Epidendrumradicans Fungal Community during Ex Situ Germination and Isolation of Germination-Enhancing Fungi
Abstract
1. Introduction
2. Materials and methods
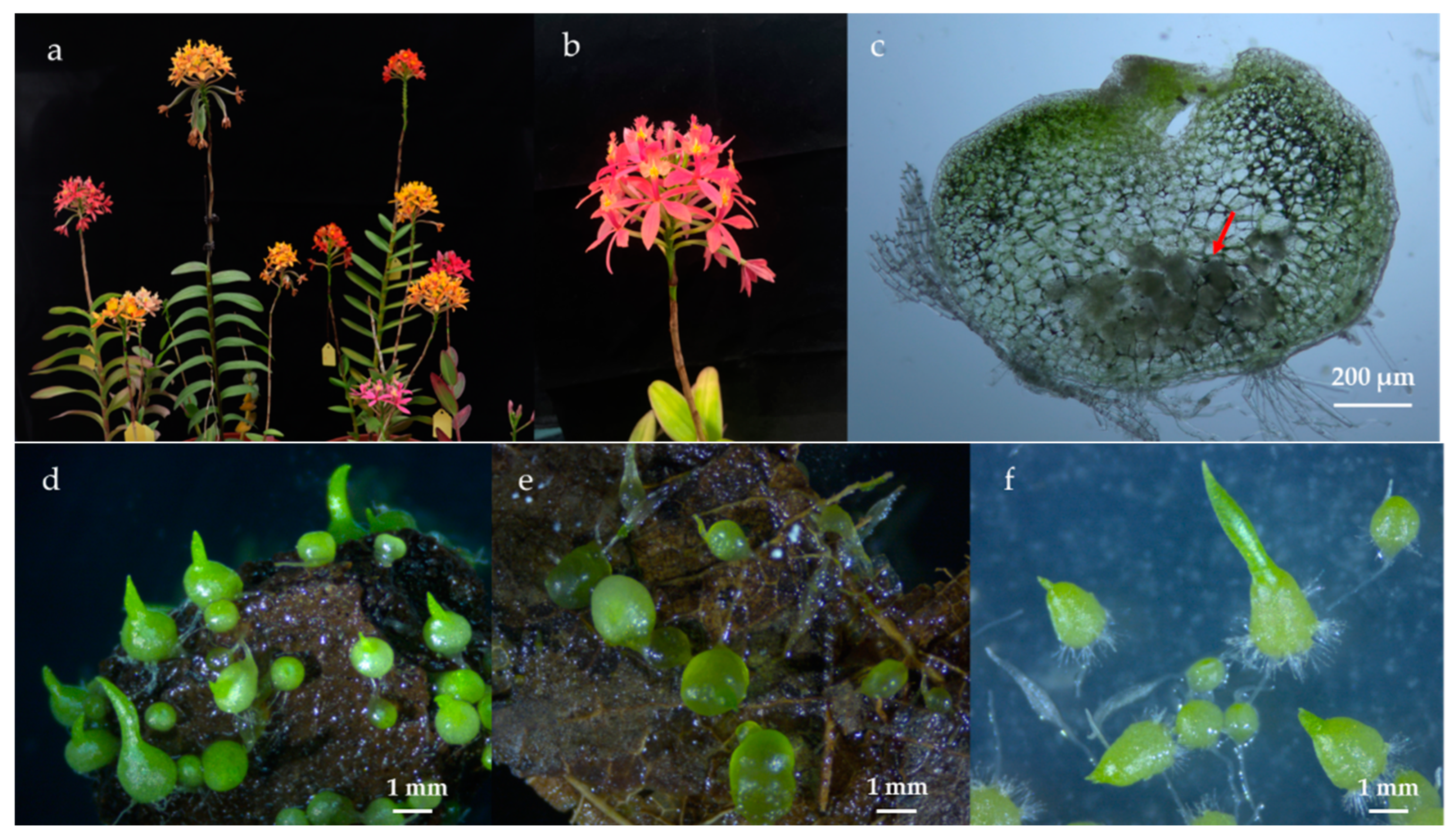
2.1. Study Materials
2.2. Seed Ex Situ Germination and Sampling
2.3. Assessment of Fungal Communities in E. radicans Protocorms
2.4. Fungal Isolation and Identification
2.5. Testing Fungal Promotion of Seed Germination
3. Results
3.1. Efficiency of Ex Situ Germination on Different Substrates
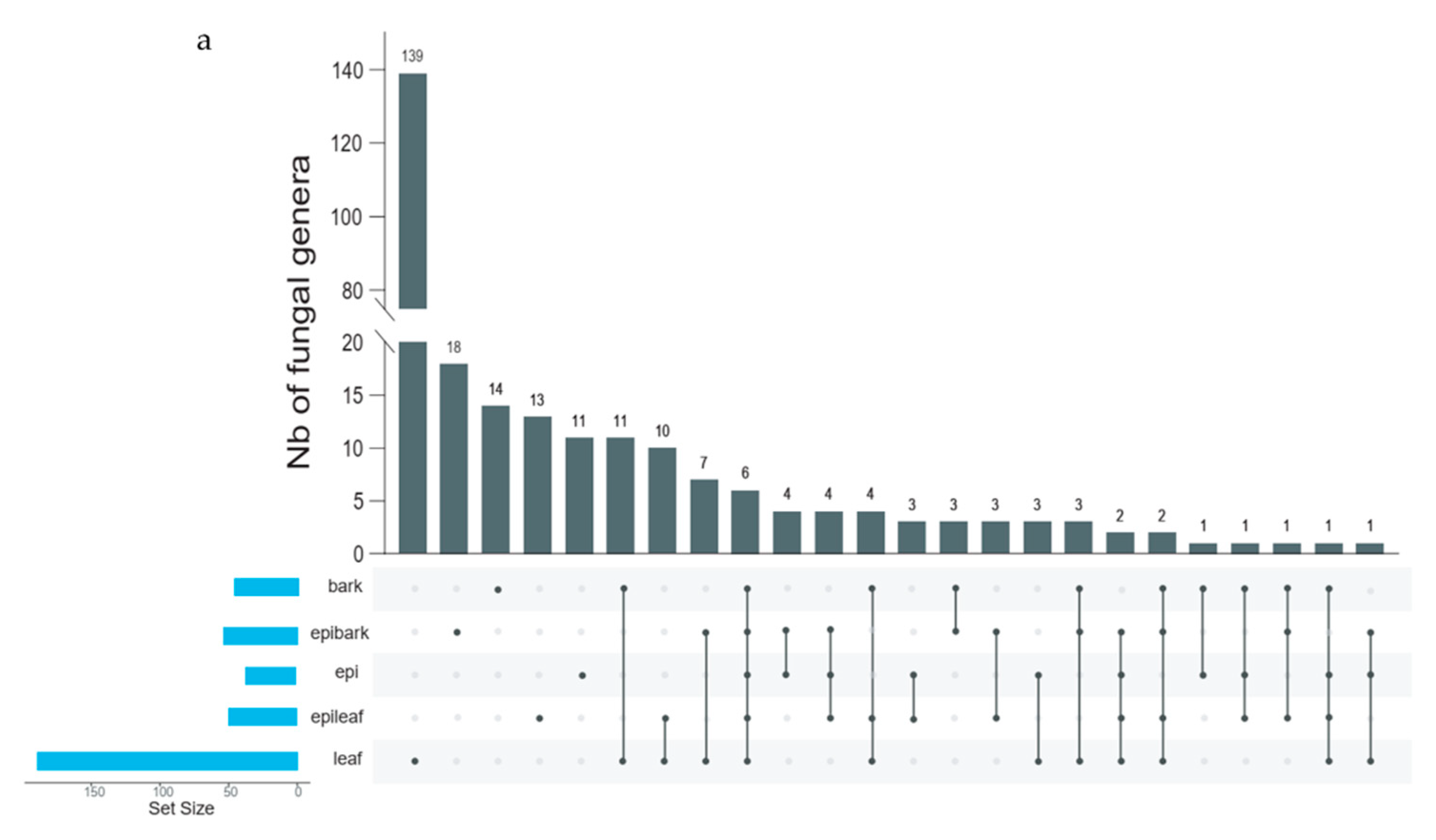
3.2. Fungal Community Composition
3.3. Diversity of Fungal Communities
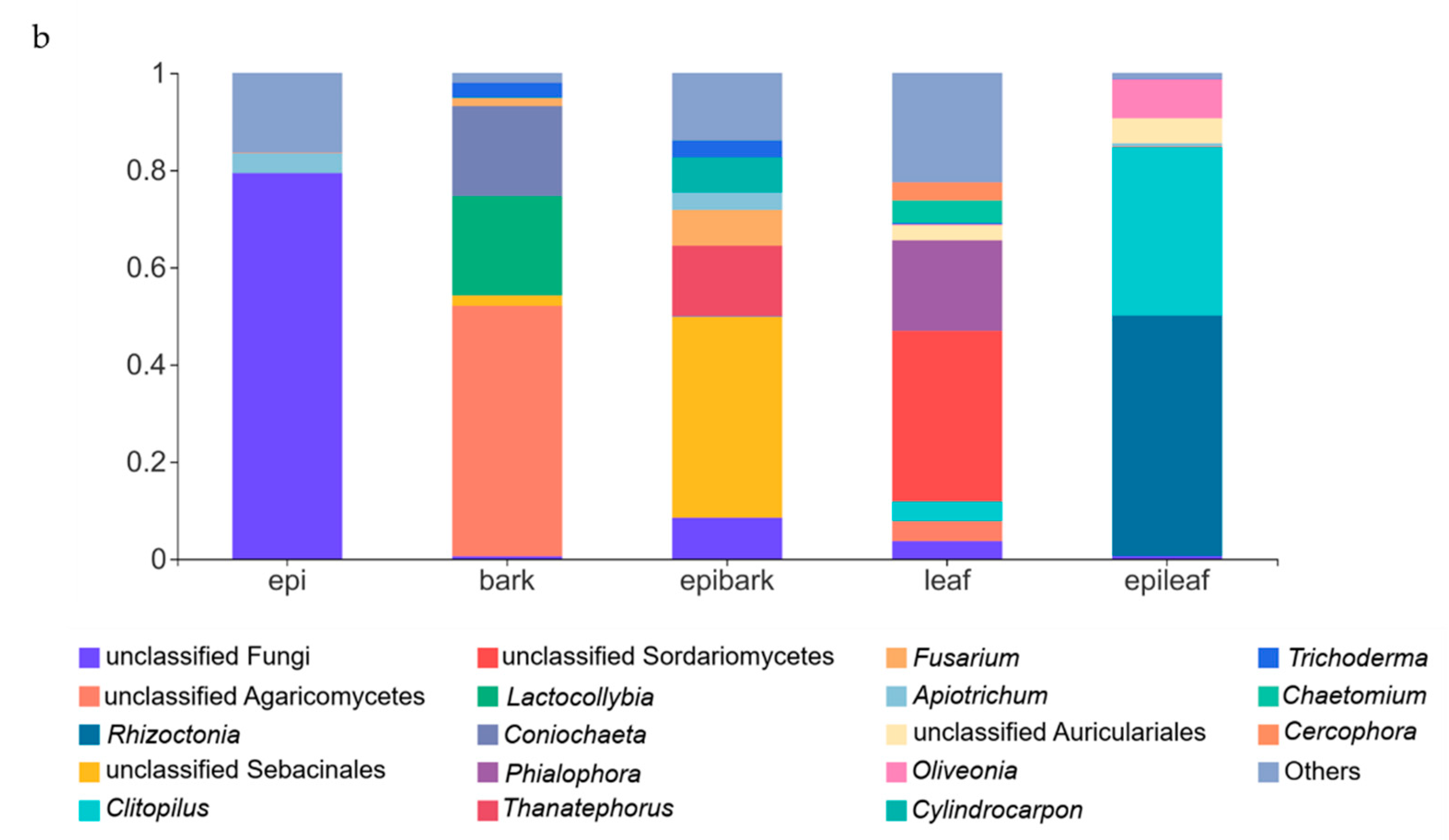
3.4. Taxon Composition of Fungal Communities in Protocorms
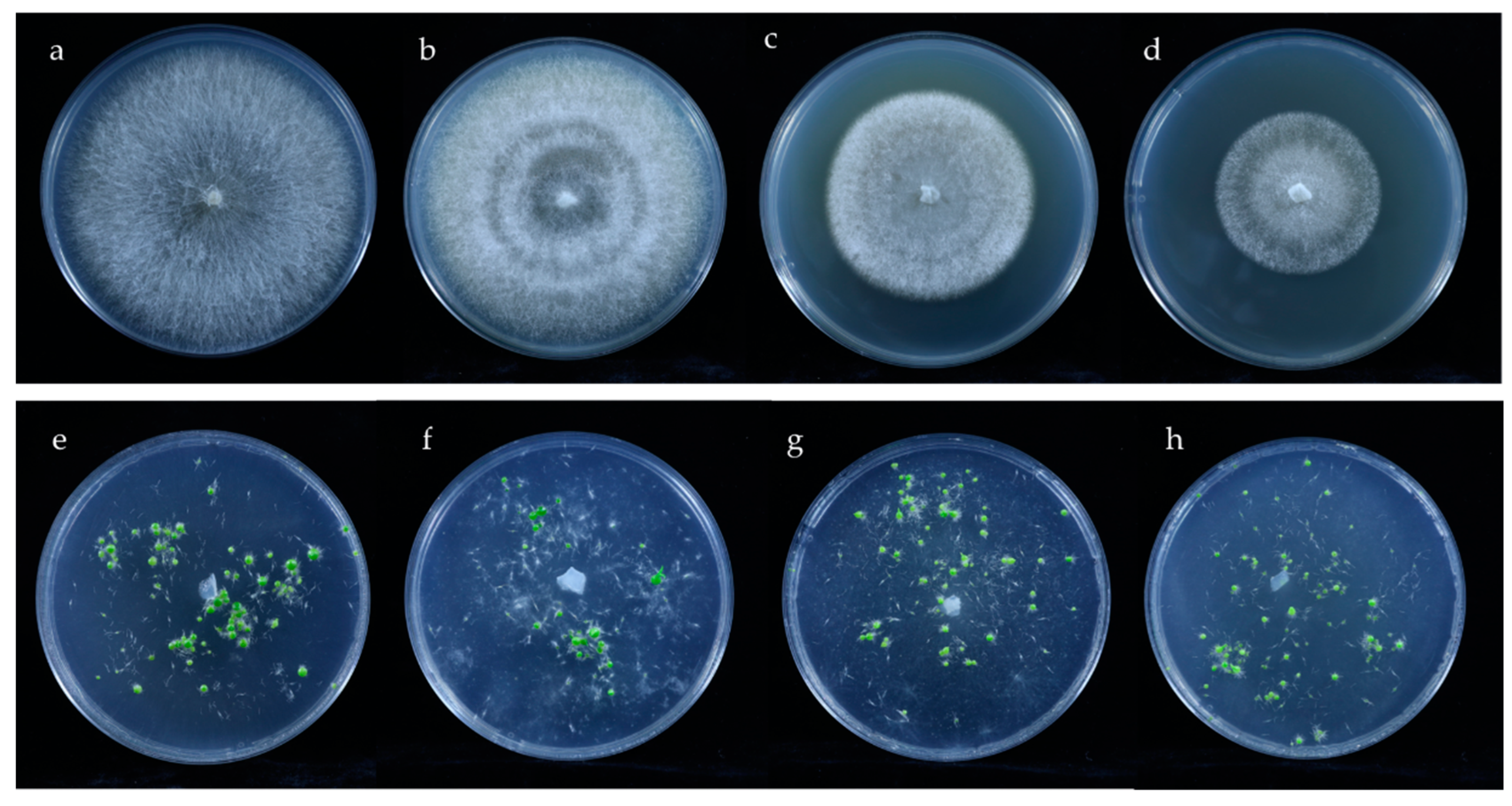
3.5. Identification of Isolated Fungi and In Vitro Symbiotic Germination
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Brundrett, M.C.; Tedersoo, L. Evolutionary history of mycorrhizal symbioses and global host plant diversity. New Phytol. 2018, 220, 1108–1115. [Google Scholar] [CrossRef]
- Arditti, J.; Ghani, A.K.A. Tansley review no. 110. Numerical and physical properties of orchid seeds and their biological implications. New Phytol. 2000, 145, 367–421. [Google Scholar] [CrossRef]
- Rasmussen, H.N.; Whigham, D.F. Seed ecology of dust seeds in situ: A new study technique and its application in terrestrial orchids. Am. J. Bot. 1993, 80, 1374–1378. [Google Scholar] [CrossRef]
- Zi, X.M.; Sheng, C.L.; Goodale, U.M.; Shao, S.C.; Gao, J.Y. In situ seed baiting to isolate germination-enhancing fungi for an epiphytic orchid, Dendrobium aphyllum (Orchidaceae). Mycorrhiza 2014, 24, 487–499. [Google Scholar] [CrossRef]
- Mccormick, M.K.; Taylor, D.L.; Juhaszova, K.; Burnett, R.K.; Whigham, D.F.; O’Neill, J.P. Limitations on orchid recruitment: Not a simple picture. Mol. Ecol. 2012, 21, 1511–1523. [Google Scholar] [CrossRef]
- Rochefort, A.; Simonin, M.; Marais, C.; Guillerm-Erckelboudt, A.-Y.; Barret, M.; Sarniguet, A. Transmission of seed and soil microbiota to seedling. mSystems 2021, 6, e0044621. [Google Scholar] [CrossRef]
- Pecoraro, L.; Rasmussen, H.N.; Gomes, S.I.F.; Wang, X.; Merckx, V.S.F.T.; Cai, L.; Rasmussen, F.N. Fungal diversity driven by bark features affects phorophyte preference in epiphytic orchids from southern China. Sci. Rep. 2021, 11, 11287. [Google Scholar] [CrossRef]
- Zarate-García, A.M.; Noguera-Savelli, E.; Andrade-Canto, S.B.; Zavaleta-Mancera, H.A.; Gauthier, A.; Alatorre-Cobos, F. Bark water storage capacity influences epiphytic orchid preference for host trees. Am. J. Bot. 2020, 107, 726–734. [Google Scholar] [CrossRef]
- Wang, M.; Eyre, A.W.; Thon, M.R.; Oh, Y.; Dean, R.A. Dynamic changes in the microbiome of rice during shoot and root growth derived from seeds. Front. Microbiol. 2020, 11, 559728. [Google Scholar] [CrossRef]
- Abdelfattah, A.; Wisniewski, M.; Schena, L.; Tack, A.J.M. Experimental evidence of microbial inheritance in plants and transmission routes from seed to phyllosphere and root. Environ. Microbiol. 2021, 23, 2199–2214. [Google Scholar] [CrossRef]
- Li, T.Q.; Yang, W.K.; Wu, S.M.; Selosse, M.; Gao, J.Y. Progress and prospects of mycorrhizal fungal diversity in orchids. Front. Plant Sci. 2021, 12, 646325. [Google Scholar]
- Chen, J.H.; Wang, H.; Liu, S.S.; Li, Y.Y.; Guo, S.X. Ultrastructure of symbiotic germination of the orchid Dendrobium officinale with its mycobiont, Sebacina sp. Aust. J. Bot. 2014, 62, 229–234. [Google Scholar]
- Chen, D.Y.; Wang, X.J.; Li, T.Q.; Li, N.Q.; Gao, J.Y. In situ seedling baiting to isolate plant growth-promoting fungi from Dendrobium officinale, an over-collected medicinal orchid in China. Glob. Ecol. Conserv. 2021, 28, e01659. [Google Scholar]
- Huang, H.; Zi, X.M.; Lin, H.; Gao, J.Y. Host-specificity of symbiotic mycorrhizal fungi for enhancing seed germination, protocorm formation and seedling development of over-collected medicinal orchid, Dendrobium devonianum. J. Microbiol. 2018, 56, 42–48. [Google Scholar]
- Meng, Y.Y.; Shao, S.C.; Liu, S.J.; Gao, J.Y. Do the fungi associated with roots of adult plants support seed germination? A case study on Dendrobium exile (Orchidaceae). Glob. Ecol. Conserv. 2019, 17, e00582. [Google Scholar]
- Yang, W.K.; Li, T.Q.; Wu, S.M.; Finnegan, P.M.; Gao, J.Y. Ex situ seed baiting to isolate germination-enhancing fungi for assisted colonization in Paphiopedilum spicerianum, a critically endangered orchid in China. Glob. Ecol. Conserv. 2020, 23, e01147. [Google Scholar]
- Rammitsu, K.; Yagame, T.; Yamashita, Y.; Yukawa, T.; Isshiki, S.; Ogura-Tsujita, Y. A leafless epiphytic orchid, Taeniophyllum glandulosum Blume (Orchidaceae), is specifically associated with the Ceratobasidiaceae family of basidiomycetous fungi. Mycorrhiza 2019, 29, 159–166. [Google Scholar]
- Wang, X.J.; Wu, Y.H.; Ming, X.J.; Wang, G.; Gao, J.Y. Isolating ecological-specific fungi and creating fungus-seed bags for epiphytic orchid conservation. Glob. Ecol. Conserv. 2021, 28, e01714. [Google Scholar]
- Kendon, J.P.; Yokoya, K.; Zettler, L.W.; Jacob, A.S.; McDiarmid, F.; Bidartondo, M.I.; Sarasan, V. Recovery of mycorrhizal fungi from wild collected protocorms of Madagascan endemic orchid Aerangis ellisii (B.S. Williams) Schltr. and their use in seed germination in vitro. Mycorrhiza 2020, 30, 567–576. [Google Scholar]
- Tesitelova, T.; Jersakova, J.; Roy, M.; Kubatova, B.; Tesitel, J.; Urfus, T.; Travnıcek, P.; Suda, J. Ploidy-specific symbiotic interactions: Divergence of mycorrhizal fungi between cytotypes of the Gymnadenia conopsea group (Orchidaceae). New Phytol. 2013, 199, 1022–1033. [Google Scholar]
- Tesitelova, T.; Kotilınek, M.; Jersakova, J.; Joly, F.X.; Kosnar, J.; Tatarenko, I.; Selosse, M.A. Two widespread green Neottia species (Orchidaceae) show mycorrhizal preference for Sebacinales in various habitats and ontogenetic stages. Mol. Ecol. 2015, 24, 1122–1134. [Google Scholar] [PubMed]
- Hardoim, P.R.; van Overbeek, L.S.; Berg, G.; Pirttilä, A.M.; Compant, S.; Campisano, A.; Döring, M.; Sessitsch, A. The hidden world within plants: Ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol. Mol. Biol. Rev. 2015, 79, 293–320. [Google Scholar] [PubMed]
- Jia, M.; Chen, L.; Xin, H.L.; Zheng, C.J.; Rahman, K.; Han, T.; Qin, L.P. A friendly relationship between endophytic fungi and medicinal plants: A systematic review. Front. Microbiol. 2016, 7, 906. [Google Scholar] [PubMed]
- Brader, G.; Compant, S.; Vescio, K.; Mitter, B.; Trognitz, F.; Ma, L.J.; Sessitsch, A. Ecology and genomic insights into plant-pathogenic and plant-nonpathogenic endophytes. Annu. Rev. Phytopathol. 2017, 55, 61–83. [Google Scholar] [PubMed]
- Voyron, S.; Ercole, E.; Ghignone, S.; Perotto, S.; Girlanda, M. Fine-scale spatial distribution of orchid mycorrhizal fungi in the soil of host-rich grasslands. New Phytol. 2017, 213, 1428–1439. [Google Scholar] [PubMed]
- Chen, L.; Wang, Y.C.; Qin, L.Y.; He, H.Y.; Yu, X.L.; Yang, M.Z.; Zhang, H.B. Dynamics of fungal communities during Gastrodia elata growth. BMC Microbiol. 2019, 19, 158. [Google Scholar]
- Pateli, P.; Papafotiou, M.; Chronopoulos, J. Influence of in vitro culture medium on Epidendrum radicans seed germination, seeding growth and ex vitro establishment. Acta Hortic. 2003, 616, 189–192. [Google Scholar]
- Riofrio, M.L.; Cruz, D.; Torres, E.; Cruz, M.D.L.; Iriondo, J.M.; Suarez, J.P. Mycorrhizal preferences and fine spatial structure of the epiphytic orchid Epidendrum rhopalostele. Am. J. Bot. 2013, 100, 2339–2348. [Google Scholar]
- Pereira, M.C.; Coelho, I.S.; Valadares, R.B.S.; Oliveira, S.F.; Bocayuva, M.; Pereira, O.L.; Araújo, E.F.; Kasuya, M.C.M. Morphological and molecular characterization of Tulasnella spp. fungi isolated from the roots of Epidendrum secundum, a widespread Brazilian orchid. Symbiosis 2014, 62, 111–121. [Google Scholar]
- Pereira, O.L.; Kasuya, M.C.M.; Borges, A.C.; Araújo, E.F. Morphological and molecular characterization of mycorrhizal fungi isolated from neotropical orchids in Brazil. Botany 2005, 83, 54–65. [Google Scholar]
- Pereira, O.L.; Rollemberg, C.L.; Borges, A.C.; Matsuoka, K.; Kasuya, M.C.M.; Matsuoka, K. Epulorhiza epiphytica sp. nov. isolated from mycorrhizal roots of epiphytic orchids in Brazil. Mycoscience 2003, 44, 153–155. [Google Scholar]
- Durán-López, M.E.; Caroca-Cáceres, R.; Jahreis, K.; Narváez-Vera, M.; Cazar, M.E. The mycorrhizal fungi Ceratobasidium sp. and Sebacina vermifera promote seed germination and seedling development of the terrestrial orchid Epidendrum secundum Jacq. South Afr. J. Bot. 2019, 125, 54–61. [Google Scholar]
- Brundrett, M.C.; Scade, A.; Batty, A.L.; Dixon, K.W.; Sivasithamparam, K. Development of in situ and ex situ seed baiting techniques to detect mycorrhizal fungi from terrestrial orchid habitats. Mycol. Res. 2003, 107, 1210–1220. [Google Scholar]
- Tian, B.L.; Pei, Y.C.; Huang, W.; Ding, J.Q.; Siemann, E. Increasing flavonoid concentrations in root exudates enhance associations between arbuscular mycorrhizal fungi and an invasive plant. ISME J. 2021, 15, 1919–1930. [Google Scholar] [PubMed]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar]
- Magoč, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods. 2013, 10, 996–998. [Google Scholar]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar]
- Nilsson, R.H.; Larsson, K.H.; Taylor, A.F.S.; Bengtsson-Palme, J.; Jeppesen, T.S.; Schigel, D.; Kennedy, P.; Picard, K.; Glöckner, F.O.; Tedersoo, L.; et al. The UNITE database for molecular identification of fungi: Handling dark taxa and parallel taxonomic classifications. Nucleic. Acids Res. 2019, 47, 259–264. [Google Scholar]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package; R Package Version 2.5-6. 2019. Available online: https://CRAN.R-project.org/package=vegan (accessed on 5 July 2022).
- Chen, T.; Liu, Y.X.; Huang, L.Q. ImageGP: An easy-to-use data visualization web server for scientific researchers. iMeta 2022, 1, e5. [Google Scholar]
- Wei, J.C. Identification Manual for Fungi; Shanghai Scientific & Technical Publishers: Shanghai, China, 1979. [Google Scholar]
- White, T.J.; Bruns, T.D.; Lee, S.; Taylor, J.W. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar]
- Ji, X.; Wu, D.M.; Song, C.G.; Liu, S.; Si, J.; Cui, B.K. Two new Neofomitella species (Polyporaceae, Basidiomycota) based on morphological and molecular evidence. Mycol. Prog. 2019, 18, 593–602. [Google Scholar] [CrossRef]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bioassay with tobacco tissue culture. Physiol. Plantarum. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Arditti, J. Factors affecting the germination of orchid seeds. Bot. Rev. 1967, 33, 1–97. [Google Scholar]
- Meng, Y.Y.; Zhang, W.L.; Selosse, M.A.; Gao, J.Y. Are fungi from adult orchid roots the best symbionts at germination? A case study. Mycorrhiza 2019, 29, 541–547. [Google Scholar]
- Oja, J.; Kohout, P.; Tedersoo, L.; Kull, T.; Koljalg, U. Temporal patterns of orchid mycorrhizal fungi in meadows and forests as revealed by 454 pyrosequencing. New Phytol. 2015, 205, 1608–1618. [Google Scholar] [PubMed]
- Waud, M.; Brys, R.; Landuyt, W.V.; Lievens, B.; Jacquemyn, H. Mycorrhizal specificity does not limit the distribution of an endangered orchid species. Mol. Ecol. 2017, 26, 1687–1701. [Google Scholar] [PubMed]
- Tobias, T.B.; Farrer, E.C.; Rosales, A.; Sinsabaugh, R.L.; Suding, K.N.; Porras-Alfaro, A. Seed-associated fungi in the alpine tundra: Both mutualists and pathogens could impact plant recruitment. Fungal Ecol. 2017, 30, 10–18. [Google Scholar] [CrossRef]
- Qin, Y.; Pan, X.; Yuan, Z. Seed endophytic microbiota in a coastal plant and phytobeneficial properties of the fungus Cladosporium cladosporioides. Fungal Ecol. 2016, 24, 53–60. [Google Scholar]
- Lu, M. Endophytes Isolated from Dendrobium Seeds and Their Effects on Growth and High Temperature Resistance of Dendrobium Seedlings. Master Dissertation, Southwest University, Chongqing, China, 2018. [Google Scholar]
- Miller, S.L.; Buyck, B. Molecular phylogeny of the genus Russula in Europe with a comparison of modern infrageneric classifications. Mycol. Res. 2002, 106, 259–276. [Google Scholar]
- Gardes, M.; Bruns, T.D. Community structure of ectomycorrhizal fungi in a Pinus muricata forest: Above- and below-ground views. Can. J. Bot. 1996, 74, 1572–1583. [Google Scholar] [CrossRef]
- Pecoraro, L.; Wang, X.; Venturella, G.; Gao, W.; Gupta, V.K. Molecular evidence supports simultaneous association of the achlorophyllous orchid Chamaegastrodia inverta with ectomycorrhizal Ceratobasidiaceae and Russulaceae. BMC Microbiol. 2020, 20, 236. [Google Scholar] [CrossRef] [PubMed]
- Suetsugu, K.; Yamato, M.; Matsubayashi, J.; Tayasu, I. Comparative study of nutritional mode and mycorrhizal fungi in green and albino variants of Goodyera velutina, an orchid mainly utilizing saprotrophic rhizoctonia. Mol. Ecol. 2019, 28, 4290–4299. [Google Scholar] [CrossRef] [PubMed]
- Jacquemyn, H.; Waud, M.; Brys, R.; Lallemand, F.; Courty, P.-E.; Robionek, A.; Selosse, M.-A. Mycorrhizal associations and trophic modes incoexisting orchids: An ecological continuum between auto and mixotrophy. Front. Plant Sci. 2017, 8, 1497. [Google Scholar] [CrossRef] [PubMed]
- Kaur, J.; Andrews, L.; Sharma, J. High specificity of a rare terrestrial orchid toward a rare fungus within the north American tallgrass prairie. Fungal Biol. 2019, 123, 895–904. [Google Scholar] [CrossRef]
- Han, J.Y.; Xiao, H.F.; Gao, J.Y. Seasonal dynamics of mycorrhizal fungi in Paphiopedilum spicerianum (Rchb f) Pfitzer—A critically endangered orchid from China. Glob. Ecol. Conserv. 2016, 6, 327–338. [Google Scholar] [CrossRef]
- Waud, M.; Busschaert, P.; Lievens, B.; Jacquemyn, H. Specificity and localised distribution of mycorrhizal fungi in the soil may contribute to co-existence of orchid species. Fungal Ecol. 2016, 20, 55–165. [Google Scholar] [CrossRef]
- Southworth, D. Oaks and mycorrhizal fungi. In Oak: Ecology, Types and Management; Environmental Science, Engineering and Technology Series; Chuteira, C.A., Grao, A.B., Eds.; Nova Science Publishers Inc.: New York, NY, USA, 2013; pp. 207–218. [Google Scholar]
- Howe, V.K. Mycorrhizal Fungi of White Oak. Ph.D. Dissertation, Iowa State University of Science and Technology, Ames, IA, USA, 1964. [Google Scholar]
- Keizer, P.J.; Arnolds, E. Succession of ectomycorrhizal fungi in roadside verges planted with common oak (Quercus robur L.) in Drenthe, The Netherlands. Mycorrhiza 1994, 4, 147–159. [Google Scholar] [CrossRef]
- Peng, L.; Shan, X.L.; Yang, Y.Z.; Wang, Y.C.; Druzhinina, I.S.; Pan, X.Y.; Jin, W.; He, X.H.; Wang, X.Y.; Zhang, X.G.; et al. Facultative symbiosis with a saprotrophic soil fungus promotes potassium uptake in American sweetgum trees. Plant Cell Environ. 2021, 44, 2793–2809. [Google Scholar] [CrossRef]
- Jiang, J.H.; Lee, Y.I.; Cubeta, M.A.; Chen, L.C. Characterization and colonization of endomycorrhizal Rhizoctonia fungi in the medicinal herb Anoectochilus formosanus (Orchidaceae). Mycorrhiza 2015, 25, 431–445. [Google Scholar] [CrossRef][Green Version]
- Yang, Q.; Xu, L.; Xia, W.; Liang, L.; Bai, X.; Li, L.; Xu, L.; Liu, L. Mycorrhizal compatibility and germination-Promoting activity of Tulasnella species in two species of orchid (Cymbidium mannii and Epidendrum radicans). Horticulturae 2021, 7, 472. [Google Scholar] [CrossRef]
- Ma, J.S.; Yue, H.M.; Li, H.Q.; Zhang, J.; Zhang, Y.H.; Wang, X.L.; Gong, S.; Liu, G.Q. Selective delignification of poplar wood with a newly isolated white-rot basidiomycete Peniophora incarnata T-7 by submerged fermentation to enhance saccharification. Biotechnol. Biofuels. 2021, 14, 135. [Google Scholar] [CrossRef] [PubMed]
- Brglez, A.; Pikur, B.; Ogris, N. In vitro interactions between Eutypella parasitica and some frequently isolated fungi from the wood of the dead branches of young sycamore maple (Acer pseudoplatanus). Forests 2020, 11, 1072. [Google Scholar] [CrossRef]


| Isolate Number | Substrate | Closest Match in GenBank (Accession Number) | Taxonomic Affiliation | Accession Number | Identity | Corresponding OTU |
|---|---|---|---|---|---|---|
| Epi124 | Pine bark | KM211337 | Uncultured Tulasnellaceae | ON629737 | 99.65% | Not found |
| Epi221 | Pine bark | MK842111 | Peniophora incarnata | ON629738 | 99.83% | Not found |
| Epi1217 | Pine bark | MN959990 | Fusarium oxysporum | ON629747 | 100.00% | OTU728 |
| Epi1417 | Pine bark | MK870486 | Trichoderma spp. | ON629749 | 99.66% | OTU602 |
| Epi128 | Oak leaf | KR259886 | Ceratobasidium spp. AG-A | ON629750 | 98.78% | OTU762 |
| Epi317 | Oak leaf | LC480736 | Uncultured Tulasnellaceae | ON629753 | 99.83% | Not found |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yao, N.; Wang, T.; Cao, X. Epidendrumradicans Fungal Community during Ex Situ Germination and Isolation of Germination-Enhancing Fungi. Microorganisms 2022, 10, 1841. https://doi.org/10.3390/microorganisms10091841
Yao N, Wang T, Cao X. Epidendrumradicans Fungal Community during Ex Situ Germination and Isolation of Germination-Enhancing Fungi. Microorganisms. 2022; 10(9):1841. https://doi.org/10.3390/microorganisms10091841
Chicago/Turabian StyleYao, Na, Tao Wang, and Xiaolu Cao. 2022. "Epidendrumradicans Fungal Community during Ex Situ Germination and Isolation of Germination-Enhancing Fungi" Microorganisms 10, no. 9: 1841. https://doi.org/10.3390/microorganisms10091841
APA StyleYao, N., Wang, T., & Cao, X. (2022). Epidendrumradicans Fungal Community during Ex Situ Germination and Isolation of Germination-Enhancing Fungi. Microorganisms, 10(9), 1841. https://doi.org/10.3390/microorganisms10091841







