 | 1.949 | 0.005 | 0.031 | no | Isolated in human clinical specimens, resistance to polymyxins | [70] |
| Secundilactobacillus malefermentans | 99.78 | 0.921 | 12.892 | 0.945 | no | Food fermentation at low temperatures (sauerkraut), possible contribution to the aroma development | [56,57] |
| Loigolactobacillus coryniformis subsp. torquens | 100 | 1.127 | 1.285 | 0.008 | no | Isolated from Turkish cheese and goat cheese, antibacterial properties | [54,55] |
| Carnobacterium maltaromaticum | 100 | 2.633 | 0.007 | 0.637 | no | Psychotropic bacterium isolated in French cheeses, food protection against spoilage and pathogenic bacteria, major role in cheese ripening (favorable aroma) | [64,65,66,84] |
| Enterobacter hormaechei subsp. hoffmannii | 100 | 2.773 | 0.027 | 0.026 | no | Poor hygiene indicator of cheese production, potential contribution to cheese flavour | [61,67,68] |
| Acinetobacter albensis | 99.78 | 0.742 | 0.009 | 0.001 | no | Isolated in traditional Brazilian cheeses, possible involvement in spoilage and shelf-life (carp fillets) | [7,85] |
| Staphylococcus aureus subsp. aureus | 99.78 | 0.487 | 0.004 | 0.109 | no | Milk contaminant associated with subclinical intramammary infections in ruminants | [86] |
| Streptococcus parauberis | 100 | 0.553 | 1.642 | 0.328 | no | Isolated in artisanal sheep and goat cheeses and Feta cheese, milk contaminant associated with intramammary infections in small ruminants | [38,58,59] |
| Marinilactibacillus psychrotolerans | 99.78 | 0.322 | 0.003 | 0.001 | no | Halotolerant, involved in cheese fermentation and ripening, possibly sea salt contaminant | [87] |
| Lacticaseibacillus rhamnosus | 99.78 | 0.323 | 0.003 | 0.323 | no | Involved in cheese ripening and flavor development, probiotic potential | [88,89] |
| Limosilactobacillus fermentum | 99.78 | 0.286 | 0.006 | 0.008 | no | Involved in nutritional value, organoleptic and technological properties and preservation of food products | [90] |
| Pseudomonas azotoformans/lactis/carnis/paracarnis/paralactis | 99.78 | 0.554 | 0.013 | 0.008 | no | Pseud. azotoformans: pigmented bacteria, causing visual spoilage in dairy foods, case of gray milk and blue pigment formation in cheese | [91] |
| Pseud. lactis/paralactis: involved in spoilage of mozzarella cheese | [92] | ||||||
| Pseud. carnis/paracarnis: N/A | - | ||||||
| Streptococcus caledonicus | 100 | 0.254 | 0.000 | 0.000 | no | Isolated from clinical specimens of sheep | [93] |
| Exiguobacterium artemiae | 100 | 0.156 | 0.000 | 0.000 | no | Isolated in Latin-style cheeses | [94] |
| Pediococcus parvulus | 100 | 0.039 | 0.620 | 0.075 | no | Antibacterial activity against Bacillus cereus | [95] |
| Bacillus mycoides/cereus/pseudomycoides/gaemokensis/bingmayongensis/toyonensis/wiedmannii/albus/paramycoides/proteolyticus | 99.78 | 0.065 | 0.000 | 0.098 | yes | B. mycoides: cheese spoilage and pink discoloration in Ricotta cheese | [96] |
| B.cereus: found in dairy products, foodborne pathogen (toxin production), spoilage | [97,98] | ||||||
| B. wiedmannii: isolated from raw milk, cytotoxic member of the B. cereus group | [99] | ||||||
| Bacillus pseudomycoides/gaemokensis/bingmayongensis/toyonensis/wiedmannii/albus/paramycoides/proteolyticus: N/A | - | ||||||
| Pseudomonas bubulae | 99.77 | 0.131 | 0.001 | 0.002 | no | Isolated from raw refrigerated processed meat of bovine origin | [100] |
| Sphingomonas paucimobilis/sanguinis/yabuuchiae/pseudosanguinis | 99.76 | 0.155 | 0.461 | 0.010 | no | Sphingomonas paucimobilis, pseudosanguinis: improving texture (viscosity) by gellan polysaccharide production | [101,102] |
| Sphingomonas yabuuchiae: improving texture (viscosity) by gellan polysaccharide production, isolated from Irish cheese | [102,103] | ||||||
| Sphingomonas sanguinis: N/A | - | ||||||
| Shewanella spp. | 99.78 | 0.053 | 0.000 | 0.000 | no | Food spoilage, opportunistic human pathogen, goat skin microbiome | [104,105,106] |
| Levilactobacillus huananensis/lindianensis | 99.55 | 0.043 | 0.000 | 0.000 | no | Isolated from traditional Chinese pickle, putative amino acid decarboxylases for biogenic amines production | [107,108] |
| Kaistella haifensis | 99.77 | 0.054 | 0.274 | 0.012 | no | Isolated from raw milk and Feta cheese from Epirus, lipolytic and proteolytic activity | [38,109,110] |
| Weissella thailandensis | 99.55 | 0.001 | 0.000 | 0.079 | no | Halotolerant, proteolytic activity, isolated from Mexican cheese | [111] |
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Di Trana, A.; Di Rosa, A.R.; Addis, M.; Fiori, M.; Di Grigoli, A.; Morittu, V.M.; Spina, A.A.; Claps, S.; Chiofalo, V.; Licitra, G.; et al. The Quality of Five Natural, Historical Italian Cheeses Produced in Different Months: Gross Composition, Fat-Soluble Vitamins, Fatty Acids, Total Phenols, Antioxidant Capacity, and Health Index. Animals 2022, 12, 199. [Google Scholar] [CrossRef] [PubMed]
- Cirne, C.T.; Tunick, M.H.; Trout, R.E. The chemical and attitudinal differences between commercial and artisanal products. NPJ Sci. Food 2019, 3, 19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Montel, M.-C.; Buchin, S.; Mallet, A.; Delbes-Paus, C.; Vuitton, D.A.; Desmasures, N.; Berthier, F. Traditional cheeses: Rich and diverse microbiota with associated benefits. Int. J. Food Microbiol. 2014, 177, 136–154. [Google Scholar] [CrossRef]
- Riquelme, C.; Câmara, S.; Dapkevicius, M.D.L.N.E.; Vinuesa, P.; da Silva, C.C.G.; Malcata, F.X.; Rego, O.A. Characterization of the bacterial biodiversity in Pico cheese (an artisanal Azorean food). Int. J. Food Microbiol. 2015, 192, 86–94. [Google Scholar] [CrossRef] [Green Version]
- Bozoudi, D.; Torriani, S.; Zdragas, A.; Litopoulou-Tzanetaki, E. Assessment of microbial diversity of the dominant microbiota in fresh and mature PDO Feta cheese made at three mountainous areas of Greece. LWT Food Sci. Technol. 2016, 72, 525–533. [Google Scholar] [CrossRef]
- Tzora, A.; Nelli, A.; Voidarou, C.; Fthenakis, G.; Rozos, G.; Theodorides, G.; Bonos, E.; Skoufos, I. Microbiota “Fingerprint” of Greek Feta Cheese through Ripening. Appl. Sci. 2021, 11, 5631. [Google Scholar] [CrossRef]
- Kothe, C.I.; Mohellibi, N.; Renault, P. Revealing the microbial heritage of traditional Brazilian cheeses through metagenomics. Food Res. Int. 2022, 157, 111265. [Google Scholar] [CrossRef] [PubMed]
- Settanni, L.; Moschetti, G. Non-starter lactic acid bacteria used to improve cheese quality and provide health benefits. Food Microbiol. 2010, 27, 691–697. [Google Scholar] [CrossRef]
- Blaya, J.; Barzideh, Z.; Lapointe, G. Symposium review: Interaction of starter cultures and nonstarter lactic acid bacteria in the cheese environment. J. Dairy Sci. 2018, 101, 3611–3629. [Google Scholar] [CrossRef]
- de Almeida, W.L.G., Jr.; da SilvaFerrari, Í.; de Souza, J.V.; da Silva, C.D.A.; da Costa, M.M.; Dias, F.S. Characterization and evaluation of lactic acid bacteria isolated from goat milk. Food Control 2015, 53, 96–103. [Google Scholar] [CrossRef]
- Câmara, S.; Dapkevicius, A.; Riquelme, C.; Elias, R.; Silva, C.; Malcata, F.; Dapkevicius, M.; Câmara, S. Potential of lactic acid bacteria from Pico cheese for starter culture development. Food Sci. Technol. Int. 2019, 25, 303–317. [Google Scholar] [CrossRef] [PubMed]
- Kanak, E.K.; Yilmaz, S. Maldi-tof mass spectrometry for the identification and detection of antimicrobial activity of lactic acid bacteria isolated from local cheeses. Food Sci. Technol. 2019, 39, 462–469. [Google Scholar] [CrossRef] [Green Version]
- Morandi, S.; Silvetti, T.; Battelli, G.; Brasca, M. Can lactic acid bacteria be an efficient tool for controlling Listeria monocytogenes contamination on cheese surface? The case of Gorgonzola cheese. Food Control 2019, 96, 499–507. [Google Scholar] [CrossRef]
- Reuben, R.; Roy, P.; Sarkar, S.; Alam, A.R.U.; Jahid, I. Characterization and evaluation of lactic acid bacteria from indigenous raw milk for potential probiotic properties. J. Dairy Sci. 2020, 103, 1223–1237. [Google Scholar] [CrossRef] [PubMed]
- Zoumpopoulou, G.; Papadimitriou, K.; Alexandraki, V.; Mavrogonatou, E.; Alexopoulou, K.; Anastasiou, R.; Georgalaki, M.; Kletsas, D.; Tsakalidou, E.; Giaouris, E. The microbiota of Kalathaki and Melichloro Greek artisanal cheeses comprises functional lactic acid bacteria. LWT 2020, 130, 109570. [Google Scholar] [CrossRef]
- Nagpal, R.; Behare, P.; Rana, R.; Kumar, A.; Kumar, M.; Arora, S.; Morotta, F.; Jain, S.; Yadav, H. Bioactive peptides derived from milk proteins and their health beneficial potentials: An update. Food Funct. 2011, 2, 18–27. [Google Scholar] [CrossRef]
- Nielsen, S.D.; Beverly, R.L.; Qu, Y.; Dallas, D.C. Milk bioactive peptide database: A comprehensive database of milk protein-derived bioactive peptides and novel visualization. Food Chem. 2017, 232, 673–682. [Google Scholar] [CrossRef]
- Sousa, Y.R.; Medeiros, L.B.; Pintado, M.M.E.; Queiroga, R.C. Goat milk oligosaccharides: Composition, analytical methods and bioactive and nutritional properties. Trends Food Sci. Technol. 2019, 92, 152–161. [Google Scholar] [CrossRef]
- van Leeuwen, S.S.; Poele, E.M.T.; Chatziioannou, A.C.; Benjamins, E.; Haandrikman, A.; Dijkhuizen, L. Goat Milk Oligosaccharides: Their Diversity, Quantity, and Functional Properties in Comparison to Human Milk Oligosaccharides. J. Agric. Food Chem. 2020, 68, 13469–13485. [Google Scholar] [CrossRef]
- Zotta, T.; Ricciardi, A.; Condelli, N.; Parente, E. Metataxonomic and metagenomic approaches for the study of undefined strain starters for cheese manufacture. Crit. Rev. Food Sci. Nutr. 2022, 62, 3898–3912. [Google Scholar] [CrossRef]
- Ercolini, D. High-Throughput Sequencing and Metagenomics: Moving Forward in the Culture-Independent Analysis of Food Microbial Ecology. Appl. Environ. Microbiol. 2013, 79, 3148–3155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Afshari, R.; Pillidge, C.J.; Dias, D.A.; Osborn, A.M.; Gill, H. Cheesomics: The future pathway to understanding cheese flavour and quality. Crit. Rev. Food Sci. Nutr. 2020, 60, 33–47. [Google Scholar] [CrossRef] [PubMed]
- Aldrete-Tapia, A.; Escobar-Ramírez, M.C.; Tamplin, M.L.; Hernández-Iturriaga, M. High-throughput sequencing of microbial communities in Poro cheese, an artisanal Mexican cheese. Food Microbiol. 2014, 44, 136–141. [Google Scholar] [CrossRef]
- De Filippis, F.; La Storia, A.; Stellato, G.; Gatti, M.; Ercolini, D. A Selected Core Microbiome Drives the Early Stages of Three Popular Italian Cheese Manufactures. PLoS ONE 2014, 9, e89680. [Google Scholar] [CrossRef] [PubMed]
- Delcenserie, V.; Taminiau, B.; Delhalle, L.; Nezer, C.; Doyen, P.; Crevecoeur, S.; Roussey, D.; Korsak, N.; Daube, G. Microbiota characterization of a Belgian protected designation of origin cheese, Herve cheese, using metagenomic analysis. J. Dairy Sci. 2014, 97, 6046–6056. [Google Scholar] [CrossRef] [Green Version]
- De Filippis, F.; Genovese, A.; Ferranti, P.; Gilbert, J.A.; Ercolini, D. Metatranscriptomics reveals temperature-driven functional changes in microbiome impacting cheese maturation rate. Sci. Rep. 2016, 6, 21871. [Google Scholar] [CrossRef] [Green Version]
- Papadimitriou, K.; Anastasiou, R.; Georgalaki, M.; Bounenni, R.; Paximadaki, A.; Charmpi, C.; Alexandraki, V.; Kazou, M.; Tsakalidou, E. Comparison of the Microbiome of Artisanal Homemade and Industrial Feta Cheese through Amplicon Sequencing and Shotgun Metagenomics. Microorganisms 2022, 10, 1073. [Google Scholar] [CrossRef]
- Tsigkrimani, M.; Bakogianni, M.; Paramithiotis, S.; Bosnea, L.; Pappa, E.; Drosinos, E.H.; Skandamis, P.N.; Mataragas, M. Microbial Ecology of Artisanal Feta and Kefalograviera Cheeses, Part I: Bacterial Community and Its Functional Characteristics with Focus on Lactic Acid Bacteria as Determined by Culture-Dependent Methods and Phenotype Microarrays. Microorganisms 2022, 10, 161. [Google Scholar] [CrossRef]
- Sepe, L.; Argüello, A. Recent advances in dairy goat products. Asian Australas. J. Anim. Sci. 2019, 32, 1306–1320. [Google Scholar] [CrossRef] [Green Version]
- Verruck, S.; Dantas, A.; Prudencio, E.S. Functionality of the components from goat’s milk, recent advances for functional dairy products development and its implications on human health. J. Funct. Foods 2019, 52, 243–257. [Google Scholar] [CrossRef]
- Nayik, G.A.; Jagdale, Y.D.; Gaikwad, S.A.; Devkatte, A.N.; Dar, A.H.; Dezmirean, D.S.; Bobis, O.; Ranjha, M.M.A.N.; Ansari, M.J.; Hemeg, H.A.; et al. Recent Insights into Processing Approaches and Potential Health Benefits of Goat Milk and Its Products: A Review. Front. Nutr. 2021, 8, 789117. [Google Scholar] [CrossRef] [PubMed]
- Giorgio, D.; Di Trana, A.; Di Napoli, M.; Sepe, L.; Cecchini, S.; Rossi, R.; Claps, S. Comparison of cheeses from goats fed 7 forages based on a new health index. J. Dairy Sci. 2019, 102, 6790–6801. [Google Scholar] [CrossRef] [PubMed]
- Food and Agriculture Organization of the United Nations; FAOSTAT. FAOSTAT Statistical Database. Available online: http://www.fao.org/faostat (accessed on 20 June 2022).
- Litopoulou-Tzanetaki, E.; Tzanetakis, N. Microbiological characteristics of Greek traditional cheeses. Small Rumin. Res. 2011, 101, 17–32. [Google Scholar] [CrossRef]
- Tsafrakidou, P.; Bozoudi, D.; Pavlidou, S.; Kotzamanidis, C.; Hatzikamari, M.; Zdragas, A.; Litopoulou-Tzanetaki, E. Technological, phenotypic and genotypic characterization of lactobacilli from Graviera Kritis PDO Greek cheese, manufactured at two traditional dairies. LWT Food Sci. Technol. 2016, 68, 681–689. [Google Scholar] [CrossRef]
- Gantzias, C.; Lappa, I.K.; Aerts, M.; Georgalaki, M.; Manolopoulou, E.; Papadimitriou, K.; De Brandt, E.; Tsakalidou, E.; Vandamme, P. MALDI-TOF MS profiling of non-starter lactic acid bacteria from artisanal cheeses of the Greek island of Naxos. Int. J. Food Microbiol. 2020, 323, 108586. [Google Scholar] [CrossRef]
- Spyrelli, E.; Stamatiou, A.; Tassou, C.; Nychas, G.-J.; Doulgeraki, A. Microbiological and Metagenomic Analysis to Assess the Effect of Container Material on the Microbiota of Feta Cheese during Ripening. Fermentation 2020, 6, 12. [Google Scholar] [CrossRef] [Green Version]
- Papadakis, P.; Konteles, S.; Batrinou, A.; Ouzounis, S.; Tsironi, T.; Halvatsiotis, P.; Tsakali, E.; Van Impe, J.F.M.; Vougiouklaki, D.; Strati, I.F.; et al. Characterization of Bacterial Microbiota of P.D.O. Feta Cheese by 16S Metagenomic Analysis. Microorganisms 2021, 9, 2377. [Google Scholar] [CrossRef]
- Tzora, A.; Nelli, A.; Kritikou, A.S.; Katsarou, D.; Giannenas, I.; Lagkouvardos, I.; Thomaidis, N.S.; Skoufos, I. The “Crosstalk” between Microbiota and Metabolomic Profile of Kefalograviera Cheese after the Innovative Feeding Strategy of Dairy Sheep by Omega-3 Fatty Acids. Foods 2022, 11, 3164. [Google Scholar] [CrossRef]
- Samelis, J.; Kakouri, A. Major technological differences between an industrial-type and five artisan-type Greek PDO Galotyri market cheeses as revealed by great variations in their lactic acid microbiota. AIMS Agric. Food 2019, 4, 685–710. [Google Scholar] [CrossRef]
- Moatsou, G.; Govaris, A. White brined cheeses: A diachronic exploitation of small ruminants milk in Greece. Small Rumin. Res. 2011, 101, 113–121. [Google Scholar] [CrossRef]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of General 16S Ribosomal RNA Gene PCR Primers for Classical and Next-Generation Sequencing-Based Diversity Studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lagkouvardos, I.; Fischer, S.; Kumar, N.; Clavel, T. Rhea: A transparent and modular R pipeline for microbial profiling based on 16S rRNA gene amplicons. Peerj 2017, 5, e2836. [Google Scholar] [CrossRef] [Green Version]
- Reitmeier, S.; Hitch, T.C.A.; Treichel, N.; Fikas, N.; Hausmann, B.; Ramer-Tait, A.E.; Neuhaus, K.; Berry, D.; Haller, D.; Lagkouvardos, I.; et al. Handling of spurious sequences affects the outcome of high-throughput 16S rRNA gene amplicon profiling. ISME Commun. 2021, 1, 31. [Google Scholar] [CrossRef]
- Kamilari, E.; Anagnostopoulos, D.A.; Papademas, P.; Kamilaris, A.; Tsaltas, D. Characterizing Halloumi cheese’s bacterial communities through metagenomic analysis. LWT 2020, 126, 109298. [Google Scholar] [CrossRef] [Green Version]
- Yeluri Jonnala, B.R.; McSweeney, P.L.H.; Sheehan, J.J.; Cotter, P.D. Sequencing of the Cheese Microbiome and Its Relevance to Industry. Front. Microbiol. 2018, 9, 1020. [Google Scholar] [CrossRef] [Green Version]
- Tilocca, B.; Costanzo, N.; Morittu, V.M.; Spina, A.A.; Soggiu, A.; Britti, D.; Roncada, P.; Piras, C. Milk microbiota: Characterization methods and role in cheese production. J. Proteom. 2020, 210, 103534. [Google Scholar] [CrossRef]
- Quigley, L.; O’Sullivan, O.; Beresford, T.P.; Ross, R.P.; Fitzgerald, G.F.; Cotter, P.D. High-Throughput Sequencing for Detection of Subpopulations of Bacteria Not Previously Associated with Artisanal Cheeses. Appl. Environ. Microbiol. 2012, 78, 5717–5723. [Google Scholar] [CrossRef] [Green Version]
- Penland, M.; Falentin, H.; Parayre, S.; Pawtowski, A.; Maillard, M.-B.; Thierry, A.; Mounier, J.; Coton, M.; Deutsch, S.-M. Linking Pélardon artisanal goat cheese microbial communities to aroma compounds during cheese-making and ripening. Int. J. Food Microbiol. 2021, 345, 109130. [Google Scholar] [CrossRef]
- Tilocca, B.; Soggiu, A.; Iavarone, F.; Greco, V.; Putignani, L.; Ristori, M.V.; Macari, G.; Spina, A.A.; Morittu, V.M.; Ceniti, C.; et al. The Functional Characteristics of Goat Cheese Microbiota from a One-Health Perspective. Int. J. Mol. Sci. 2022, 23, 14131. [Google Scholar] [CrossRef]
- Monnet, C.; Bleicher, A.; Neuhaus, K.; Sarthou, A.-S.; Leclercq-Perlat, M.-N.; Irlinger, F. Assessment of the anti-listerial activity of microfloras from the surface of smear-ripened cheeses. Food Microbiol. 2010, 27, 302–310. [Google Scholar] [CrossRef] [PubMed]
- de Paula, A.C.L.; Medeiros, J.D.; Fernandes, G.D.R.; da Silva, V.L.; Diniz, C.G. Microbiome of industrialized Minas Frescal Cheese reveals high prevalence of putative bacteria: A concern in the One Health context. LWT 2021, 139, 110791. [Google Scholar] [CrossRef]
- Martín, R.; Olivares, M.; Marín, M.; Xaus, J.; Fernández, L.; Rodríguez, J. Characterization of a reuterin-producing Lactobacillus coryniformis strain isolated from a goat’s milk cheese. Int. J. Food Microbiol. 2005, 104, 267–277. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, S.; Çon, A.H. Isolation and characterization of potential probiotic lactic acid bacteria from traditional cheese. LWT 2021, 152, 112319. [Google Scholar] [CrossRef]
- Nigatu, A.; Ahrné, S.; Molin, G. Temperature-Dependent Variation in API 50 CH Fermentation Profiles of Lactobacillus Species. Curr. Microbiol. 2000, 41, 21–26. [Google Scholar] [CrossRef]
- Tlais, A.Z.A.; Junior, W.J.F.L.; Filannino, P.; Campanaro, S.; Gobbetti, M.; Di Cagno, R. How Microbiome Composition Correlates with Biochemical Changes during Sauerkraut Fermentation: A Focus on Neglected Bacterial Players and Functionalities. Microbiol. Spectr. 2022, 10, e00168-00122. [Google Scholar] [CrossRef]
- Fuka, M.M.; Wallisch, S.; Engel, M.; Welzl, G.; Havranek, J.; Schloter, M. Dynamics of Bacterial Communities during the Ripening Process of Different Croatian Cheese Types Derived from Raw Ewe’s Milk Cheeses. PLoS ONE 2013, 8, e80734. [Google Scholar] [CrossRef] [Green Version]
- Rosa, N.M.; Penati, M.; Fusar-Poli, S.; Addis, M.F.; Tola, S. Species identification by MALDI-TOF MS and gap PCR–RFLP of non-aureus Staphylococcus, Mammaliicoccus, and Streptococcus spp. associated with sheep and goat mastitis. Vet. Res. 2022, 53, 84. [Google Scholar] [CrossRef]
- Fragkou, I.A.; Skoufos, J.; Cripps, P.J.; Kyriazakis, I.; Papaioannou, N.; Boscos, C.M.; Tzora, A.; Fthenakis, G.C. Differences in susceptibility to Mannheimia haemolytica-associated mastitis between two breeds of dairy sheep. J. Dairy Res. 2007, 74, 349–355. [Google Scholar] [CrossRef]
- Pangallo, D.; Šaková, N.; Koreňová, J.; Puškárová, A.; Kraková, L.; Valík, L.; Kuchta, T. Microbial diversity and dynamics during the production of May bryndza cheese. Int. J. Food Microbiol. 2014, 170, 38–43. [Google Scholar] [CrossRef]
- Kümmel, J.; Stessl, B.; Gonano, M.; Walcher, G.; Bereuter, O.; Fricker, M.; Grunert, T.; Wagner, M.; Ehling-Schulz, M. Staphylococcus aureus Entrance into the Dairy Chain: Tracking S. aureus from Dairy Cow to Cheese. Front. Microbiol. 2016, 7, 1603. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silva, G.; Castro, R.; Oliveira, L.; Sant’Anna, F.; Barbosa, C.; Sandes, S.; Silva, R.; Resende, M.; Lana, A.; Nunes, A.; et al. Viability of Staphylococcus aureus and expression of its toxins (SEC and TSST-1) in cheeses using Lactobacillus rhamnosus D1 or Weissella paramesenteroides GIR16L4 or both as starter cultures. J. Dairy Sci. 2020, 103, 4100–4108. [Google Scholar] [CrossRef] [PubMed]
- Afzal, M.I.; Delaunay, S.; Paris, C.; Borges, F.; Revol-Junelles, A.-M.; Cailliez-Grimal, C. Identification of metabolic pathways involved in the biosynthesis of flavor compound 3-methylbutanal from leucine catabolism by Carnobacterium maltaromaticum LMA 28. Int. J. Food Microbiol. 2012, 157, 332–339. [Google Scholar] [CrossRef] [PubMed]
- Afzal, M.I.; Ariceaga, C.C.G.; Lhomme, E.; Ali, N.K.; Payot, S.; Burgain, J.; Gaiani, C.; Borges, F.; Revol-Junelles, A.-M.; Delaunay, S.; et al. Characterization of Carnobacterium maltaromaticum LMA 28 for its positive technological role in soft cheese making. Food Microbiol. 2013, 36, 223–230. [Google Scholar] [CrossRef]
- Hammi, I.; Delalande, F.; Belkhou, R.; Marchioni, E.; Cianferani, S.; Ennahar, S. Maltaricin CPN, a new class IIa bacteriocin produced by Carnobacterium maltaromaticum CPN isolated from mould-ripened cheese. J. Appl. Microbiol. 2016, 121, 1268–1274. [Google Scholar] [CrossRef] [Green Version]
- Fuka, M.M.; Engel, M.; Skelin, A.; Redžepović, S.; Schloter, M. Bacterial communities associated with the production of artisanal Istrian cheese. Int. J. Food Microbiol. 2010, 142, 19–24. [Google Scholar] [CrossRef]
- Martelli, F.; Bancalari, E.; Neviani, E.; Bottari, B. Novel insights on pink discoloration in cheese: The case of Pecorino Toscano. Int. Dairy J. 2020, 111, 104829. [Google Scholar] [CrossRef]
- Ramos-Vivas, J.; Tapia, O.; Elexpuru-Zabaleta, M.; Pifarre, K.T.; Diaz, Y.A.; Battino, M.; Giampieri, F. The Molecular Weaponry Produced by the Bacterium Hafnia alvei in Foods. Molecules 2022, 27, 5585. [Google Scholar] [CrossRef]
- Nemec, A.; Radolfova-Krizova, L.; Maixnerova, M.; Sedo, O. Acinetobacter colistiniresistens sp. nov. (formerly genomic species 13 sensu Bouvet and Jeanjean and genomic species 14 sensu Tjernberg and Ursing), isolated from human infections and characterized by intrinsic resistance to polymyxins. Int. J. Syst. Evol. Microbiol. 2017, 67, 2134–2141. [Google Scholar] [CrossRef]
- Markakiou, S.; Gaspar, P.; Johansen, E.; Zeidan, A.A.; Neves, A.R. Harnessing the metabolic potential of Streptococcus thermophilus for new biotechnological applications. Curr. Opin. Biotechnol. 2020, 61, 142–152. [Google Scholar] [CrossRef]
- Pappa, E.C.; Kondyli, E.; Samelis, J. Microbiological and biochemical characteristics of Kashkaval cheese produced using pasteurised or raw milk. Int. Dairy J. 2019, 89, 60–67. [Google Scholar] [CrossRef]
- Tadjine, D.; Boudalia, S.; Bousbia, A.; Gueroui, Y.; Symeon, G.; Boudechiche, L.M.; Tadjine, A.; Chemmam, M. Milk heat treatment affects microbial characteristics of cows’ and goats’ “Jben” traditional fresh cheeses. Food Sci. Technol. 2021, 41, 136–143. [Google Scholar] [CrossRef]
- Mataragas, M. Investigation of genomic characteristics and carbohydrates’ metabolic activity of Lactococcus lactis subsp. lactis during ripening of a Swiss-type cheese. Food Microbiol. 2020, 87, 103392. [Google Scholar] [CrossRef] [PubMed]
- Stachelska, M.A.; Foligni, R. Development of a time-effective and highly specific quantitative real-time polymerase chain reaction assay for the identification of Lactobacillus delbrueckii subsp. bulgaricus and Streptococcus thermophilus in artisanal raw cow’s milk cheese. Acta Vet. Brno 2018, 87, 301–308. [Google Scholar] [CrossRef] [Green Version]
- Albano, C.; Silvetti, T.; Brasca, M. Screening of lactic acid bacteria producing folate and their potential use as adjunct cultures for cheese bio-enrichment. FEMS Microbiol. Lett. 2020, 367, fnaa059. [Google Scholar] [CrossRef]
- Margalho, L.P.; Feliciano, M.D.; Silva, C.E.; Abreu, J.S.; Piran, M.V.F.; Sant’Ana, A.S. Brazilian artisanal cheeses are rich and diverse sources of nonstarter lactic acid bacteria regarding technological, biopreservative, and safety properties—Insights through multivariate analysis. J. Dairy Sci. 2020, 103, 7908–7926. [Google Scholar] [CrossRef]
- Ma, Y.; Hu, C.; Yan, W.; Jiang, H.; Liu, G. Lactobacillus pentosus Increases the Abundance of Akkermansia and Affects the Serum Metabolome to Alleviate DSS-Induced Colitis in a Murine Model. Front. Cell Dev. Biol. 2020, 8, 591408. [Google Scholar] [CrossRef]
- Syrokou, M.K.; Paramithiotis, S.; Drosinos, E.H.; Bosnea, L.; Mataragas, M. A Comparative Genomic and Safety Assessment of Six Lactiplantibacillus plantarum subsp. argentoratensis Strains Isolated from Spontaneously Fermented Greek Wheat Sourdoughs for Potential Biotechnological Application. Int. J. Mol. Sci. 2022, 23, 2487. [Google Scholar] [CrossRef]
- Liu, D.D.; Gu, C.T. Lactobacillus pingfangensis sp. nov., Lactobacillus daoliensis sp. nov., Lactobacillus nangangensis sp. nov., Lactobacillus daowaiensis sp. nov., Lactobacillus dongliensis sp. nov., Lactobacillus songbeiensis sp. nov. and Lactobacillus kaifaensis sp. nov., isolated from traditional Chinese pickle. Int. J. Syst. Evol. Microbiol. 2019, 69, 3237–3247. [Google Scholar] [CrossRef]
- Molina, F.; Simancas, A.; Tabla, R.; Gómez, A.; Roa, I.; Rebollo, J.E. Diversity and Local Coadaptation of Escherichia coli and Coliphages From Small Ruminants. Front. Microbiol. 2020, 11, 564522. [Google Scholar] [CrossRef] [PubMed]
- Selover, B.; Johnson, J.; Waite-Cusic, J.G. Population dynamics of coliforms in a commercial Cheddar cheese production facility. J. Dairy Sci. 2021, 104, 7480–7488. [Google Scholar] [CrossRef] [PubMed]
- Pogačić, T.; Maillard, M.-B.; Leclerc, A.; Hervé, C.; Chuat, V.; Valence, F.; Thierry, A. Lactobacillus and Leuconostoc volatilomes in cheese conditions. Appl. Microbiol. Biotechnol. 2016, 100, 2335–2346. [Google Scholar] [CrossRef] [PubMed]
- Cailliez-Grimal, C.; Edima, H.; Revol-Junelles, A.-M.; Millière, J.-B. Short Communication: Carnobacterium maltaromaticum: The Only Carnobacterium Species in French Ripened Soft Cheeses as Revealed by Polymerase Chain Reaction Detection. J. Dairy Sci. 2007, 90, 1133–1138. [Google Scholar] [CrossRef] [PubMed]
- Kaszab, E.; Farkas, M.; Radó, J.; Micsinai, A.; Nyírő-Fekete, B.; Szabó, I.; Kriszt, B.; Urbányi, B.; Szoboszlay, S. Novel members of bacterial community during a short-term chilled storage of common carp (Cyprinus carpio). Folia Microbiol. 2022, 67, 299–310. [Google Scholar] [CrossRef] [PubMed]
- Contreras, A.; Sierra, D.; Sánchez, A.; Corrales, J.; Marco, J.; Paape, M.; Gonzalo, C. Mastitis in small ruminants. Small Rumin. Res. 2007, 68, 145–153. [Google Scholar] [CrossRef]
- Ishikawa, M.; Kodama, K.; Yasuda, H.; Okamoto-Kainuma, A.; Koizumi, K.; Yamasato, K. Presence of halophilic and alkaliphilic lactic acid bacteria in various cheeses. Lett. Appl. Microbiol. 2007, 44, 308–313. [Google Scholar] [CrossRef]
- Mathipa-Mdakane, M.G.; Thantsha, M.S. Lacticaseibacillus rhamnosus: A Suitable Candidate for the Construction of Novel Bioengineered Probiotic Strains for Targeted Pathogen Control. Foods 2022, 11, 785. [Google Scholar] [CrossRef] [PubMed]
- Lazzi, C.; Povolo, M.; Locci, F.; Bernini, V.; Neviani, E.; Gatti, M. Can the development and autolysis of lactic acid bacteria influence the cheese volatile fraction? The case of Grana Padano. Int. J. Food Microbiol. 2016, 233, 20–28. [Google Scholar] [CrossRef]
- Ale, E.C.; Rojas, M.F.; Reinheimer, J.A.; Binetti, A.G. Lactobacillus fermentum: Could EPS production ability be responsible for functional properties? Food Microbiol. 2020, 90, 103465. [Google Scholar] [CrossRef]
- Makarov, D.A.; Ivanova, O.E.; Pomazkova, A.V.; Egoreva, M.A.; Prasolova, O.V.; Lenev, S.V.; Gergel, M.A.; Bukova, N.K.; Karabanov, S.Y. Antimicrobial resistance of commensal Enterococcus faecalis and Enterococcus faecium from food-producing animals in Russia. Vet. World 2022, 15, 611–621. [Google Scholar] [CrossRef]
- Quintieri, L.; Caputo, L.; De Angelis, M.; Fanelli, F. Genomic Analysis of Three Cheese-Borne Pseudomonas lactis with Biofilm and Spoilage-Associated Behavior. Microorganisms 2020, 8, 1208. [Google Scholar] [CrossRef] [PubMed]
- Foster, G.; Kirchner, M.; Muchowski, J.; Duggett, N.; Randall, L.; Knight, H.I.; Whatmore, A.M. Streptococcus caledonicus sp. nov., isolated from sheep. Int. J. Syst. Evol. Microbiol. 2020, 70, 2611–2615. [Google Scholar] [CrossRef] [PubMed]
- Lusk, T.S.; Ottesen, A.R.; White, J.R.; Allard, M.W.; Brown, E.W.; Kase, J.A. Characterization of microflora in Latin-style cheeses by next-generation sequencing technology. BMC Microbiol. 2012, 12, 254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Immerstrand, T.; Paul, C.J.; Rosenquist, A.; Deraz, S.; Mårtensson, O.B.; Ljungh, Å.; Blücher, A.; Öste, R.; Holst, O.; Karlsson, E.N. Characterization of the Properties of Pediococcus parvulus for Probiotic or Protective Culture Use. J. Food Prot. 2010, 73, 960–966. [Google Scholar] [CrossRef]
- Sattin, E.; Andreani, N.; Carraro, L.; Fasolato, L.; Balzan, S.; Novelli, E.; Squartini, A.; Telatin, A.; Simionati, B.; Cardazzo, B. Microbial dynamics during shelf-life of industrial Ricotta cheese and identification of a Bacillus strain as a cause of a pink discolouration. Food Microbiol. 2016, 57, 8–15. [Google Scholar] [CrossRef]
- Montone, A.M.I.; Capuano, F.; Mancusi, A.; Di Maro, O.; Peruzy, M.F.; Proroga, Y.T.R.; Cristiano, D. Exposure to Bacillus cereus in Water Buffalo Mozzarella Cheese. Foods 2020, 9, 1899. [Google Scholar] [CrossRef]
- Vidic, J.; Chaix, C.; Manzano, M.; Heyndrickx, M. Food Sensing: Detection of Bacillus cereus Spores in Dairy Products. Biosensors 2020, 10, 15. [Google Scholar] [CrossRef] [Green Version]
- Miller, R.A.; Beno, S.M.; Kent, D.J.; Carroll, L.; Martin, N.H.; Boor, K.; Kovac, J. Bacillus wiedmannii sp. nov., a psychrotolerant and cytotoxic Bacillus cereus group species isolated from dairy foods and dairy environments. Int. J. Syst. Evol. Microbiol. 2016, 66, 4744–4753. [Google Scholar] [CrossRef]
- Lick, S.; Kröckel, L.; Wibberg, D.; Winkler, A.; Blom, J.; Goesmann, A.; Kalinowski, J. Pseudomonas bubulae sp. nov., isolated from beef. Int. J. Syst. Evol. Microbiol. 2020, 70, 292–301. [Google Scholar] [CrossRef]
- Fialho, A.M.; Martins, L.O.; Donval, M.-L.; Leitão, J.H.; Ridout, M.J.; Jay, A.J.; Morris, V.J.; Sá-Correia, I. Structures and Properties of Gellan Polymers Produced by Sphingomonas paucimobilis ATCC 31461 from Lactose Compared with Those Produced from Glucose and from Cheese Whey. Appl. Environ. Microbiol. 1999, 65, 2485–2491. [Google Scholar] [CrossRef]
- Raghunandan, K.; Kumar, A.; Kumar, S.; Permaul, K.; Singh, S. Production of gellan gum, an exopolysaccharide, from biodiesel-derived waste glycerol by Sphingomonas spp. 3 Biotech 2018, 8, 71. [Google Scholar] [CrossRef] [PubMed]
- Kamilari, E.; Tsaltas, D.; Stanton, C.; Ross, R.P. Metataxonomic Mapping of the Microbial Diversity of Irish and Eastern Mediterranean Cheeses. Foods 2022, 11, 2483. [Google Scholar] [CrossRef] [PubMed]
- Pushpam, P.L.; Rajesh, T.; Gunasekaran, P. Identification and characterization of alkaline serine protease from goat skin surface metagenome. AMB Express 2011, 1, 3. [Google Scholar] [CrossRef] [Green Version]
- Yousfi, K.; Bekal, S.; Usongo, V.; Touati, A. Current trends of human infections and antibiotic resistance of the genus Shewanella. Eur. J. Clin. Microbiol. Infect. Dis. 2017, 36, 1353–1362. [Google Scholar] [CrossRef]
- Palevich, N.; Palevich, F.P.; Gardner, A.; Brightwell, G.; Mills, J. Genome collection of Shewanella spp. isolated from spoiled lamb. Front. Microbiol. 2022, 13, 976152. [Google Scholar] [CrossRef]
- Long, G.Y.; Gu, C.T. Lactobacillus jixianensis sp. nov., Lactobacillus baoqingensis sp. nov., Lactobacillus jiayinensis sp. nov., Lactobacillus zhaoyuanensis sp. nov., Lactobacillus lindianensis sp. nov., Lactobacillus huananensis sp. nov., Lactobacillus tangyuanensis sp. nov., Lactobacillus fuyuanensis sp. nov., Lactobacillus tongjiangensis sp. nov., Lactobacillus fujinensis sp. nov. and Lactobacillus mulengensis sp. nov., isolated from Chinese traditional pickle. Int. J. Syst. Evol. Microbiol. 2019, 69, 2340–2353. [Google Scholar] [CrossRef] [PubMed]
- Berthoud, H.; Wechsler, D.; Irmler, S. Production of Putrescine and Cadaverine by Paucilactobacillus wasatchensis. Front. Microbiol. 2022, 13, 842403. [Google Scholar] [CrossRef]
- Hantsis-Zacharov, E.; Halpern, M. Chryseobacterium haifense sp. nov., a psychrotolerant bacterium isolated from raw milk. Int. J. Syst. Evol. Microbiol. 2007, 57, 2344–2348. [Google Scholar] [CrossRef]
- Hantsis-Zacharov, E.; Halpern, M. Culturable Psychrotrophic Bacterial Communities in Raw Milk and Their Proteolytic and Lipolytic Traits. Appl. Environ. Microbiol. 2007, 73, 7162–7168. [Google Scholar] [CrossRef] [Green Version]
- Morales, F.; Morales, J.I.; Hernández, C.H.; Hernández-Sánchez, H. Isolation and Partial Characterization of Halotolerant Lactic Acid Bacteria from Two Mexican Cheeses. Appl. Biochem. Biotechnol. 2011, 164, 889–905. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nelli, A.; Venardou, B.; Skoufos, I.; Voidarou, C.; Lagkouvardos, I.; Tzora, A. An Insight into Goat Cheese: The Tales of Artisanal and Industrial Gidotyri Microbiota. Microorganisms 2023, 11, 123. https://doi.org/10.3390/microorganisms11010123
Nelli A, Venardou B, Skoufos I, Voidarou C, Lagkouvardos I, Tzora A. An Insight into Goat Cheese: The Tales of Artisanal and Industrial Gidotyri Microbiota. Microorganisms. 2023; 11(1):123. https://doi.org/10.3390/microorganisms11010123
Chicago/Turabian StyleNelli, Aikaterini, Brigkita Venardou, Ioannis Skoufos, Chrysoula (Chrysa) Voidarou, Ilias Lagkouvardos, and Athina Tzora. 2023. "An Insight into Goat Cheese: The Tales of Artisanal and Industrial Gidotyri Microbiota" Microorganisms 11, no. 1: 123. https://doi.org/10.3390/microorganisms11010123
APA StyleNelli, A., Venardou, B., Skoufos, I., Voidarou, C., Lagkouvardos, I., & Tzora, A. (2023). An Insight into Goat Cheese: The Tales of Artisanal and Industrial Gidotyri Microbiota. Microorganisms, 11(1), 123. https://doi.org/10.3390/microorganisms11010123







