Evidence, Challenges, and Knowledge Gaps Regarding Latent Tuberculosis in Animals
Abstract
:1. Introduction
2. The Epidemiology of TB at the Domestic–Wildlife–Human Interfaces
3. Response of Different Hosts to MTBC Infection?
3.1. Human TB Infection Stages
3.2. Domestic and Wildlife Infection Stages of TB
3.3. Diagnostic Challenges in Differential Stages of TB
4. The Influence of MTBC Characteristics on TB Stages
5. Candidate Models to Improve Understanding of TB Stages in Animals
5.1. In Vitro Models
5.2. In Vivo Models
6. Candidate Host and Pathogen Biomarkers to Improve Understanding of TB Stages
6.1. Candidate Host Markers
6.2. Candidate Pathogen Biomarkers
7. How Can Researchers Bridge Existing LTB Knowledge Gaps in Animals?
7.1. Factors Contributing to Existing Knowledge Gaps
- A paucity of studies describing and comparing the pathogenesis of M. bovis infection and other MTBC species in different animal species.
- Limited availability of sensitive and specific diagnostic tools for detection and differentiation of MTBC infection stages in domestic animals and wildlife, especially antemortem tests.
- Incomplete information on the diversity of MTBC virulent clinical strains, primarily M. bovis, and their influence on pathogenesis.
- A limited understanding of the role and variability in immune responses to mycobacterial infection in different animal species.
- A poor understanding of the genetic, metabolic, and physiological characteristics of M. bovis could promote persister bacilli formation.
- A lack of clarity on how TB stages vary in different hosts (humans, domestic and wild animals).
- A lack of well-characterised in vitro and in vivo models of M. bovis infection to simulate different stages of infection, including LTB.
7.2. Recommendations for Future Research
- Developing a consensus on the definition of latency in domestic and wild animals and identifying a model that could be used to find biomarkers for this state.
- Identification of blood-based host and pathogen biomarkers that can differentiate between ATB and different stages of M. bovis infection in different animal species.
- Utilising available tools to study the phenotypic state of persister bacilli at a single-cell level to understand the physiological, phenotypic, and molecular features of different strains.
- Comparing the pathogenesis of M. bovis and other MTBC in different animal species to characterise the chronic asymptomatic state in infected hosts.
- Exploring host–pathogen similarities and differences of host–pathogen interactions to elucidate factors leading to LTB and susceptibility of different species to latent infections.
8. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- WHO. Global Tuberculosis Report. 2021. Available online: https://www.who.int/publications/i/item/9789240037021 (accessed on 9 May 2022).
- Tadesse, T.; Lelo, U.; Markos, T.; Tadesse, T.; Birhan, F.; Tona, T. Review on the Epidemiology, Public Health and Economic Importance of Bovine Tuberculosis. J. Biol. Agric. Healthc. 2017, 7, 16–23. [Google Scholar]
- Lesslie, I.W.; Magnus, K.; Stewart, C.J. The Prevalence of Bovine Type Tuberculous Infection in Man in the English Rural Population. Tubercle 1972, 53, 198–204. [Google Scholar] [CrossRef]
- Torres-Gonzalez, P.; Soberanis-Ramos, O.; Martinez-Gamboa, A.; Chavez-Mazari, B.; Barrios-Herrera, M.T.; Torres-Rojas, M.; Cruz-Hervert, L.P.; Garcia-Garcia, L.; Singh, M.; Gonzalez-Aguirre, A.; et al. Prevalence of Latent and Active Tuberculosis among Dairy Farm Workers Exposed to Cattle Infected by Mycobacterium bovis. PLoS Negl. Trop. Dis. 2013, 7, e2177. [Google Scholar] [CrossRef]
- Kanabalan, R.D.; Lee, L.J.; Lee, T.Y.; Chong, P.P.; Hassan, L.; Ismail, R.; Chin, V.K. Human Tuberculosis and Mycobacterium tuberculosis Complex: A Review on Genetic Diversity, Pathogenesis and Omics Approaches in Host Biomarkers Discovery. Microbiol. Res. 2021, 246, 126674. [Google Scholar] [CrossRef]
- Miller, M.A.; Lyashchenko, K.P. 15 Mycobacterial Infections in Other Zoo Animals. In Tuberculosis, Leprosy and Mycobacterial Diseases of Man and Animals: The Many Hosts of Mycobacteria; CABI: London, UK, 2015. [Google Scholar] [CrossRef]
- Zanardi, G.; Boniotti, M.B.; Gaffuri, A.; Casto, B.; Zanoni, M.; Pacciarini, M.L. Tuberculosis Transmission by Mycobacterium bovis in a Mixed Cattle and Goat Herd. Res. Vet. Sci. 2013, 95, 430–433. [Google Scholar] [CrossRef]
- Pesciaroli, M.; Alvarez, J.; Boniotti, M.B.; Cagiola, M.; Di Marco, V.; Marianelli, C.; Pacciarini, M.; Pasquali, P. Tuberculosis in Domestic Animal Species. Res. Vet. Sci. 2014, 97, S78–S85. [Google Scholar] [CrossRef] [PubMed]
- Muñoz Mendoza, M.; de Juan, L.; Menéndez, S.; Ocampo, A.; Mourelo, J.; Sáez, J.L.; Domínguez, L.; Gortázar, C.; García Marín, J.F.; Balseiro, A. Tuberculosis Due to Mycobacterium bovis and Mycobacterium caprae in Sheep. Vet. J. 2012, 191, 267–269. [Google Scholar] [CrossRef]
- Hang’ombe, M.B.; Munyeme, M.; Nakajima, C.; Fukushima, Y.; Suzuki, H.; Matandiko, W.; Ishii, A.; Mweene, A.S.; Suzuki, Y. Mycobacterium bovis Infection at the Interface between Domestic and Wild Animals in Zambia. BMC Vet. Res. 2012, 8, 221. [Google Scholar] [CrossRef]
- Olivier, T.T.; Viljoen, I.M.; Hofmeyr, J.; Hausler, G.A.; Goosen, W.J.; Tordiffe, A.S.W.; Buss, P.; Loxton, A.G.; Warren, R.M.; Miller, M.A.; et al. Development of a Gene Expression Assay for the Diagnosis of Mycobacterium bovis Infection in African Lions (Panthera leo). Transbound. Emerg. Dis. 2017, 64, 774–781. [Google Scholar] [CrossRef]
- Bernitz, N.; Kerr, T.J.; Goosen, W.J.; Chileshe, J.; Higgitt, R.L.; Roos, E.O.; Meiring, C.; Gumbo, R.; de Waal, C.; Clarke, C.; et al. Review of Diagnostic Tests for Detection of Mycobacterium bovis Infection in South African Wildlife. Front. Vet. Sci. 2021, 8, 588697. [Google Scholar] [CrossRef]
- Malone, K.M.; Gordon, S.V. Mycobacterium tuberculosis Complex Members Adapted to Wild and Domestic Animals. Adv. Exp. Med. Biol. 2017, 1019, 135–154. [Google Scholar] [CrossRef] [PubMed]
- Mostowy, S.; Cousins, D.; Behr, M.A. Genomic Interrogation of the Dassie Bacillus Reveals It as a Unique RD1 Mutant within the Mycobacterium tuberculosis Complex. J. Bacteriol. 2004, 186, 104–109. [Google Scholar] [CrossRef] [PubMed]
- Brites, D.; Loiseau, C.; Menardo, F.; Borrell, S.; Boniotti, M.B.; Warren, R.; Dippenaar, A.; Parsons, S.D.C.; Beisel, C.; Behr, M.A.; et al. A New Phylogenetic Framework for the Animal-Adapted Mycobacterium tuberculosis Complex. Front. Microbiol. 2018, 9, 2820. [Google Scholar] [CrossRef] [PubMed]
- Coscolla, M.; Lewin, A.; Metzger, S.; Maetz-Rennsing, K.; Calvignac-Spencer, S.; Nitsche, A.; Dabrowski, P.W.; Radonic, A.; Niemann, S.; Parkhill, J.; et al. Novel Mycobacterium tuberculosis Complex Isolate from a Wild Chimpanzee. Emerg. Infect. Dis. 2013, 19, 969. [Google Scholar] [CrossRef]
- Gummow, B. A Survey of Zoonotic Diseases Contracted by South African Veterinarians. J. S. Afr. Vet. Assoc. 2003, 74, 72–76. [Google Scholar] [CrossRef]
- Sichewo, P.R.; Vander Kelen, C.; Thys, S.; Michel, A.L. Risk Practices for Bovine Tuberculosis Transmission to Cattle and Livestock Farming Communities Living at Wildlife-Livestock-Human Interface in Northern Kwazulu Natal, South Africa. PLoS Negl. Trop. Dis. 2020, 14, 7681. [Google Scholar] [CrossRef]
- Moyo, M.; Lebina, L.; Milovanovic, M.; MacPherson, P.; Michel, A.; Martinson, N. Tuberculosis Patients at the Human-Animal Interface: Potential Zooanthroponotic and Zoonotic Transmission. One Health 2021, 13, 100319. [Google Scholar] [CrossRef]
- Behr, M.A.; Edelstein, P.H.; Ramakrishnan, L. Revisiting the Timetable of Tuberculosis. BMJ 2018, 362, k2738. [Google Scholar] [CrossRef]
- Gormley, E.; Corner, L.A.L. Pathogenesis of Mycobacterium bovis Infection: The Badger Model as a Paradigm for Understanding Tuberculosis in Animals. Front. Vet. Sci. 2018, 4, 247. [Google Scholar] [CrossRef]
- Michel, A.L.; Lane, E.P.; de Klerk-Lorist, L.M.; Hofmeyr, M.; van der Heijden, E.M.D.L.; Botha, L.; van Helden, P.; Miller, M.; Buss, P. Experimental Mycobacterium bovis Infection in Three White Rhinoceroses (Ceratotherium simum): Susceptibility, Clinical and Anatomical Pathology. PLoS ONE 2017, 12, e0179943. [Google Scholar] [CrossRef]
- Palmer, M.V.; Kanipe, C.; Boggiatto, P.M. The Bovine Tuberculoid Granuloma. Pathogens 2022, 11, 61. [Google Scholar] [CrossRef]
- Álvarez, A.H.; Estrada-Chávez, C.; Flores-Valdez, M.A. Molecular Findings and Approaches Spotlighting Mycobacterium bovis Persistence in Cattle. Vet. Res. 2009, 40, 22. [Google Scholar] [CrossRef] [PubMed]
- Pollock, J.M.; Neill, S.D. Mycobacterium bovis Infection and Tuberculosis in Cattle. Vet. J. 2002, 163, 115–127. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, A.H.; Gutiérrez-Ortega, A.; Gómez-Entzin, V.; Pérez-Mayorga, G.; Naranjo-Bastién, J.; González-Martínez, V.; Milián-Suazo, F.; Martínez-Velázquez, M.; Herrera-Rodríguez, S.; Hinojoza-Loza, E. Assessment of Antigenic Supplementation of Bovine Purified Protein Derivative for Diagnosis of Subclinical Infection with Mycobacterium bovis in Cattle. Microb. Pathog. 2017, 108, 114–121. [Google Scholar] [CrossRef] [PubMed]
- García, J.S.Y.; Bigi, M.M.; Klepp, L.I.; García, E.A.; Blanco, F.C.; Bigi, F. Does Mycobacterium bovis Persist in Cattle in a Non-Replicative Latent State as Mycobacterium tuberculosis in Human Beings? Vet. Microbiol. 2020, 247, 108758. [Google Scholar] [CrossRef] [PubMed]
- Mukundan, H.; Chambers, M.; Waters, R.; Larsen, M. Tuberculosis, Leprosy and Mycobacterial Diseases of Man and Animals: The Many Hosts of Mycobacteria; CABI: London, UK, 2015. [Google Scholar] [CrossRef]
- Brosch, R.; Gordon, S.V.; Marmiesse, M.; Brodin, P.; Buchrieser, C.; Eiglmeier, K.; Garnier, T.; Gutierrez, C.; Hewinson, G.; Kremer, K.; et al. A New Evolutionary Scenario for the Mycobacterium tuberculosis Complex. Proc. Natl. Acad. Sci. USA 2002, 99, 3684–3689. [Google Scholar] [CrossRef]
- Niroula, N.; Lim, Z.L.; Walker, S.; Huang, Y.; Gerdts, V.; Zriba, S.; Drever, K.; Chen, J.M. Domestic Pigs Experimentally Infected with Mycobacterium bovis and Mycobacterium tuberculosis Exhibit Different Disease Outcomes. Tuberculosis 2022, 133, 102167. [Google Scholar] [CrossRef]
- Villarreal-Ramos, B.; Berg, S.; Whelan, A.; Holbert, S.; Carreras, F.; Salguero, F.J.; Khatri, B.L.; Malone, K.; Rue-Albrecht, K.; Shaughnessy, R.; et al. Experimental Infection of Cattle with Mycobacterium tuberculosis Isolates Shows the Attenuation of the Human Tubercle Bacillus for Cattle. Sci. Rep. 2018, 8, 894. [Google Scholar] [CrossRef]
- Barry, C.E.; Boshoff, H.; Dartois, V.; Dick, T.; Ehrt, S.; Flynn, J.; Schnappinger, D.; Wilkinson, R.J.; Young, D. The Spectrum of Latent Tuberculosis: Rethinking the Goals of Prophylaxis. Nat. Rev. Microbiol. 2009, 7, 845–855. [Google Scholar] [CrossRef]
- Jenkins, H.E.; Woodroffe, R.; Donnelly, C.A.; Cox, D.R.; Johnston, W.T.; Bourne, F.J.; Cheeseman, C.L.; Clifton-Hadley, R.S.; Gettinby, G.; Gilks, P.; et al. Effects of Culling on Spatial Associations of Mycobacterium bovis Infections in Badgers and Cattle. J. Appl. Ecol. 2007, 44, 897–908. [Google Scholar] [CrossRef]
- Van Tonder, A.J.; Thornton, M.J.; Conlan, A.J.K.; Jolley, K.A.; Goolding, L.; Mitchell, A.P.; Dale, J.; Palkopoulou, E.; Hogarth, P.J.; Hewinson, R.G.; et al. Inferring Mycobacterium bovis Transmission between Cattle and Badgers Using Isolates from the Randomised Badger Culling Trial. PLoS Pathog. 2021, 17, e1010075. [Google Scholar] [CrossRef] [PubMed]
- Gormley, E.; Corner, L.A.L. Wild Animal Tuberculosis: Stakeholder Value Systems and Management of Disease. Front. Vet. Sci. 2018, 5, 327. [Google Scholar] [CrossRef] [PubMed]
- Smith, R.L.; Tauer, L.W.; Schukken, Y.H.; Lu, Z.; Grohn, Y.T. Minimization of Bovine Tuberculosis Control Costs in US Dairy Herds. Prev. Vet. Med. 2013, 112, 266–275. [Google Scholar] [CrossRef]
- Karolemeas, K.; de la Rua-Domenech, R.; Cooper, R.; Goodchild, A.V.; Clifton-Hadley, R.S.; Conlan, A.J.K.; Mitchell, A.P.; Hewinson, R.G.; Donnelly, C.A.; Wood, J.L.N.; et al. Estimation of the Relative Sensitivity of the Comparative Tuberculin Skin Test in Tuberculous Cattle Herds Subjected to Depopulation. PLoS ONE 2012, 7, e43217. [Google Scholar] [CrossRef]
- Smith, R.L.; YH, S.; Lu, Z.; RM, M.; YT, G. Development of a Model to Simulate Infection Dynamics of Mycobacterium bovis in Cattle Herds in the United States. J. Am. Vet. Med. Assoc. 2013, 243, 411–423. [Google Scholar] [CrossRef]
- Miller, M.A.; Buss, P.; Parsons, S.D.C.; Roos, E.; Chileshe, J.; Goosen, W.J.; van Schalkwyk, L.; de Klerk-Lorist, L.M.; Hofmeyr, M.; Hausler, G.; et al. Conservation of White Rhinoceroses Threatened by Bovine Tuberculosis, South Africa, 2016–2017. Emerg. Infect. Dis. 2018, 24, 2373–2375. [Google Scholar] [CrossRef] [PubMed]
- Michel, A.L.; Bengis, R.G.; Keet, D.F.; Hofmeyr, M.; De Klerk, L.M.; Cross, P.C.; Jolles, A.E.; Cooper, D.; Whyte, I.J.; Buss, P.; et al. Wildlife Tuberculosis in South African Conservation Areas: Implications and Challenges. Vet. Microbiol. 2006, 112, 91–100. [Google Scholar] [CrossRef]
- Gompo, T.R.; Shrestha, A.; Ranjit, E.; Gautam, B.; Ale, K.; Shrestha, S.; Bhatta, D.D. Risk Factors of Tuberculosis in Human and Its Association with Cattle TB in Nepal: A One Health Approach. One Health 2020, 10, 100156. [Google Scholar] [CrossRef]
- Silva, M.R.; Rocha, A.D.S.; Araújo, F.R.; Fonseca-Júnior, A.A.; de Alencar, A.P.; Suffys, P.N.; da Costa, R.R.; Moreira, M.A.S.; Guimarães, M.D.C. Risk Factors for Human Mycobacterium bovis Infections in an Urban Area of Brazil. Mem. Inst. Oswaldo Cruz 2018, 113, e170445. [Google Scholar] [CrossRef] [Green Version]
- Sichewo, P.R.; Michel, A.L.; Musoke, J.; Etter, E.M.C. Risk Factors for Zoonotic Tuberculosis at the Wildlife–Livestock–Human Interface in South Africa. Pathogens 2019, 8, 101. [Google Scholar] [CrossRef]
- Bernitz, N.; Clarke, C.; Roos, E.O.; Goosen, W.J.; Cooper, D.; van Helden, P.D.; Parsons, S.D.C.; Miller, M.A. Detection of Mycobacterium bovis Infection in African Buffaloes (Syncerus caffer) Using QuantiFERON®-TB Gold (QFT) Tubes and the Qiagen Cattletype® IFN-Gamma ELISA. Vet. Immunol. Immunopathol. 2018, 196, 48–52. [Google Scholar] [CrossRef] [PubMed]
- Clarke, C.; Cooper, D.; Goosen, W.J.; McFadyen, R.; Warren, R.M.; van Helden, P.D.; Parsons, S.D.C.; Miller, M.A. Antigen-Specific Interferon-Gamma Release Is Decreased Following the Single Intradermal Comparative Cervical Skin Test in African Buffaloes (Syncerus caffer). Vet. Immunol. Immunopathol. 2018, 201, 12–15. [Google Scholar] [CrossRef]
- Miller, M.; Buss, P.; Hofmeyr, J.; Olea-Popelka, F.; Parsons, S.; van Helden, P. Antemortem Diagnosis of Mycobacterium bovis Infection in Free-Ranging African Lions (Panthera Leo) and Implications for Transmission. J. Wildl. Dis. 2015, 51, 493–497. [Google Scholar] [CrossRef] [PubMed]
- Van Der Merwe, P.; Saayman, M. Determining the Economic Value of Game Farm Tourism. Koedoe 2003, 46, 103–112. [Google Scholar] [CrossRef]
- Herrera, M.; Vera, C.; Keynan, Y.; Rueda, Z.V. Gaps in Study Design for Immune Parameter Research for Latent Tuberculosis Infection: A Systematic Review. J. Immunol. Res. 2020, 2020, 8074183. [Google Scholar] [CrossRef]
- Miller, M. Tuberculosis in South African Wildlife: Why Is It Important? 2015. Available online: http://www.sun.ac.za/english/Inaugurallectures/Inaugurallectures/InauguralLectureProfMiller.pdf (accessed on 17 January 2019).
- Siddique, A.B.; Hussain, R.; Jamal, A.; Hossain, M.B.; Ahmad, Z.; Mansoor, M.K.; Khan, I.; Zahra, K.; Khan, A. Histopathological Investigations and Molecular Confirmation Reveal Mycobacterium bovis in One-Horned Rhinoceros (Rhinoceros Unicorns). BioMed Res. Int. 2022, 2022, 5816986. [Google Scholar] [CrossRef]
- Alvarez, A.H. Revisiting Tuberculosis Screening: An Insight to Complementary Diagnosis and Prospective Molecular Approaches for the Recognition of the Dormant TB Infection in Human and Cattle Hosts. Microbiol. Res. 2021, 252, 126853. [Google Scholar] [CrossRef] [PubMed]
- Lopes, B.C.; dos Reis, E.M.; de Bitencourt, F.B.R.; Loiko, M.R.; Bezerra, A.V.A.; Bueno, T.S.; Lape, I.T.; Cerva, C.; Coppola, M.D.M.; Rodrigues, R.O.; et al. A Molecular Strategy to Optimize Bovine Tuberculosis Post-Mortem Diagnosis and the Exposure to Mycobacterium tuberculosis Variant Bovis. Mol. Biol. Rep. 2020, 47, 7291–7296. [Google Scholar] [CrossRef]
- Meurens, F.; Dunoyer, C.; Fourichon, C.; Gerdts, V.; Haddad, N.; Kortekaas, J.; Lewandowska, M.; Monchatre-Leroy, E.; Summerfield, A.; Wichgers Schreur, P.J.; et al. Animal Board Invited Review: Risks of Zoonotic Disease Emergence at the Interface of Wildlife and Livestock Systems. Animal 2021, 15, 100241. [Google Scholar] [CrossRef]
- Odoi, A.; Chavez-Lindell, T.L.; Moncayo, A.L.; Fernanda, M.; Veloz, V. An Exploratory Assessment of Human and Animal Health Concerns of Smallholder Farmers in Rural Communities of Chimborazo, Ecuador. PeerJ 2022, 9, e12208. [Google Scholar] [CrossRef]
- Hlokwe, T.M.; Jenkins, A.O.; Streicher, E.M.; Venter, E.H.; Cooper, D.; Godfroid, J.; Michel, A.L. Molecular Characterisation of Mycobacterium bovis Isolated from African Buffaloes (Syncerus Caffer) in Hluhluwe-IMfolozi Park in KwaZulu-Natal, South Africa. Onderstepoort J. Vet. Res. 2011, 78, 6. [Google Scholar] [CrossRef] [PubMed]
- Roug, A.; Muse, E.A.; Clifford, D.L.; Paul, G.; Mpanduji, D.; Makingi, G.; Magesa, W.; Josephat, E.; Mazet, J.; Bird, B.; et al. Health of African Buffalos (Syncerus caffer) in Ruaha National Park, Tanzania. J. Wildl. Dis. 2020, 56, 495–498. [Google Scholar] [CrossRef]
- Shah, Y.; Paudel, S. Protect Elephants from Tuberculosis. Science 2021, 374, 832–833. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.A.; Finnegan, M.; Storms, T.; Garner, M.; Lyashchenko, K.P. Outbreak of Mycobacterium tuberculosis in a Herd of Captive Asian Elephants (Elephas maximus): Antemortem Diagnosis, Treatment, and Lessons Learned. J. Zoo Wildl. Med. 2018, 49, 748–754. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.A.; Hogan, J.N.; Meehan, C.L. Housing and Demographic Risk Factors Impacting Foot and Musculoskeletal Health in African Elephants [Loxodonta africana] and Asian Elephants [Elephas Maximus] in North American Zoos. PLoS ONE 2016, 11, e0155223. [Google Scholar] [CrossRef]
- Glaeser, S.S.; Edwards, K.L.; Paris, S.; Scarlata, C.; Lee, B.; Wielebnowski, N.; Finnell, S.; Somgird, C.; Brown, J.L. Characterization of Longitudinal Testosterone, Cortisol, and Musth in Male Asian Elephants (Elephas maximus), Effects of Aging, and Adrenal Responses to Social Changes and Health Events. Animals 2022, 12, 1332. [Google Scholar] [CrossRef] [PubMed]
- Ruetten, M.; Steinmetz, H.W.; Thiersch, M.; Kik, M.; Vaughan, L.; Altamura, S.; Muckenthaler, M.U.; Gassmann, M. Iron Regulation in Elderly Asian Elephants (Elephas maximus) Chronically Infected With Mycobacterium tuberculosis. Front. Vet. Sci. 2020, 7, 596379. [Google Scholar] [CrossRef]
- Mikota, S.K.; Gairhe, K.; Giri, K.; Hamilton, K.; Miller, M.; Paudel, S.; Lyashchenko, K.; Larsen, R.S.; Payeur, J.B.; Waters, W.R.; et al. Tuberculosis Surveillance of Elephants (Elephas maximus) in Nepal at the Captive-Wild Interface. Eur. J. Wildl. Res. 2015, 61, 221–229. [Google Scholar] [CrossRef]
- Katale, B.Z.; Mbugi, E.V.; Kendal, S.; Fyumagwa, R.D.; Kibiki, G.S.; Godfrey-Faussett, P.; Keyyu, J.D.; van Helden, P.; Matee, M.I. Bovine Tuberculosis at the Human-Livestock-Wildlife Interface: Is It a Public Health Problem in Tanzania?: A Review. Onderstepoort J. Vet. Res. 2012, 79, 8. [Google Scholar] [CrossRef] [PubMed]
- Ayele, W.; Neill, S.; Zinsstag, J.; Weiss, M.; Pavlik, I. Bovine Tuberculosis: An Old Disease but a New Threat to Africa. Int. J. Tuberc. Lung Dis. 2004, 8, 924–937. [Google Scholar] [CrossRef]
- Kaneene, J.B.; Miller, R.; de Kantor, I.N.; Thoen, C.O. Tuberculosis in Wild Animals. Int. J. Tuberc. Lung Dis. 2010, 14, 1508–1512. [Google Scholar] [PubMed]
- Fitzgerald, S.D.; Kaneene, J.B. Wildlife Reservoirs of Bovine Tuberculosis Worldwide: Hosts, Pathology, Surveillance, and Control. Vet. Pathol. 2013, 50, 488–499. [Google Scholar] [CrossRef]
- Goodchild, A.V.; Clifton-Hadley, R.S. Cattle-to-Cattle Transmission of Mycobacterium bovis. Tuberculosis 2001, 81, 23–41. [Google Scholar] [CrossRef] [PubMed]
- Sanou, A.; Tarnagda, Z.; Kanyala, E.; Zingué, D.; Nouctara, M.; Ganamé, Z.; Combary, A.; Hien, H.; Dembele, M.; Kabore, A.; et al. Mycobacterium bovis in Burkina Faso: Epidemiologic and Genetic Links between Human and Cattle Isolates. PLoS Negl. Trop. Dis. 2014, 8, 3142. [Google Scholar] [CrossRef] [PubMed]
- Neill, S.D.; Cassidy, J.; Hanna, J.; Mackie, D.P.; Pollock, J.M.; Clements, A.; Walton, E.; Bryson, D.G. Detection of Mycobacterium bovis Infection in Skin Test-Negative Cattle with an Assay for Bovine Interferon-Gamma. Vet. Rec. 1994, 135, 134–135. [Google Scholar] [CrossRef]
- De Vos, V.; Bengis, R.G.; Kriek, N.P.; Michel, A.; Keet, D.F.; Raath, J.P.; Huchzermeyer, H.F. The Epidemiology of Tuberculosis in Free-Ranging African Buffalo (Syncerus caffer) in the Kruger National Park, South Africa. Onderstepoort J. Vet. Res. 2001, 68, 119–130. [Google Scholar]
- Keet, D.F.; Kriek, N.P.J.; Bengis, R.G.; Grobler, D.G.; Michel, A. The Rise and Fall of Tuberculosis in a Free-Ranging Chacma Baboon Troop in the Kruger National Park. Onderstepoort J. Vet. Res. 2000, 67, 115–122. [Google Scholar]
- Keet, D.F.; Kriek, N.P.J.; Bengis, R.G.; Michel, A.L. Tuberculosis in Kudus (Tragelaphus strepsiceros) in the Kruger National Park. Onderstepoort J. Vet. Res. 2001, 68, 225–230. [Google Scholar]
- Paine, R.; Martinaglia, G. Tuberculosis in Wild Buck Living Under Natural Conditions. J. Comp. Pathol. Ther. 1929, 42, 1–8. [Google Scholar] [CrossRef]
- Goosen, W.J.; Kleynhans, L.; Kerr, T.J.; van Helden, P.D.; Buss, P.; Warren, R.M.; Miller, M.A. Improved Detection of Mycobacterium tuberculosis and M. Bovis in African Wildlife Samples Using Cationic Peptide Decontamination and Mycobacterial Culture Supplementation. J. Vet. Diagnostic Investig. 2022, 34, 61–67. [Google Scholar] [CrossRef]
- Hlokwe, T.M.; Id, O.; Article, O. First Detection of Mycobacterium bovis Infection in Giraffe (Giraffa camelopardalis) in the Greater Kruger National Park Complex: Role and Implications. Transbound Emerg. Dis. 2019, 66, 2264–2270. [Google Scholar] [CrossRef]
- Gavier-Widen, D.; Chambers, M.A.; Palmer, N.; Newell, D.G.; Hewinson, R.G. Pathology of Natural Mycobacterium bovis Infection in European Badgers (Meles meles) and Its Relationship with Bacterial Excretion. Vet. Rec. 2001, 148, 299–304. [Google Scholar] [CrossRef] [PubMed]
- Martin, L.E.R.; Byrne, A.W.; O’Keeffe, J.; Miller, M.A.; Olea-Popelka, F.J. Weather Influences Trapping Success for Tuberculosis Management in European Badgers (Meles meles). Eur. J. Wildl. Res. 2017, 63, 30. [Google Scholar] [CrossRef]
- Jolma, E.R.; Delahay, R.J.; Smith, F.; Drewe, J.A. Serologic Responses Correlate with Current but Not Future Bacterial Shedding in Badgers Naturally Infected with Mycobacterium bovis. Transbound. Emerg. Dis. 2021, 69, 1922–1932. [Google Scholar] [CrossRef] [PubMed]
- Infantes-Lorenzo, J.A.; Dave, D.; Moreno, I.; Anderson, P.; Lesellier, S.; Gormley, E.; Dominguez, L.; Balseiro, A.; Gortázar, C.; Dominguez, M.; et al. New Serological Platform for Detecting Antibodies against Mycobacterium tuberculosis Complex in European Badgers. Vet. Med. Sci. 2019, 5, 61–69. [Google Scholar] [CrossRef]
- Katale, B.Z.; Mbugi, E.V.; Siame, K.K.; Keyyu, J.D.; Kendall, S.; Kazwala, R.R.; Dockrell, H.M.; Fyumagwa, R.D.; Michel, A.L.; Rweyemamu, M.; et al. Isolation and Potential for Transmission of Mycobacterium bovis at Human–Livestock–Wildlife Interface of the Serengeti Ecosystem, Northern Tanzania. Transbound. Emerg. Dis. 2017, 64, 815–825. [Google Scholar] [CrossRef]
- Michel, A.L.; Coetzee, M.L.; Keet, D.F.; Maré, L.; Warren, R.; Cooper, D.; Bengis, R.G.; Kremer, K.; van Helden, P. Molecular Epidemiology of Mycobacterium bovis Isolates from Free-Ranging Wildlife in South African Game Reserves. Vet. Microbiol. 2009, 133, 335–343. [Google Scholar] [CrossRef]
- Bouts, T.; Vordermeier, M.; Flach, E.; Routh, A. Positive Skin and Serologic Test Results of Diagnostic Assays for Bovine Tuberculosis and Subsequent Isolation of Mycobacterium interjectum in a Pygmy Hippopotamus (Hexaprotodon liberiensis). J. Zoo Wildl. Med. 2009, 40, 536–542. [Google Scholar] [CrossRef]
- Kerr, T.J.; Goosen, W.J.; Gumbo, R.; de Klerk-Lorist, L.M.; Pretorius, O.; Buss, P.E.; Kleynhans, L.; Lyashchenko, K.P.; Warren, R.M.; van Helden, P.D.; et al. Diagnosis of Mycobacterium bovis Infection in Free-Ranging Common Hippopotamus (Hippopotamus amphibius). Transbound. Emerg. Dis. 2022, 69, 378–384. [Google Scholar] [CrossRef]
- De Vos, V.; McCully, R.M.; van Niekerk, C.A.W.J. Mycobacteriosis in the Kruger National Park. Koedoe 1977, 20, a928. [Google Scholar] [CrossRef]
- Tanner, M.; Michel, A.L. Investigation of the Viability of Mycobacterium bovis under Different Environmental Conditions in the Kruger National Park. Onderstepoort J. Vet. Res. 1999, 66, 185–190. [Google Scholar]
- Hlokwe, T.M.; Mogano, R.M. Utility of Xpert® MTB/RIF Ultra Assay in the Rapid Diagnosis of Bovine Tuberculosis in Wildlife and Livestock Animals from South Africa. Prev. Vet. Med. 2020, 177, 104980. [Google Scholar] [CrossRef]
- Palmer, M.V. Mycobacterium bovis: Characteristics of Wildlife Reservoir Hosts. Transbound. Emerg. Dis. 2013, 60 (Suppl. S1), 1–13. [Google Scholar] [CrossRef]
- Miller, M.A.; Buss, P.; Sylvester, T.T.; Lyashchenko, K.P.; Deklerk-Lorist, L.-M.; Bengis, R.; Hofmeyr, M.; Hofmeyr, J.; Mathebula, N.; Hausler, G.; et al. Mycobacterium bovis in Free-Ranging Lions (Panthera leo)—Evaluation of Serological and Tuberculin Skin Tests for Detection of Infection and Disease. J. Zoo Wildl. Med. 2019, 50, 7. [Google Scholar] [CrossRef] [PubMed]
- Gumbo, R.; Sylvester, T.T.; Goosen, W.J.; Buss, P.E.; de Klerk-Lorist, L.-M.; van Schalkwyk, O.L.; McCall, A.; Warren, R.M.; van Helden, P.D.; Miller, M.A.; et al. Adaptation and Diagnostic Potential of a Commercial Cat Interferon Gamma Release Assay for the Detection of Mycobacterium bovis Infection in African Lions (Panthera leo). Pathogens 2022, 11, 765. [Google Scholar] [CrossRef] [PubMed]
- Brüns, A.C.; Brüns, B.; Tanner, M.; Williams, M.C.; Botha, L.; O’brien, A.; Fosgate, G.T.; Van Helden, P.D.; Clarke, J.; Michel, A.L. Diagnosis and Implications of Mycobacterium bovis Infection in Banded Mongooses (Mungos mungo) in the Kruger National Park, South Africa. J. Wildl. Dis. 2017, 53, 19–29. [Google Scholar] [CrossRef]
- Palmer, M.V.; Thacker, T.C.; Rabideau, M.M.; Jones, G.J.; Kanipe, C.; Vordermeier, H.M.; Waters, W.R. Veterinary Immunology and Immunopathology Biomarkers of Cell-Mediated Immunity to Bovine Tuberculosis. Vet. Immunol. Immunopathol. 2020, 220, 109988. [Google Scholar] [CrossRef]
- Thomas, J.; Balseiro, A.; Gortázar, C.; Risalde, M.A. Diagnosis of Tuberculosis in Wildlife: A Systematic Review. Vet. Res. 2021, 52, 31. [Google Scholar] [CrossRef]
- Miller, M.; Chavey, P.S.; Hofmeyr, J.; Mathebula, N.; Doering, A.; Buss, P.; Olea-Popelka, F. Evaluation of Serum Ferritin and Serum Iron in Free-Ranging Black Rhinoceros (Diceros bicornis) as a Tool to Understand Factors Affecting Iron-Overload Disorder. J. Zoo Wildl. Med. 2016, 47, 820–826. [Google Scholar] [CrossRef]
- Miller, M.A.; Greenwald, R.; Lyashchenko, K.P. Potential for Serodiagnosis of Tuberculosis in Black Rhinoceros (Diceros bicornis). J. Zoo Wildl. Med. 2015, 46, 100–104. [Google Scholar] [CrossRef]
- Dwyer, R.A.; Witte, C.; Buss, P.; Goosen, W.J.; Miller, M.; Caron, A. Epidemiology of Tuberculosis in Multi-Host Wildlife Systems: Implications for Black (Diceros bicornis) and White (Ceratotherium simum) Rhinoceros. Front. Vet. Sci. 2020, 7, 580476. [Google Scholar] [CrossRef] [PubMed]
- Richomme, C.; Réveillaud, E.; Moyen, J.-L.; Sabatier, P.; De Cruz, K.; Michelet, L.; Boschiroli, M.L. Mycobacterium bovis Infection in Red Foxes in Four Animal Tuberculosis Endemic Areas in France. Microorganisms 2020, 8, 1070. [Google Scholar] [CrossRef]
- Roos, E.O.; Buss, P.; de Klerk-Lorist, L.M.; Hewlett, J.; Hausler, G.A.; Rossouw, L.; McCall, A.J.; Cooper, D.; van Helden, P.D.; Parsons, S.D.C.; et al. Test Performance of Three Serological Assays for the Detection of Mycobacterium bovis Infection in Common Warthogs (Phacochoerus africanus). Vet. Immunol. Immunopathol. 2016, 182, 79–84. [Google Scholar] [CrossRef] [PubMed]
- Roos, E.O.; Olea-Popelka, F.; Buss, P.; Hausler, G.A.; Warren, R.; Van Helden, P.D.; Parsons, S.D.C.; De Klerk-Lorist, L.M.; Miller, M.A. Measuring Antigen-Specific Responses in Mycobacterium bovis-Infected Warthogs (Phacochoerus africanus) Using the Intradermal Tuberculin Test. BMC Vet. Res. 2018, 14, 360. [Google Scholar] [CrossRef] [PubMed]
- Roos, E.O.; Olea-Popelka, F.; Buss, P.; de Klerk-Lorist, L.M.; Cooper, D.; van Helden, P.D.; Parsons, S.D.C.; Miller, M.A. Seroprevalence of Mycobacterium bovis Infection in Warthogs (Phacochoerus africanus) in Bovine Tuberculosis-Endemic Regions of South Africa. Transbound. Emerg. Dis. 2018, 65, 1182–1189. [Google Scholar] [CrossRef]
- Campana, M.G.; Parker, L.D.; Hawkins, M.T.R.; Young, H.S.; Helgen, K.M.; Szykman Gunther, M.; Woodroffe, R.; Maldonado, J.E.; Fleischer, R.C. Genome Sequence, Population History, and Pelage Genetics of the Endangered African Wild Dog (Lycaon pictus). BMC Genom. 2016, 17, 1. [Google Scholar] [CrossRef] [PubMed]
- Meiring, C.; Schurz, H.; van Helden, P.; Hoal, E.; Tromp, G.; Kinnear, C.; Kleynhans, L.; Glanzmann, B.; van Schalkwyk, L.; Miller, M.; et al. African Wild Dogs (Lycaon pictus) from the Kruger National Park, South Africa Are Currently Not Inbred but Have Low Genomic Diversity. Sci. Rep. 2022, 12, 14979. [Google Scholar] [CrossRef]
- Overton, J.M.C.; Elizalde Castells, D.; Figueira Fernandes Elizalde, S.R.; Valério, H.M.; Alexandre Zumbo, M.N.; Groom, R.J.; Durant, S.M. Endangered African Wild Dogs (Lycaon pictus Temm.) in Angola: Filling a 50-Year Gap of Knowledge with Findings from Two National Parks. Afr. J. Ecol. 2020, 58, 582–587. [Google Scholar] [CrossRef]
- Higgitt, R.L.; Van Schalkwyk, O.L.; De Klerk-Lorist, L.M.; Buss, P.E.; Caldwell, P.; Rossouw, L.; Manamela, T.; Hausler, G.A.; Hewlett, J.; Mitchell, E.P.; et al. Mycobacterium bovis Infection in African Wild Dogs, Kruger National Park, South Africa. Emerg. Infect. Dis. 2019, 25, 1425–1427. [Google Scholar] [CrossRef]
- Kock, R.A.; Woodford, M.H.; Rossiter, P.B. Disease Risks Associated with the Translocation of Wildlife. OIE Rev. Sci. Tech. 2010, 29, 329–350. [Google Scholar] [CrossRef]
- Hlokwe, T.M.; van Helden, P.; Michel, A.L. Evidence of Increasing Intra and Inter-Species Transmission of Mycobacterium bovis in South Africa: Are We Losing the Battle? Prev. Vet. Med. 2014, 115, 10–17. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.; Michel, A.; van Helden, P.; Buss, P. Tuberculosis in Rhinoceros: An Underrecognized Threat? Transbound. Emerg. Dis. 2017, 64, 1071–1078. [Google Scholar] [CrossRef] [PubMed]
- Monies, B.; de la Rua, R.; Jahans, K. Bovine Tuberculosis in Cats. Vet. Rec. 2006, 158, 490–491. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, K.; Chauhan, D.S.; Gupta, P.; Singh, H.B.; Sharma, V.D.; Yadav, V.S.; Sreekumaran; Thakral, S.S.; Dharamdheeran, J.S.; Nigam, P.; et al. Isolation of Mycobacterium bovis & Mycobacterium tuberculosis from Cattle of Some Farms in North India-Possible Relevance in Human Health. Indian J. Med. Res. 2008, 128, 26–31. [Google Scholar]
- Sweetline Anne, N.; Ronald, B.S.M.; Kumar, T.M.A.S.; Kannan, P.; Thangavelu, A. Molecular Identification of Mycobacterium tuberculosis in Cattle. Vet. Microbiol. 2017, 198, 81–87. [Google Scholar] [CrossRef]
- Ghodbane, R.; Drancourt, M. Non-Human Sources of Mycobacterium tuberculosis. Tuberculosis 2013, 93, 589–595. [Google Scholar] [CrossRef]
- Ravindran, R.; Krishnan, V.V.; Dhawan, R.; Wunderlich, M.L.; Lerche, N.W.; Flynn, J.L.; Luciw, P.A.; Khan, I.H. Plasma Antibody Profiles in Non-Human Primate Tuberculosis. J. Med. Primatol. 2014, 43, 59–71. [Google Scholar] [CrossRef]
- Parsons, S.D.C.; Warren, R.M.; Ottenhoff, T.H.M.; Gey van Pittius, N.C.; van Helden, P.D. Detection of Mycobacterium tuberculosis Infection in Dogs in a High-Risk Setting. Res. Vet. Sci. 2012, 92, 414–419. [Google Scholar] [CrossRef]
- Haydock, L.A.J.; Abrams-Ogg, A.C.G.; Weese, J.S.; Goldstein, M.R.; Clifford, A.B.; Sebastian, A.; Rea, E.H.; Jamieson, F.B.; Duncan, C.; Andrievskaia, O.; et al. Diagnostic and Public Health Investigation of Mycobacterium tuberculosis Infection in a Dog in Ontario, Canada. J. Vet. Diagn. Investig. 2022, 34, 292–297. [Google Scholar] [CrossRef]
- Marfil, M.J.; Barandiaran, S.; Zumárraga, M.J.; Germani, L.; Faccini, T.; Linares, M.; Capra, S.; Gramajo, L.; Martínez Vivot, M.; Falzoni, E. Mycobacterium tuberculosis Infection in a Free-Ranging Urban Dog from Argentina. Vet. Res. Commun. 2022, 46, 781–788. [Google Scholar] [CrossRef]
- Une, Y.; Mori, T. Tuberculosis as a Zoonosis from a Veterinary Perspective. Comp. Immunol. Microbiol. Infect. Dis. 2007, 30, 415–425. [Google Scholar] [CrossRef] [PubMed]
- Montali, R.J.; Mikota, S.K.; Cheng, L.I. Mycobaterium tuberculosis in Zoo and Wildlife Species. Sci. Tech. Rev. Int. Off. Epizoot. 2001, 20, 291–303. [Google Scholar] [CrossRef] [PubMed]
- Rosen, L.E.; Hanyire, T.G.; Dawson, J.; Foggin, C.M.; Michel, A.L.; Huyvaert, K.P.; Miller, M.A.; Olea-Popelka, F.J. Tuberculosis Serosurveillance and Management Practices of Captive African Elephants (Loxodonta africana) in the Kavango-Zambezi Transfrontier Conservation Area. Transbound. Emerg. Dis. 2018, 65, e344–e354. [Google Scholar] [CrossRef] [PubMed]
- Landolfi, J.A.; Terio, K.A.; Miller, M.; Junecko, B.F.; Reinhart, T. Pulmonary Tuberculosis in Asian Elephants (Elephas maximus): Histologic Lesions with Correlation to Local Immune Responses. Vet. Pathol. 2015, 52, 535–542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sarad, P.; Tsubota, T. Tuberculosis in Elephants: A Zoonotic Disease at the Human-Elephant Interface. Japanese J. Zoo Wildl. Med. 2016, 21, 65–69. [Google Scholar] [CrossRef]
- Bezos, J.; Casal, C.; Díez-Delgado, I.; Romero, B.; Liandris, E.; Álvarez, J.; Sevilla, I.A.; de Juan, L.; Domínguez, L.; Gortázar, C. Goats Challenged with Different Members of the Mycobacterium tuberculosis Complex Display Different Clinical Pictures. Vet. Immunol. Immunopathol. 2015, 167, 185–189. [Google Scholar] [CrossRef]
- Cadmus, S.I.B.; Jenkins, A.O.; Godfroid, J.; Osinusi, K.; Adewole, I.F.; Murphy, R.L.; Taiwo, B.O. Mycobacterium tuberculosis and Mycobacterium africanum in Stools from Children Attending an Immunization Clinic in Ibadan, Nigeria. Int. J. Infect. Dis. 2009, 13, 740–744. [Google Scholar] [CrossRef]
- Silva, M.L.; Cá, B.; Osório, N.S.; Rodrigues, P.N.S.; Maceiras, A.R.; Saraiva, M. Tuberculosis Caused by Mycobacterium africanum: Knowns and Unknowns. PLoS Pathog. 2022, 18, e1010490. [Google Scholar] [CrossRef]
- Gonzalo-Asensio, J.; Astarie-Dequeker, C.; Malaga, W.; Brosch, R.; Moreau, F.; Passemar, C.; Martin, C.; Laval, F.; Daffe, M.; Guilhot, C.; et al. Evolutionary History of Tuberculosis Shaped by Conserved Mutations in the PhoPR Virulence Regulator. Proc. Natl. Acad. Sci. USA 2014, 111, 11491–11496. [Google Scholar] [CrossRef]
- Rahim, Z.; Möllers, M.; Te Koppele-Vije, A.; De Beer, J.; Zaman, K.; Matin, M.A.; Kamal, M.; Raquib, R.; Van Soolingen, D.; Baqi, M.A.; et al. Characterization of Mycobacterium africanum Subtype 1 among Cows in a Dairy Farm in Bangladesh Using Spoligotyping. Southeast Asian J. Trop. Med. Public Health 2007, 38, 706–713. [Google Scholar]
- Gudan, A.; Artuković, B.; Cvetnić, Ž.; Špičić, S.; Beck, A.; Hohšteter, M.; Naglić, T.; Bata, I.; Grabarević, Ž. Disseminated Tuberculosis in Hyrax (Procavia capensis) Caused by Mycobacterium africanum. J. Zoo Wildl. Med. 2008, 39, 386–391. [Google Scholar] [CrossRef] [PubMed]
- Cousins, D.V.; Bastida, R.; Cataldi, A.; Quse, V.; Redrobe, S.; Dow, S.; Duignan, P.; Murray, A.; Dupont, C.; Ahmed, N.; et al. Tuberculosis in Seals Caused by a Novel Member of the Mycobacterium tuberculosis Complex: Mycobacterium pinnipedii Sp. Nov. Int. J. Syst. Evol. Microbiol. 2003, 53, 1305–1314. [Google Scholar] [CrossRef]
- Kiers, A.; Klarenbeek, A.; Mendelts, B.; Van Soolingen, D.; Koëter, G. Transmission of Mycobacterium pinnipedii to Humans in a Zoo with Marine Mammals. Int. J. Tuberc. Lung Dis. 2008, 12, 1469–1473. [Google Scholar] [PubMed]
- Macedo, R.; Isidro, J.; Gomes, M.C.; Botelho, A.; Albuquerque, T.; Sogorb, A.; Bernardino, R.; Fernandes, T.L.; Mourato, T.; Durval, M.; et al. Animal-to-Human Transmission of Mycobacterium pinnipedii. Eur. Respir. J. 2020, 56, 2000371. [Google Scholar] [CrossRef]
- Loeffler, S.H.; de Lisle, G.W.; Neill, M.A.; Collins, D.M.; Price-Carter, M.; Paterson, B.; Crews, K.B. The Seal Tuberculosis Agent, Mycobacterium pinnipedii, Infects Domestic Cattle in New Zealand: Epidemiologic Factors and DNA Strain Typing. J. Wildl. Dis. 2014, 50, 180–187. [Google Scholar] [CrossRef] [PubMed]
- Aranaz, A.; Liebana, E.; Gomez-Mampaso, E.; Galán, J.C.; Cousins, D.; Ortega, A.; Blázquez, J.; Baquero, F.; Mateos, A.; Súarez, G.; et al. Mycobacterium tuberculosis Subsp. caprae subsp. nov.: A Taxonomic Study of a New Member of the Mycobacterium tuberculosis Complex Isolated from Goats in Spain. Int. J. Syst. Bacteriol. 1999, 49, 1263–1273. [Google Scholar] [CrossRef] [PubMed]
- Gormley, E.; Corner, L.A.L.; Costello, E.; Rodriguez-Campos, S. Bacteriological Diagnosis and Molecular Strain Typing of Mycobacterium bovis and Mycobacterium caprae. Res. Vet. Sci. 2014, 97, S30–S43. [Google Scholar] [CrossRef]
- Shrestha, A.; Picoy, J.; Torres, A.; Moore, D.A.; Gilman, R.H.; Coronel, J.; Grandjean, L. A Case Report of Transmission and Disease Caused by Mycobacterium caprae and Mycobacterium bovis in Lima, Peru. BMC Infect. Dis. 2021, 21, 4–9. [Google Scholar] [CrossRef]
- Huitema, H.; Jaartsveld, F.H.J. Mycobacterium microti Infection in a Cat and Some Pigs. Antonie Van Leeuwenhoek 1967, 33, 209–212. [Google Scholar] [CrossRef]
- Wells, A.Q.; Robb-Smith, A.H.T. Murine Type of Tubercle Bacillus (the Vole Acid-Fast Bacillus). 1946. Available online: https://agris.fao.org/agris-search/search.do?recordID=US201300343701 (accessed on 11 August 2022).
- Van Soolingen, D.; Van Der Zanden, A.G.M.; De Haas, P.E.W.; Noordhoek, G.T.; Kiers, A.; Foudraine, N.A.; Portaels, F.; Kolk, A.H.J.; Kremer, K.; Van Embden, J.D.A. Diagnosis of Mycobacterium microti Infections among Humans by Using Novel Genetic Markers. J. Clin. Microbiol. 1998, 36, 1840–1845. [Google Scholar] [CrossRef]
- van Ingen, J.; Brosch, R.; van Soolingen, D. Characterization of Mycobacterium orygis. Emerg. Infect. Dis. 2013, 19, 521–522. [Google Scholar] [CrossRef] [PubMed]
- Van Soolingen, D.; De Haas, P.E.W.; Haagsma, J.; Eger, T.; Hermans, P.W.M.; Ritacco, V.; Alito, A.; Va Embdenl, J.D.A. Use of Various Genetic Markers in Differentiation of Mycobacterium bovis Strains from Animals and Humans and for Studying Epidemiology of Bovine Tuberculosis. J. Clin. Microbiol. 1994, 32, 2425–2433. [Google Scholar] [CrossRef] [PubMed]
- Parsons, S.D.C. Mycobacterium orygis: A Zoonosis, Zooanthroponosis, or Both? Lancet Microbe 2020, 1, e240. [Google Scholar] [CrossRef]
- Pfyffer, G.E.; Auckenthaler, R.; Van Embden, J.D.A.; Van Soolingen, D. Mycobacterium canettii, the Smooth Variant of Mycobacterium tuberculosis, Isolated from a Swiss Patient Exposed in Africa. Emerg. Infect. Dis. 1998, 4, 631–634. [Google Scholar] [CrossRef] [PubMed]
- Van Soolingen, D.; Hoogenboezem, T.; De Haas, P.E.W.; Hermans, P.W.M.; Koedam, M.A.; Teppema, K.S.; Brennan, P.J.; Besra, G.S.; Portaels, F.; Top, J.; et al. A Novel Pathogenic Taxon of the Mycobacterium tuberculosis Complex, Canetti: Characterization of an Exceptional Isolate from Africa. Int. J. Syst. Bacteriol. 1997, 47, 1236–1245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Supply, P.; Brosch, R. The Biology and Epidemiology of Mycobacterium canettii. Adv. Exp. Med. Biol. 2017, 1019, 27–41. [Google Scholar] [CrossRef]
- Alexander, K.A.; Laver, P.N.; Michel, A.L.; Williams, M.; van Helden, P.D.; Warren, R.M.; van Pittius, N.C.G. Novel Mycobacterium tuberculosis Complex Pathogen, M. Mungi. Emerg. Infect. Dis. 2010, 16, 1296–1299. [Google Scholar] [CrossRef]
- Dippenaar, A.; Parsons, S.D.C.; Sampson, S.L.; Van Der Merwe, R.G.; Drewe, J.A.; Abdallah, A.M.; Siame, K.K.; Gey Van Pittius, N.C.; Van Helden, P.D.; Pain, A.; et al. Whole Genome Sequence Analysis of Mycobacterium suricattae. Tuberculosis 2015, 95, 682–688. [Google Scholar] [CrossRef]
- Clarke, C.; Van Helden, P.; Miller, M.; Parsons, S. Animal-Adapted Members of the Mycobacterium tuberculosis Complex Endemic to the Southern African Subregion. J. S. Afr. Vet. Assoc. 2016, 87, 1322. [Google Scholar] [CrossRef]
- Parsons, S.D.C.; Drewe, J.A.; van Pittius, N.C.G.; Warren, R.M.; van Helden, P.D. Novel Cause of Tuberculosis in Meerkats, South Africa. Emerg. Infect. Dis. 2013, 19, 2004–2007. [Google Scholar] [CrossRef]
- Glaziou, P.; Floyd, K.; Raviglione, M.C. Global Epidemiology of Tuberculosis. Semin. Respir. Crit. Care Med. 2015, 39, 271–285. [Google Scholar] [CrossRef] [PubMed]
- Morris, R.S.; Pfeiffer, D.U.; Jackson, R. The Epidemiology of Mycobacterium bovis Infections. Vet. Microbiol. 1994, 40, 153–177. [Google Scholar] [CrossRef]
- Phipps, E.; McPhedran, K.; Edwards, D.; Russell, K.; O’Connor, C.M.; Gunn-Moore, D.A.; O’Halloran, C.; Roberts, T.; Morris, J. Bovine Tuberculosis in Working Foxhounds: Lessons Learned from a Complex Public Health Investigation. Epidemiol. Infect. 2019, 147, E24. [Google Scholar] [CrossRef] [PubMed]
- Haddad, N.; Ostyn, A.; Karoui, C.; Masselot, M.; Thorel, M.F.; Hughes, S.L.; Inwald, J.; Hewinson, R.G.; Durand, B. Spoligotype Diversity of Mycobacterium bovis Strains Isolated in France from 1979 to 2000. J. Clin. Microbiol. 2001, 39, 3623–3632. [Google Scholar] [CrossRef] [PubMed]
- Almaw, G.; Mihret, A.; Abebe, T.; Ameni, G.; Gumi, B.; Olani, A.; Tamiru, M.; Koran, T.; Aliy, A.; Sombo, M.; et al. Spoligotype Analysis of Mycobacterium bovis Isolates from Cattle and Assessment of Zoonotic TB Transmission among Individuals Working in Bovine TB- Infected Dairy Farms in Ethiopia. Zoonoses Public Health 2022, 69, 663–672. [Google Scholar] [CrossRef]
- Branger, M.; Loux, V.; Cochard, T.; Boschiroli, M.L.; Biet, F.; Michelet, L. The Complete Genome Sequence of Mycobacterium bovis Mb3601, a SB0120 Spoligotype Strain Representative of a New Clonal Group. Infect. Genet. Evol. 2020, 82, 104309. [Google Scholar] [CrossRef]
- Boniotti, M.B.; Goria, M.; Loda, D.; Garrone, A.; Benedetto, A.; Mondo, A.; Tisato, E.; Zanoni, M.; Zoppi, S.; Dondo, A.; et al. Molecular Typing of Mycobacterium bovis Strains Isolated in Italy from 2000 to 2006 and Evaluation of Variable-Number Tandem Repeats for Geographically Optimized Genotyping. J. Clin. Microbiol. 2009, 47, 636–644. [Google Scholar] [CrossRef]
- Hlokwe, T.M.; Van Helden, P.; Michel, A. Evaluation of the Discriminatory Power of Variable Number of Tandem Repeat Typing of Mycobacterium bovis Isolates from Southern Africa. Transbound. Emerg. Dis. 2013, 60 (Suppl. S1), 111–120. [Google Scholar] [CrossRef]
- Amato, B.; Di Marco Lo Presti, V.; Gerace, E.; Capucchio, M.T.; Vitale, M.; Zanghì, P.; Pacciarini, M.L.; Marianelli, C.; Boniotti, M.B. Molecular Epidemiology of Mycobacterium tuberculosis Complex Strains Isolated from Livestock and Wild Animals in Italy Suggests the Need for a Different Eradication Strategy for Bovine Tuberculosis. Transbound. Emerg. Dis. 2018, 65, e416–e424. [Google Scholar] [CrossRef]
- Broeckl, S.; Krebs, S.; Varadharajan, A.; Straubinger, R.K.; Blum, H.; Buettner, M. Investigation of Intra-Herd Spread of Mycobacterium caprae in Cattle by Generation and Use of a Whole-Genome Sequence. Vet. Res. Commun. 2017, 41, 113–128. [Google Scholar] [CrossRef]
- Orloski, K.; Robbe-Austerman, S.; Tod Stuber, B.H.; Schoenbaum, M. Whole Genome Sequencing of Mycobacterium bovis Isolated from Livestock in the United States, 1989–2018. Front. Vet. Sci. 2018, 5, 253. [Google Scholar] [CrossRef]
- Valcheva, V.; Perea, C.; Savova-Lalkovska, T.; Dimitrova, A.; Radulski, L.; Mokrousov, I.; Marinov, K.; Najdenski, H.; Bonovska, M. Mycobacterium bovis and Mycobacterium caprae in Bulgaria: Insight into Transmission and Phylogeography Gained through Whole-Genome Sequencing. BMC Vet. Res. 2022, 18, 148. [Google Scholar] [CrossRef] [PubMed]
- Sandoval-Azuara, S.E.; Muñiz-Salazar, R.; Perea-Jacobo, R.; Robbe-Austerman, S.; Perera-Ortiz, A.; López-Valencia, G.; Bravo, D.M.; Sanchez-Flores, A.; Miranda-Guzmán, D.; Flores-López, C.A.; et al. Whole Genome Sequencing of Mycobacterium bovis to Obtain Molecular Fingerprints in Human and Cattle Isolates from Baja California, Mexico. Int. J. Infect. Dis. 2017, 63, 48–56. [Google Scholar] [CrossRef] [PubMed]
- Price-Carter, M.; Brauning, R.; de Lisle, G.W.; Livingstone, P.; Neill, M.; Sinclair, J.; Paterson, B.; Atkinson, G.; Knowles, G.; Crews, K.; et al. Whole Genome Sequencing for Determining the Source of Mycobacterium bovis Infections in Livestock Herds and Wildlife in New Zealand. Front. Vet. Sci. 2018, 5, 272. [Google Scholar] [CrossRef] [PubMed]
- de la Rua-Domenech, R.; Goodchild, A.T.; Vordermeier, H.M.; Hewinson, R.G.; Christiansen, K.H.; Clifton-Hadley, R.S. Ante Mortem Diagnosis of Tuberculosis in Cattle: A Review of the Tuberculin Tests, γ-Interferon Assay and Other Ancillary Diagnostic Techniques. Res. Vet. Sci. 2006, 81, 190–210. [Google Scholar] [CrossRef]
- Roos, E.O.; Olea-Popelka, F.; Buss, P.; de Klerk-Lorist, L.M.; Cooper, D.; Warren, R.M.; van Helden, P.D.; Parsons, S.D.C.; Miller, M.A. IP-10: A Potential Biomarker for Detection of Mycobacterium bovis Infection in Warthogs (Phacochoerus africanus). Vet. Immunol. Immunopathol. 2018, 201, 43–48. [Google Scholar] [CrossRef]
- Roos, E.O. Detection and Characterization of Mycobacterial Infections Occurring in Phacochoerus Africanus (Gmelin, 1788) (Common Warthog); Stellenbosch University: Stellenbosch, South Africa, 2018. [Google Scholar]
- Majoor, C.J.; Magis-Escurra, C.; van Ingen, J.; Boeree, M.J.; van Soolingen, D. Epidemiology of Mycobacterium bovis Disease in Humans, The Netherlands, 1993–2007. Emerg. Infect. Dis. 2011, 17, 457–463. [Google Scholar] [CrossRef]
- Rabaa, M.A.; Tue, N.T.; Phuc, T.M.; Carrique-Mas, J.; Saylors, K.; Cotten, M.; Bryant, J.E.; Nghia, H.D.T.; Van Cuong, N.; Pham, H.A.; et al. The Vietnam Initiative on Zoonotic Infections (VIZIONS): A Strategic Approach to Studying Emerging Zoonotic Infectious Diseases. Ecohealth 2015, 12, 726–735. [Google Scholar] [CrossRef]
- Durr, P.A.; Hewinson, R.G.; Clifton-Hadley, R.S. Molecular Epidemiology of Bovine Tuberculosis. I. Mycobacterium bovis Genotyping. Rev. Sci. Tech. 2000, 19, 675–688. [Google Scholar] [CrossRef]
- Mostowy, S.; Behr, M.A. Comparative Genomics in the Fight against Tuberculosis: Diagnostics, Epidemiology, and BCG Vaccination. Am. J. Pharm. 2002, 2, 189–196. [Google Scholar] [CrossRef]
- Campbell, E.L.; Byrne, A.W.; Menzies, F.D.; Milne, G.; McBride, K.R.; McCormick, C.M.; Scantlebury, D.M.; Reid, N. Quantifying Intraherd Cattle Movement Metrics: Implications for Disease Transmission Risk. Prev. Vet. Med. 2020, 185, 106192. [Google Scholar] [CrossRef] [PubMed]
- Ghebremariam, M.K.; Michel, A.L.; Vernooij, J.C.M.; Nielen, M.; Rutten, V.P.M.G. Prevalence of Bovine Tuberculosis in Cattle, Goats, and Camels of Traditional Livestock Raising Communities in Eritrea. BMC Vet. Res. 2018, 14, 73. [Google Scholar] [CrossRef]
- O’Brien, D.J.; Schmitt, S.M.; Fierke, J.S.; Hogle, S.A.; Winterstein, S.R.; Cooley, T.M.; Moritz, W.E.; Diegel, K.L.; Fitzgerald, S.D.; Berry, D.E.; et al. Epidemiology of Mycobacterium bovis in Free-Ranging White-Tailed Deer, Michigan, USA, 1995–2000. Prev. Vet. Med. 2002, 54, 47–63. [Google Scholar] [CrossRef]
- Zimpel, C.K.; Patané, J.S.L.; Guedes, A.C.P.; de Souza, R.F.; Silva-Pereira, T.T.; Camargo, N.C.S.; de Souza Filho, A.F.; Ikuta, C.Y.; Neto, J.S.F.; Setubal, J.C.; et al. Global Distribution and Evolution of Mycobacterium bovis Lineages. Front. Microbiol. 2020, 11, 843. [Google Scholar] [CrossRef] [PubMed]
- Galagan, J.E. Genomic Insights into Tuberculosis. Nat. Rev. Genet. 2014, 15, 307–320. [Google Scholar] [CrossRef]
- Coscolla, M.; Gagneux, S. Consequences of Genomic Diversity in Mycobacterium tuberculosis. Semin. Immunol. 2014, 26, 431–444. [Google Scholar] [CrossRef]
- Otchere, I.D.; van Tonder, A.J.; Asante-Poku, A.; Sánchez-Busó, L.; Coscollá, M.; Osei-Wusu, S.; Asare, P.; Aboagye, S.Y.; Ekuban, S.A.; Yahayah, A.I.; et al. Molecular Epidemiology and Whole Genome Sequencing Analysis of Clinical Mycobacterium bovis from Ghana. PLoS ONE 2019, 14, e0209395. [Google Scholar] [CrossRef]
- De Jong, B.C.; Adetifa, I.; Walther, B.; Hill, P.C.; Antonio, M.; Ota, M.; Adegbola, R.A. Differences between Tuberculosis Cases Infected with Mycobacterium africanum, West African Type 2, Relative to Euro-American Mycobacterium tuberculosis: An Update. FEMS Immunol. Med. Microbiol. 2010, 58, 102–105. [Google Scholar] [CrossRef]
- Palisson, A.; Courcoul, A.; Durand, B. Role of Cattle Movements in Bovine Tuberculosis Spread in France between 2005 and 2014. PLoS ONE 2016, 11, e0152578. [Google Scholar] [CrossRef]
- Pereira, A.C.; Reis, A.C.; Ramos, B.; Cunha, M.V. Animal Tuberculosis: Impact of Disease Heterogeneity in Transmission, Diagnosis and Control. Transbound. Emerg. Dis. 2020, 2019, 13539. [Google Scholar] [CrossRef]
- Jones, B.A.; Grace, D.; Kock, R.; Alonso, S.; Rushton, J.; Said, M.Y.; McKeever, D.; Mutua, F.; Young, J.; McDermott, J.; et al. Zoonosis Emergence Linked to Agricultural Intensification and Environmental Change. Proc. Natl. Acad. Sci. USA 2013, 110, 8399–8404. [Google Scholar] [CrossRef] [PubMed]
- Gopal, R.; Goodchild, A.; Hewinson, G.; de la Rua Domenech, R.; Clifton-Hadley, R. Introduction of Bovine Tuberculosis to North-East England by Bought-in Cattle. Vet. Rec. 2006, 159, 265–271. [Google Scholar] [CrossRef] [PubMed]
- Brooks-Pollock, E.; Roberts, G.O.; Keeling, M.J. A Dynamic Model of Bovine Tuberculosis Spread and Control in Great Britain. Nature 2014, 511, 228–231. [Google Scholar] [CrossRef]
- Reynolds, D. A Review of Tuberculosis Science and Policy in Great Britain. Vet. Microbiol. 2006, 112, 119–126. [Google Scholar] [CrossRef] [PubMed]
- Mosavari, N.; Feizabadi, M.M.; Jamshidian, M.; Shahpouri, M.R.S.; Forbes, K.J.; Pajoohi, R.A.; Keshavarz, R.; Taheri, M.M.; Tadayon, K. Molecular Genotyping and Epidemiology of Mycobacterium bovis Strains Obtained from Cattle in Iran. Vet. Microbiol. 2011, 151, 148–152. [Google Scholar] [CrossRef]
- Pozo, P.; VanderWaal, K.; Grau, A.; de la Cruz, M.L.; Nacar, J.; Bezos, J.; Perez, A.; Minguez, O.; Alvarez, J. Analysis of the Cattle Movement Network and Its Association with the Risk of Bovine Tuberculosis at the Farm Level in Castilla y Leon, Spain. Transbound. Emerg. Dis. 2019, 66, 327–340. [Google Scholar] [CrossRef]
- Sylvester, T.T.; Martin, L.E.R.; Buss, P.; Loxton, A.G.; Hausler, G.A.; Rossouw, L.; van Helden, P.; Parsons, S.D.C.; Olea-Popelka, F.; Miller, M.A. Prevalence and Risk Factors for Mycobacterium bovis Infection in African Lions (Panthera leo) in the Kruger National Park. J. Wildl. Dis. 2017, 53, 372–376. [Google Scholar] [CrossRef]
- Botha, L.; Gey van Pittius, N.C.; van Helden, P.D. Mycobacteria and Disease in Southern Africa. Transbound. Emerg. Dis. 2013, 60 (Suppl. S1), 147–156. [Google Scholar] [CrossRef]
- Corner, L.A.L.; Murphy, D.; Gormley, E. Mycobacterium bovis Infection in the Eurasian Badger (Meles meles): The Disease, Pathogenesis, Epidemiology and Control. J. Comp. Pathol. 2011, 144, 1–24. [Google Scholar] [CrossRef]
- Corner, L.A.L.; O’Meara, D.; Costello, E.; Lesellier, S.; Gormley, E. The Distribution of Mycobacterium bovis Infection in Naturally Infected Badgers. Vet. J. 2012, 194, 166–172. [Google Scholar] [CrossRef]
- Turgenbayev, K.A.; Borsynbayeva, A.M.; Plazun, A.A.; Turgenbayev, R.K. Tuberculosis Prevalence in Animals and Humans in the Republic of Kazakhstan. Vet. World 2021, 14, 2362–2370. [Google Scholar] [CrossRef] [PubMed]
- Luciano, S.A.; Roess, A. Human Zoonotic Tuberculosis and Livestock Exposure in Low- and Middle-Income Countries: A Systematic Review Identifying Challenges in Laboratory Diagnosis. Zoonoses Public Health 2020, 67, 97–111. [Google Scholar] [CrossRef]
- Sichewo, P.R.; Etter, E.M.C.C.; Michel, A.L. Wildlife-Cattle Interactions Emerge as Drivers of Bovine Tuberculosis in Traditionally Farmed Cattle. Prev. Vet. Med. 2020, 174, 104847. [Google Scholar] [CrossRef]
- Sichewo, P.R.; Hlokwe, T.M.; Etter, E.M.C.C.; Michel, A.L. Tracing Cross Species Transmission of Mycobacterium bovis at the Wildlife / Livestock Interface in South Africa. BMC Microbiol. 2020, 20, 49. [Google Scholar] [CrossRef]
- Etter, E.; Donado, P.; Jori, F.; Caron, A.; Goutard, F.; Roger, F. Risk Analysis and Bovine Tuberculosis, a Re-Emerging Zoonosis. Ann. N. Y. Acad. Sci. 2006, 1081, 61–73. [Google Scholar] [CrossRef]
- Thoen, C.; LoBue, P.; De Kantor, I. The Importance of Mycobacterium bovis as a Zoonosis. Vet. Microbiol. 2006, 112, 339–345. [Google Scholar] [CrossRef]
- Awah Ndukum, J.; Caleb Kudi, A.; Bradley, G.; Ane-Anyangwe, I.N.; Fon-Tebug, S.; Tchoumboue, J. Prevalence of Bovine Tuberculosis in Abattoirs of the Littoral and Western Highland Regions of Cameroon: A Cause for Public Health Concern. Vet. Med. Int. 2010, 2010, 495015. [Google Scholar] [CrossRef]
- Ali, Z.I.; Hanafy, M.; Hansen, C.; Saudi, A.M.; Talaat, A.M. Genotypic Analysis of Nontuberculous Mycobacteria Isolated from Raw Milk and Human Cases in Wisconsin. J. Dairy Sci. 2020, 104, 211–220. [Google Scholar] [CrossRef]
- Collins, Á.B.; Floyd, S.; Gordon, S.V.; More, S.J. Prevalence of Mycobacterium bovis in Milk on Dairy Cattle Farms: An International Systematic Literature Review and Meta-Analysis. Tuberculosis 2022, 132, 102166. [Google Scholar] [CrossRef] [PubMed]
- Bapat, P.R.; Dodkey, R.S.; Shekhawat, S.D.; Husain, A.A.; Nayak, A.R.; Kawle, A.P.; Daginawala, H.F.; Singh, L.K.; Kashyap, R.S. Prevalence of Zoonotic Tuberculosis and Associated Risk Factors in Central Indian Populations. J. Epidemiol. Glob. Health 2017, 7, 277–283. [Google Scholar] [CrossRef] [PubMed]
- Desta, G.B.; Jena, P.K.; Hassen, H.M.; Behera, M.R. Factors Associated with Zoonosis and Reverse Zoonosis of Mycobacterium tuberculosis and Mycobacterium bovis in Ethiopia: A Systematic Review and Meta-Analysis. Int. J. Health Sci. 2022, 6, 1630–1653. [Google Scholar] [CrossRef]
- Tober, A.V.; Govender, D.; Russo, I.-R.M.; Cable, J. The Microscopic Five of the Big Five: Managing Zoonotic Diseases within and beyond African Wildlife Protected Areas. Adv. Parasitol. 2022, 117, 1–46. [Google Scholar] [CrossRef] [PubMed]
- Dürr, S.; Müller, B.; Alonso, S.; Hattendorf, J.; Laisse, C.J.M.; van Helden, P.D.; Zinsstag, J. Differences in Primary Sites of Infection between Zoonotic and Human Tuberculosis: Results from a Worldwide Systematic Review. PLoS Negl. Trop. Dis. 2013, 7, e2399. [Google Scholar] [CrossRef] [PubMed]
- Thoen, C.O.; LoBue, P.A.; Enarson, D.A. Tuberculosis in Animals and Humans: An Overview. Int. J. Tuberc. Lung Dis. 2010, 14, 1075–1078. [Google Scholar] [CrossRef]
- Carruth, L.; Roess, A.A.; Mekonnen, Y.T.; Melaku, S.K.; Nichter, M.; Salman, M. Zoonotic Tuberculosis in Africa: Challenges and Ways Forward. Lancet 2016, 388, 2460–2461. [Google Scholar] [CrossRef]
- Kock, R.; Michel, A.L.; Yeboah-Manu, D.; Azhar, E.I.; Torrelles, J.B.; Cadmus, S.I.; Brunton, L.; Chakaya, J.M.; Marais, B.; Mboera, L.; et al. Zoonotic Tuberculosis–The Changing Landscape. Int. J. Infect. Dis. 2021, 113, S68–S72. [Google Scholar] [CrossRef]
- van den Brom, R.; de Jong, A.; van Engelen, E.; Heuvelink, A.; Vellema, P. Zoonotic Risks of Pathogens from Sheep and Their Milk Borne Transmission. Small Rumin. Res. 2020, 189, 106123. [Google Scholar] [CrossRef]
- Drain, P.K.; Bajema, K.L.; Dowdy, D.; Dheda, K.; Naidoo, K.; Schumacher, S.G.; Ma, S.; Meermeier, E.; Lewinsohn, D.M.; Sherman, D.R.; et al. Incipient and Subclinical Tuberculosis: A Clinical Review of Early Stages and Progression of Infection. Clin. Microbiol. Rev. 2018, 31, e00021-18. [Google Scholar] [CrossRef]
- Dutta, N.K.; Karakousis, P.C. Latent Tuberculosis Infection: Myths, Models, and Molecular Mechanisms. Microbiol. Mol. Biol. Rev. 2014, 78, 343–371. [Google Scholar] [CrossRef]
- Furin, J.; Cox, H.; Pai, M. Tuberculosis. Lancet 2019, 393, 1642–1656. [Google Scholar] [CrossRef]
- Esmail, H.; Barry, C.E.; Wilkinson, R.J. Understanding Latent Tuberculosis: The Key to Improved Diagnostic and Novel Treatment Strategies. Drug Discov. Today 2012, 17, 514–521. [Google Scholar] [CrossRef] [PubMed]
- Suárez, I.; Fünger, S.M.; Kröger, S.; Rademacher, J.; Gerd Fätkenheuer, J.; Rybniker, A. The Diagnosis and Treatment of Tuberculosis. Dtsch. Arztebl. Int. 2019, 116, 729–735. [Google Scholar] [CrossRef] [PubMed]
- Zumla, A.I.; Lawn, S.D. Tuberculosis. Lancet 2011, 378, 57–72. [Google Scholar] [CrossRef] [PubMed]
- Faryar, K.A.; Braun, R.; Ancona, R.M.; Ajayi, E.; Bryant, W.; Rehman, S.; Sall, H.; Lyons, M.S.; Huaman, M.A. Emergency Department Screening for Latent Tuberculosis Infection. Am. J. Emerg. Med. 2021, 54, 323.e5–323.e8. [Google Scholar] [CrossRef]
- Houben, R.M.G.J.; Dodd, P.J. The Global Burden of Latent Tuberculosis Infection: A Re-Estimation Using Mathematical Modelling. PLoS Med. 2016, 13, e1002152. [Google Scholar] [CrossRef] [Green Version]
- Bañuls, A.L.; Sanou, A.; Van Anh, N.T.; Godreuil, S. Mycobacterium tuberculosis: Ecology and Evolution of a Human Bacterium. J. Med. Microbiol. 2015, 64, 1261–1269. [Google Scholar] [CrossRef]
- Hershberg, R.; Lipatov, M.; Small, P.M.; Sheffer, H.; Niemann, S.; Homolka, S.; Roach, J.C.; Kremer, K.; Petrov, D.A.; Feldman, M.W.; et al. High Functional Diversity in Mycobacterium tuberculosis Driven by Genetic Drift and Human Demography. PLoS Biol. 2008, 6, 2658–2671. [Google Scholar] [CrossRef]
- Parbhoo, T.; Sampson, S.L.; Mouton, J.M. Recent Developments in the Application of Flow Cytometry to Advance Our Understanding of Mycobacterium tuberculosis Physiology and Pathogenesis. Cytom. Part A 2020, 97, 683–693. [Google Scholar] [CrossRef]
- Gengenbacher, M.; Kaufmann, S.H.E. Mycobacterium tuberculosis: Success through Dormancy. FEMS Microbiol. Rev. 2012, 36, 514–532. [Google Scholar] [CrossRef]
- Borah, K.; Xu, Y.; McFadden, J. Dissecting Host-Pathogen Interactions in TB Using Systems-Based Omic Approaches. Front. Immunol. 2021, 12, 762315. [Google Scholar] [CrossRef]
- Gagneux, S.; DeRiemer, K.; Van, T.; Kato-Maeda, M.; De Jong, B.C.; Narayanan, S.; Nicol, M.; Niemann, S.; Kremeri, K.; Gutierrez, M.C.; et al. Variable Host-Pathogen Compatibility in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 2006, 103, 2869–2873. [Google Scholar] [CrossRef]
- Manina, G.; Dhar, N.; McKinney, J.D. Stress and Host Immunity Amplify Mycobacterium tuberculosis Phenotypic Heterogeneity and Induce Nongrowing Metabolically Active Forms. Cell Host Microbe 2015, 17, 32–46. [Google Scholar] [CrossRef] [PubMed]
- Laureillard, D.; Marcy, O.; Madec, Y.; Chea, S.; Chan, S.; Borand, L.; Fernandez, M.; Prak, N.; Kim, C.; Dim, B.; et al. Paradoxical Tuberculosis-Associated Immune Reconstitution Inflammatory Syndrome after Early Initiation of Antiretroviral Therapy in a Randomized Clinical Trial. Aids 2013, 27, 2577–2586. [Google Scholar] [CrossRef] [PubMed]
- Varela-Castro, L.; Barral, M.; Arnal, M.C.; Luco, D.F.D.; Gortázar, C.; Garrido, J.M.; Sevilla, I.A. Beyond Tuberculosis: Diversity and Implications of Non-Tuberculous Mycobacteria at the Wildlife-Livestock Interface. Transbound. Emerg. Dis. 2022. [Google Scholar] [CrossRef]
- Keet, D.F.; Michel, A.L.; Bengis, R.G.; Becker, P.; van Dyk, D.S.; van Vuuren, M.; Rutten, V.P.M.G.; Penzhorn, B.L. Intradermal Tuberculin Testing of Wild African Lions (Panthera leo) Naturally Exposed to Infection with Mycobacterium bovis. Vet. Microbiol. 2010, 144, 384–391. [Google Scholar] [CrossRef] [PubMed]
- Riska, P.F.; Carleton, S. Latent Tuberculosis: Models, Mechanisms, and Novel Prospects for Eradication. Semin. Pediatr. Infect. Dis. 2002, 13, 263–272. [Google Scholar] [CrossRef]
- Meier, N.R.; Jacobsen, M.; Ottenhoff, T.H.M.; Ritz, N. A Systematic Review on Novel Mycobacterium tuberculosis Antigens and Their Discriminatory Potential for the Diagnosis of Latent and Active Tuberculosis. Front. Immunol. 2018, 9, 2476. [Google Scholar] [CrossRef]
- Helaine, S.; AM, C.; KG, W.; LM, F.; SA, M.; DW, H.; Helaine, S.; Cheverton, A.M.; Watson, K.G.; Faure, L.M.; et al. Internalization of Salmonella by Macrophages Induces Formation of Nonreplicating Persisters. Science 2014, 343, 204–208. [Google Scholar] [CrossRef] [PubMed]
- Mouton, J.M.; Helaine, S.; Holden, D.W.; Sampson, S.L. Elucidating Population-Wide Mycobacterial Replication Dynamics at the Single-Cell Level. Microbiology 2016, 162, 966–978. [Google Scholar] [CrossRef]
- Chengalroyen, M.D.; Beukes, G.M.; Gordhan, B.G.; Streicher, E.M.; Churchyard, G.; Hafner, R.; Warren, R.; Otwombe, K.; Martinson, N.; Kana, B.D. Detection and Quantification of Differentially Culturable Tubercle Bacteria in Sputum from Patients with Tuberculosis. Am. J. Respir. Crit. Care Med. 2016, 194, 1532–1540. [Google Scholar] [CrossRef]
- Chao, M.C.; Rubin, E.J. Letting Sleeping Dos Lie: Does Dormancy Play a Role in Tuberculosis? Annu. Rev. Microbiol. 2010, 64, 293–311. [Google Scholar] [CrossRef]
- Hice, S.A.; Santoscoy, M.C.; Soupir, M.L.; Cademartiri, R. Distinguishing between Metabolically Active and Dormant Bacteria on Paper. Appl. Microbiol. Biotechnol. 2018, 102, 367–375. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y. Persisters, Persistent Infections and the Yin-Yang Model. Emerg. Microbes Infect. 2014, 3, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Brauner, A.; Fridman, O.; Gefen, O.; Balaban, N.Q. Distinguishing between Resistance, Tolerance and Persistence to Antibiotic Treatment. Nat. Rev. Microbiol. 2016, 14, 320–330. [Google Scholar] [CrossRef] [PubMed]
- Balaban, N.Q.; Gerdes, K.; Lewis, K.; McKinney, J.D. A Problem of Persistence: Still More Questions than Answers? Nat. Rev. Microbiol. 2013, 11, 587–591. [Google Scholar] [CrossRef] [PubMed]
- Balaban, N.Q.; Merrin, J.; Chait, R.; Kowalik, L.; Leibler, S. Bacterial Persistence as a Phenotypic Switch. Science 2004, 305, 1622–1625. [Google Scholar] [CrossRef]
- Li, L.; Mendis, N.; Trigui, H.; Oliver, J.D.; Sebastien, P. The Importance of the Viable but Non-Culturable State in Human Bacterial Pathogens. Front. Microbiol. 2014, 5, 258. [Google Scholar] [CrossRef]
- Buddle, B.M.; Vordermeier, H.M.; Hewinson, R.G. Experimental Infection Models of Tuberculosis in Domestic Livestock. Microbiol. Spectr. 2016, 4, 4. [Google Scholar] [CrossRef]
- Domingo, M.; Vidal, E.; Marco, A. Pathology of Bovine Tuberculosis. Res. Vet. Sci. 2014, 97, S20–S29. [Google Scholar] [CrossRef]
- Thoen, C.O. Tuberculosis in Wild and Domestic Mammals. Tuberculosis 1994, 157–162. [Google Scholar] [CrossRef]
- Courtenay, O.; Reilly, L.A.; Sweeney, F.P.; Hibberd, V.; Bryan, S.; Ul-Hassan, A.; Newman, C.; Macdonald, D.W.; Delahay, R.J.; Wilson, G.J.; et al. Is Mycobacterium bovis in the Environment Important for the Persistence of Bovine Tuberculosis? Biol. Lett. 2006, 2, 460–462. [Google Scholar] [CrossRef] [PubMed]
- Ghodbane, R.; Medie, F.M.; Lepidi, H.; Nappez, C.; Drancourt, M. Long-Term Survival of Tuberculosis Complex Mycobacteria in Soil. Microbiology 2014, 160 Pt 3, 496–501. [Google Scholar] [CrossRef] [PubMed]
- Lyashchenko, K.P.; Sikar-Gang, A.; Sridhara, A.A.; Johnathan-Lee, A.; Elahi, R.; Lambotte, P.; Esfandiari, J.; Duthi, M.; Reed, S.G.; Jones, G.; et al. Novel Polyprotein Antigens Designed for Improved Serodiagnosis of Bovine Tuberculosis. Vet. Immunol. Immunopathol. 2021, 240, 110320. [Google Scholar] [CrossRef]
- Lyashchenko, K.P.; Gortázar, C.; Miller, M.A.; Waters, W.R. Spectrum of Antibody Profiles in Tuberculous Elephants, Cervids, and Cattle. Vet. Microbiol. 2018, 214, 89–92. [Google Scholar] [CrossRef] [PubMed]
- Thomas, R.; Chambers, M. Review of Methods Used for Diagnosing Tuberculosis in Captive and Free-Ranging Non-Bovid Species (2012–2020). Pathogens 2021, 10, 584. [Google Scholar] [CrossRef] [PubMed]
- Boadella, M.; Lyashchenko, K.; Greenwald, R.; Esfandiari, J.; Jaroso, R.; Carta, T.; Garrido, J.M.; Vicente, J.; de la Fuente, J.; Gortázar, C. Serologic Tests for Detecting Antibodies against Mycobacterium bovis and Mycobacterium avium Subspecies Paratuberculosis in Eurasian Wild Boar (Sus scrofa scrofa). J. Vet. Diagn. Investig. 2011, 23, 77–83. [Google Scholar] [CrossRef] [PubMed]
- Higgitt, R.L.; Buss, P.E.; Van Helden, P.D.; Miller, M.A.; Parsons, S.D.C. Development of Gene Expression Assays Measuring Immune Responses in the Spotted Hyena (Crocuta crocuta). Afr. Zool. 2017, 52, 99–104. [Google Scholar] [CrossRef]
- Whelan, A.O.; Coad, M.; Cockle, P.J.; Hewinson, G.; Vordermeier, M.; Gordon, S.V. Revisiting Host Preference in the Mycobacterium tuberculosis Complex: Experimental Infection Shows Mycobacterium tuberculosis H37Rv to Be Avirulent in Cattle. PLoS ONE 2010, 5, e8527. [Google Scholar] [CrossRef]
- Parikka, M.; Hammarén, M.M.; Harjula, S.-K.E.; Halfpenny, N.J.A.; Oksanen, K.E.; Lahtinen, M.J.; Pajula, E.T.; Iivanainen, A.; Pesu, M.; Rämet, M. Mycobacterium marinum Causes a Latent Infection That Can Be Reactivated by Gamma Irradiation in Adult Zebrafish. PLoS Pathog. 2012, 8, e1002944. [Google Scholar] [CrossRef]
- Ramos, D.F.; Silva, P.E.A.; Dellagostin, O.A. Diagnosis of Bovine Tuberculosis: Review of Main Techniques. Braz. J. Biol. 2015, 75, 830–837. [Google Scholar] [CrossRef]
- Warren, R.M.; Gey Van Pittius, N.C.; Barnard, M.; Hesseling, A.; Engelke, E.; De Kock, M.; Gutierrez, M.C.; Chege, G.K.; Victor, T.C.; Hoal, E.G.; et al. Differentiation of Mycobacterium tuberculosis Complex by PCR Amplification of Genomic Regions of Difference. Int. J. Tuberc. Lung Dis. 2006, 10, 818–822. [Google Scholar]
- Gong, W.; Wu, X. Differential Diagnosis of Latent Tuberculosis Infection and Active Tuberculosis: A Key to a Successful Tuberculosis Control Strategy. Front. Microbiol. 2021, 12, 745592. [Google Scholar] [CrossRef]
- Bigi, F.; Zumárraga, J.; Cataldi, A.A.; Soria, M.A. Polymorphisms of Twenty Regulatory Proteins between Mycobacterium tuberculosis and Mycobacterium bovis. Microbiol. Immunol. 2016, 60, 552–560. [Google Scholar] [CrossRef]
- Li, P.; Wang, R.; Dong, W.; Hu, L.; Zong, B.; Zhang, Y.; Wang, X.; Guo, A.; Zhang, A.; Xiang, Y.; et al. Comparative Proteomics Analysis of Human Macrophages Infected with Virulent Mycobacterium bovis. Front. Cell. Infect. Microbiol. 2017, 7, 65. [Google Scholar] [CrossRef] [PubMed]
- Meiring, C.; Van Helden, P.; Goosen, W.J. TB Control in Humans and Animals in South Africa: A Perspective on Problems and Successes. Front. Vet. Sci. 2018, 5, 298. [Google Scholar] [CrossRef] [PubMed]
- Lim, A.; Steibel, J.P.; Coussens, P.M.; Grooms, D.L.; Bolin, S.R. Differential Gene Expression Segregates Cattle Confirmed Positive for Bovine Tuberculosis from Antemortem Tuberculosis Test-False Positive Cattle Originating from Herds Free of Bovine Tuberculosis. Vet. Med. Int. 2012, 2012, 192926. [Google Scholar] [CrossRef] [PubMed]
- Patrick, B. Michigan Bovine Tuberculosis Eradication Project: Activities Report. 2003. Available online: https://digitalcommons.unl.edu/michbovinetb/12/ (accessed on 8 March 2022).
- Parsons, S.D.C.; Cooper, D.; Mccall, A.J.; Mccall, W.A.; Streicher, E.M.; Le Maitre, N.C.; Müller, A.; Gey Van Pittius, N.C.; Warren, R.M.; Van Helden, P.D. Modification of the QuantiFERON-TB Gold (In-Tube) Assay for the Diagnosis of Mycobacterium bovis Infection in African buffaloes (Syncerus caffer). Vet. Immunol. Immunopathol. 2011, 142, 113–118. [Google Scholar] [CrossRef] [PubMed]
- Pai, M.; Denkinger, C.M.; Kik, S.V.; Rangaka, M.X.; Zwerling, A.; Oxlade, O.; Metcalfe, J.Z.; Cattamanchi, A.; Dowdy, D.W.; Dheda, K.; et al. Gamma Interferon Release Assays for Detection of Mycobacterium tuberculosis Infection. Clin. Microbiol. Rev. 2014, 27, 3–20. [Google Scholar] [CrossRef]
- Denis, M.; Wedlock, D.N.; McCarthy, A.R.; Parlane, N.A.; Cockle, P.J.; Vordermeier, H.M.; Hewinson, R.G.; Buddle, B.M. Enhancement of the Sensitivity of the Whole-Blood Gamma Interferon Assay for Diagnosis of Mycobacterium bovis Infections in Cattle. Clin. Vaccine Immunol. 2007, 14, 1483–1489. [Google Scholar] [CrossRef]
- Clegg, T.A.; Good, M.; Doyle, M.; Duignan, A.; More, S.J.; Gormley, E. The Performance of the Interferon Gamma Assay When Used as a Diagnostic or Quality Assurance Test in Mycobacterium bovis Infected Herds. Prev. Vet. Med. 2017, 140, 116–121. [Google Scholar] [CrossRef]
- Frederico, O.; Id, I.; Soares, P.; Yumi, C.; Id, I.; Machado, A.; Ferreira, S.; Id, N.; Id, B.H.; Acha, S.J.; et al. Evolutionary Analysis of Mycobacterium bovis Genotypes across Africa Suggests Co-Evolution with Livestock and Humans. PLoS Negl. Trop. Dis. 2020, 14, e0008081. [Google Scholar] [CrossRef]
- Sukumar, R.; Joshi, B.; Lagrange, P.H.; Ophe Longuet, C.; Verma-Kumar, S.; Abraham, D.; Dendukuri, N.; Cheeran, J.V.; Narayanaswamy Balaji, K. Serodiagnosis of Tuberculosis in Asian Elephants (Elephas maximus) in Southern India: A Latent Class Analysis. PLoS ONE 2012, 7, e49548. [Google Scholar] [CrossRef]
- Golby, P.; Hatch, K.A.; Bacon, J.; Cooney, R.; Riley, P.; Allnutt, J.; Hinds, J.; Nunez, J.; Marsh, P.D.; Hewinson, R.G.; et al. Comparative Transcriptomics Reveals Key Gene Expression Differences between the Human and Bovine Pathogens of the Mycobacterium tuberculosis Complex. Microbiology 2007, 153, 3323–3336. [Google Scholar] [CrossRef] [PubMed]
- Garnier, T.; Eiglmeier, K.; Camus, J.C.; Medina, N.; Mansoor, H.; Pryor, M.; Duthoy, S.; Grondin, S.; Lacroix, C.; Monsempe, C.; et al. The Complete Genome Sequence of Mycobacterium bovis. Proc. Natl. Acad. Sci. USA 2003, 100, 7877–7882. [Google Scholar] [CrossRef]
- Marimani, M.; Ahmad, A.; Duse, A. The Role of Epigenetics, Bacterial and Host Factors in Progression of Mycobacterium tuberculosis Infection. Tuberculosis 2018, 113, 200–214. [Google Scholar] [CrossRef]
- Keating, L.A.; Wheeler, P.R.; Mansoor, H.; Inwald, J.K.; Dale, J.; Hewinson, R.G.; Gordon, S.V. The Pyruvate Requirement of Some Members of the Mycobacterium tuberculosis Complex Is Due to an Inactive Pyruvate Kinase: Implications for in vivo Growth. Mol. Microbiol. 2005, 56, 163–174. [Google Scholar] [CrossRef]
- Sohaskey, C.D.; Modesti, L. Differences in Nitrate Reduction between Mycobacterium tuberculosis and Mycobacterium bovis Are Due to Differential Expression of Both NarGHJI and NarK2. FEMS Microbiol. Lett. 2009, 290, 129–134. [Google Scholar] [CrossRef]
- Queval, C.J.; Fearns, A.; Botella, L.; Smyth, A.; Schnettger, L.; Mitermite, M.; Wooff, E.; Villarreal-Ramos, B.; Garcia-Jimenezi, W.; Heunis, T.; et al. Macrophage-Specific Responses to Humanand Animal-Adapted Tubercle Bacilli Reveal Pathogen and Host Factors Driving Multinucleated Cell Formation. PLoS Pathog. 2021, 17, e1009410. [Google Scholar] [CrossRef]
- Cheng, G.; Hussain, T.; Sabir, N.; Ni, J.; Li, M.; Zhao, D.; Zhou, X. Comparative Study of the Molecular Basis of Pathogenicity of Mycobacterium bovis Strains in a Mouse Model. Int. J. Mol. Sci. 2019, 20, 5. [Google Scholar] [CrossRef]
- Nedeltchev, G.G.; Raghunand, T.R.; Jassal, M.S.; Lun, S.; Cheng, Q.J.; Bishai, W.R. Extrapulmonary Dissemination of Mycobacterium bovis but Not Mycobacterium tuberculosis in a Bronchoscopic Rabbit Model of Cavitary Tuberculosis. Infect. Immun. 2009, 77, 598–603. [Google Scholar] [CrossRef]
- Palmer, M.V.; Thacker, T.C.; Kanipe, C.; Boggiatto, P.M.; Palmer, M.V. Heterogeneity of Pulmonary Granulomas in Cattle Experimentally Infected with Mycobacterium bovis. Front. Vet. Sci. 2021, 8, 671460. [Google Scholar] [CrossRef] [PubMed]
- Mouton, J.M.; Heunis, T.; Dippenaar, A.; Gallant, J.L.; Kleynhans, L.; Sampson, S.L. Comprehensive Characterization of the Attenuated Double Auxotroph Mycobacterium tuberculosisΔleudΔpanCD as an Alternative to H37Rv. Front. Microbiol. 2019, 10, 1922. [Google Scholar] [CrossRef] [PubMed]
- Voskuil, M.I.; Schnappinger, D.; Visconti, K.C.; Harrell, M.I.; Dolganov, G.M.; Sherman, D.R.; Schoolnik, G.K. Inhibition of Respiration by Nitric Oxide Induces a Mycobacterium tuberculosis Dormancy Program. J. Exp. Med. 2003, 198, 705–713. [Google Scholar] [CrossRef]
- Tullius, M.V.; Harth, G.; Horwitz, M.A. Glutamine Synthetase GlnA1 Is Essential for Growth of Mycobacterium tuberculosis in Human THP-1 Macrophages and Guinea Pigs. Infect. Immun. 2003, 71, 3927–3936. [Google Scholar] [CrossRef]
- Nozaki, Y.; Hasegawa, Y.; Ichiyama, S.; Nakashima, I.; Shimokata, K. Mechanism of Nitric Oxide-Dependent Killing of Mycobacterium bovis BCG in Human Alveolar Macrophages. Infect. Immun. 1997, 65, 3644–3647. [Google Scholar] [CrossRef] [PubMed]
- Denis, M. Interferon-Gamma-Treated Murine Macrophages Inhibit Growth of Tubercle Bacilli via the Generation of Reactive Nitrogen Intermediates. Cell. Immunol. 1991, 132, 150–157. [Google Scholar] [CrossRef]
- Hutter, B.; Dick, T. Up-Regulation of NarX, Encoding a Putative “fused Nitrate Reductase” in Anaerobic Dormant Mycobacterium bovis BCG. FEMS Microbiol. Lett. 1999, 178, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Boon, C.; Dick, T. Mycobacterium bovis BCG Response Regulator Essential for Hypoxic Dormancy. J. Bacteriol. 2002, 184, 6760–6767. [Google Scholar] [CrossRef]
- Slavchev, G.; Michailova, L.; Markova, N. Stress-Induced L-Forms of Mycobacterium bovis: A Challenge to Survivability. New Microbiol. 2013, 36, 157–166. [Google Scholar]
- Rodriguez, J.G.; Burbano, C.S.; Nuñez, C.; González, C.E.; Zambrano, M.M.; García, M.J.; Del Portillo, P. Rv3134c/DevR/DevS Operon of Mycobacterium bovis BCG Is Differentially Transcribed under “in vitro” Stress Conditions. Tuberculosis 2008, 88, 273–282. [Google Scholar] [CrossRef]
- Malherbe, S.T.; Shenai, S.; Ronacher, K.; Loxton, A.G.; Dolganov, G.; Kriel, M.; Van, T.; Chen, R.Y.; Warwick, J.; Via, E.; et al. Persisting PET-CT Lesion Activity and Mycobacterium tuberculosis mRNA after Pulmonary Tuberculosis Cure. Nat. Med. 2016, 22, 1094–1100. [Google Scholar] [CrossRef] [PubMed]
- Scanga, C.A.; Mohan, V.P.; Joseph, H.; Yu, K.; Chan, J.; Flynn, J.L. Reactivation of Latent Tuberculosis: Variations on the Cornell Murine Model. Infect. Immun. 1999, 67, 4531–4538. [Google Scholar] [CrossRef] [PubMed]
- Palacios, J.J.; Navarro, Y.; Romero, B.; Penedo, A.; Menéndez González, Á.; Pérez Hernández, M.D.; Fernández-Verdugo, A.; Copano, F.; Torreblanca, A.; Bouza, E.; et al. Molecular and Epidemiological Population-Based Integrative Analysis of Human and Animal Mycobacterium bovis Infections in a Low-Prevalence Setting. Vet. Microbiol. 2016, 195, 30–36. [Google Scholar] [CrossRef]
- Larsen, M.V.; Sørensen, I.J.; Thomsen, V.; Ravn, P. Re-Activation of Bovine Tuberculosis in a Patient Treated with Infliximab. Eur. Respir. J. 2008, 32, 229–231. [Google Scholar] [CrossRef]
- Cosivi, O.; Grange, J.M.; Daborn, C.J.; Raviglione, M.C.; Fujikura, T.; Cousins, D.; Robinson, R.A.; Huchzermeyer, H.F.A.K.; De Kantor, I.; Meslin, F.X. Zoonotic Tuberculosis Due to Mycobacterium bovis in Developing Countries. Emerg. Infect. Dis. 1998, 4, 59–70. [Google Scholar] [CrossRef]
- Shahmohammadi, S.; Saffar, M.J.; Rezai, M.S. BCG-Osis after BCG Vaccination in Immunocompromised Children: Case Series and Review. J. Pediatr. Rev. 2014, 2, 62–74. [Google Scholar] [CrossRef]
- Bhola, R.K.; Sarangi, R.; Dey, P.; Samal, P. Disseminated BCG-Osis with Haemophagocytosis, Tubercular Bacteraemia, and Unusual Haematological Findings with Its Haematology Analyser-Based Expression. J. Hematop. 2018, 11, 87–92. [Google Scholar] [CrossRef] [Green Version]
- Gibson, S.E.R.; Harrison, J.; Cox, J.A.G. Modelling a Silent Epidemic: A Review of the in vitro Models of Latent Tuberculosis. Pathogens 2018, 7, 88. [Google Scholar] [CrossRef]
- Alnimr, A.M. Dormancy Models for Mycobacterium tuberculosis: A Minireview. Brazilian J. Microbiol. 2015, 46, 641–647. [Google Scholar] [CrossRef]
- Kundu, M.; Basu, J. Applications of Transcriptomics and Proteomics for Understanding Dormancy and Resuscitation in Mycobacterium tuberculosis. Front. Microbiol. 2021, 12, 642487. [Google Scholar] [CrossRef]
- Gideon, H.P.; Wilkinson, K.A.; Rustad, T.R.; Oni, T.; Guio, H.; Kozak, R.A.; Sherman, D.R.; Meintjes, G.; Behr, M.A.; Vordermeier, H.M.; et al. Hypoxia Induces an Immunodominant Target of Tuberculosis Specific T Cells Absent from Common BCG Vaccines. PLoS Pathog. 2010, 6, e1001237. [Google Scholar] [CrossRef]
- Deb, C.; Lee, C.M.; Dubey, V.S.; Daniel, J.; Abomoelak, B.; Sirakova, T.D.; Pawar, S.; Rogers, L.; Kolattukudy, P.E. A Novel in vitro Multiple-Stress Dormancy Model for Mycobacterium tuberculosis Generates a Lipid-Loaded, Drug-Tolerant, Dormant Pathogen. PLoS ONE 2009, 4, e6077. [Google Scholar] [CrossRef] [PubMed]
- Rao, M.; Streur, T.L.; Aldwell, F.E.; Cook, G.M. Intracellular PH Regulation by Mycobacterium smegmatis and Mycobacterium bovis BCG. Microbiology. 2001, 147, 1017–1024. [Google Scholar] [CrossRef] [PubMed]
- Gallant, J.L.; Viljoen, A.J.; Van Helden, P.D.; Wiid, I.J.F. Glutamate Dehydrogenase Is Required by Mycobacterium bovis BCG for Resistance to Cellular Stress. PLoS ONE 2016, 11, 147706. [Google Scholar] [CrossRef] [PubMed]
- Weiss, D.J.; Evanson, O.A.; Moritz, A.; Deng, M.Q.; Abrahamsen, M.S. Differential Responses of Bovine Macrophages to Mycobacterium avium Subsp. paratuberculosis and Mycobacterium avium Subsp. avium. Infect. Immun. 2002, 70, 5556–5561. [Google Scholar] [CrossRef] [PubMed]
- Piercy, J.; Werling, D.; Coffey, T.J. Differential Responses of Bovine Macrophages to Infection with Bovine-Specific and Non-Bovine Specific Mycobacteria. Tuberculosis 2007, 87, 415–420. [Google Scholar] [CrossRef]
- Medley, J.; Goff, A.; Bettencourt, P.J.G.; Dare, M.; Cole, L.; Cantillon, D.; Waddell, S.J. Dissecting the Mycobacterium bovis BCG Response to Macrophage Infection to Help Prioritize Targets for Anti-Tuberculosis Drug and Vaccine Discovery. Vaccines 2022, 10, 113. [Google Scholar] [CrossRef]
- Zhan, X.; Yuan, W.; Zhou, Y.; Ma, R.; Ge, Z. Small RNA Sequencing and Bioinformatics Analysis of RAW264.7-Derived Exosomes after Mycobacterium bovis Bacillus Calmette-Guérin Infection. BMC Genom. 2022, 23, 355. [Google Scholar] [CrossRef]
- Lin, W.; Xin, T. The Influence of Gene Rv3671c in Mycobacterium bovis to Its Replication and Acid Resistance. bioRxiv 2020. [Google Scholar] [CrossRef]
- Ma, R.; Farrell, D.; Gonzalez, G.; Browne, J.A.; Nakajima, C.; Suzuki, Y.; Gordon, S.V. The TbD1 Locus Mediates a Hypoxia-Induced Copper Response in Mycobacterium bovis. Front. Microbiol. 2022, 13, 817952. [Google Scholar] [CrossRef]
- Malone, K.M.; Rue-Albrecht, K.; Magee, D.A.; Conlon, K.; Schubert, O.T.; Nalpas, N.C.; Browne, J.A.; Smyth, A.; Gormley, E.; Aebersold, R.; et al. Comparative ’omics Analyses Differentiate Mycobacterium tuberculosis and Mycobacterium bovis and Reveal Distinct Macrophage Responses to Infection with the Human and Bovine Tubercle Bacilli. Microb. Genom. 2018, 4, 163. [Google Scholar] [CrossRef] [PubMed]
- Gibson, A.J.; Passmore, I.J.; Faulkner, V.; Xia, D.; Nobeli, I.; Stiens, J.; Willcocks, S.; Clark, T.G.; Sobkowiak, B.; Werling, D.; et al. Probing Differences in Gene Essentiality Between the Human and Animal Adapted Lineages of the Mycobacterium tuberculosis Complex Using TnSeq. Front. Vet. Sci. 2021, 8, 717. [Google Scholar] [CrossRef] [PubMed]
- Jones, G.J.; Pirson, C.; Gideon, H.P.; Wilkinson, K.A.; Sherman, D.R.; Wilkinson, R.J.; Hewinson, R.G.; Vordermeier, H.M. Immune Responses to the Enduring Hypoxic Response Antigen Rv0188 Are Preferentially Detected in Mycobacterium bovis Infected Cattle with Low Pathology. PLoS ONE 2011, 6, e21371. [Google Scholar] [CrossRef]
- Larsen, M.H.; Lacourciere, K.; Parker, T.M.; Kraigsley, A.; Achkar, M.; Adams, L.B.; Dupnik, K.M.; Hall-stoodley, L.; Kanipe, C.; Kurtz, S.L.; et al. The Many Hosts of Mycobacteria 8 (MHM8): A Conference Report. Tuberculosis 2020, 8, 101914. [Google Scholar] [CrossRef] [PubMed]
- León, A.D.; Zumárraga, M.J.; Jiménez Oropeza, R.; Gioffré, A.K.; Bernardelli, A.; Orozco Estévez, H.; Cataldi, A.A.; Hernández Pando, R. Mycobacterium bovis with Different Genotypes and from Different Hosts Induce Dissimilar Immunopathological Lesions in a Mouse Model of Tuberculosis. Clin. Exp. Immunol. 2009, 157, 139–147. [Google Scholar] [CrossRef] [PubMed]
- Phillips, C.J.C.; Foster, C.R.W.; Morris, P.A.; Teverson, R. The Transmission of Mycobacterium bovis Infection to Cattle. Res. Vet. Sci. 2003, 74, 1–15. [Google Scholar] [CrossRef]
- Alvarez Herrera, A. Are Cattle a Surrogate Model for Pathogenic Mycobacterial Latent Infection? Mycobact. Dis. 2014, 4, 4–5. [Google Scholar] [CrossRef] [Green Version]
- Ippolito, D.; Boniotti, M.B.; Fiasconaro, M.; Fontana, S.; Boifava, M.; Ciarello, F.P.; Amato, B.; Pacciarini, M.; Presti, V.D.M.L. Development and Evaluation of a Multi-Antigen Serological Assay for the Intra-Vitam Diagnosis of Tuberculosis Caused by Mycobacterium bovis in Pigs. J. Immunol. Methods 2022, 503, 113234. [Google Scholar] [CrossRef]
- Nol, P.; Wehtje, M.E.; Bowen, R.A.; Robbe-austerman, S.; Thacker, T.C.; Lantz, K.; Rhyan, J.C.; Baeten, L.A.; Juste, A.; Sevilla, I.A.; et al. Pathogens E Ff Ects of Inactivated Mycobacterium bovis Vaccination on Molokai-Origin Wild Pigs Experimentally Infected with Virulent M. Bovis. Pathogens 2020, 9, 199. [Google Scholar] [CrossRef]
- Bolin, C.A.; Whipple, D.L.; Khanna, K.V.; Risdahl, J.M.; Peterson, P.K.; Molitor, T.W. Infection of Swine with Mycobacterium bovis as a Model of Human Tuberculosis. J. Infect. Dis. 1997, 176, 1559–1566. [Google Scholar] [CrossRef]
- Gallagher, J.; Clifton-Hadley, R.S. Tuberculosis in Badgers; a Review of the Disease and Its Significance for Other Animals. Res. Vet. Sci. 2000, 69, 203–217. [Google Scholar] [CrossRef] [PubMed]
- Gallagher, J.; Monies, R.; Gavier-Widen, M.; Rule, B. Role of Infected, Non-Diseased Badgers in the Pathogenesis of Tuberculosis in the Badger. Vet. Rec. 1998, 142, 710–714. [Google Scholar] [CrossRef] [PubMed]
- Kupz, A.; Zedler, U.; Stäber, M.; Kaufmann, S.H.E. A Mouse Model of Latent Tuberculosis Infection to Study Intervention Strategies to Prevent Reactivation. PLoS ONE 2016, 11, e0158849. [Google Scholar] [CrossRef] [PubMed]
- Karakousis, P.C.; Yoshimatsu, T.; Lamichhane, G.; Woolwine, S.C.; Nuermberger, E.L.; Grosset, J.; Bishai, W.R. Dormancy Phenotype Displayed by Extracellular Mycobacterium tuberculosis within Artificial Granulomas in Mice. J. Exp. Med. 2004, 200, 647–657. [Google Scholar] [CrossRef]
- Zhan, L.; Tang, J.; Lin, S.; Xu, Y.; Xu, Y.; Qin, C. Prophylactic Use of Ganoderma Lucidum Extract May Inhibit Mycobacterium tuberculosis Replication in a New Mouse Model of Spontaneous Latent Tuberculosis Infection. Front. Microbiol. 2016, 6, 1490. [Google Scholar] [CrossRef]
- Rhoades, E.R. Progression of Chronic Pulmonary Tuberculosis in Mice Aerogenically Infected with Virulent Mycobacterium tuberculosis. Tuber. Lung Dis. 1997, 78, 57–66. [Google Scholar] [CrossRef]
- Lu, J.B.; Chen, B.W.; Wang, G.Z.; Fu, L.L.; Shen, X.B.; Su, C.; Du, W.X.; Yang, L.; Xu, M. Recombinant Tuberculosis Vaccine AEC/BC02 Induces Antigen-Specific Cellular Responses in Mice and Protects Guinea Pigs in a Model of Latent Infection. J. Microbiol. Immunol. Infect. 2015, 48, 597–603. [Google Scholar] [CrossRef] [Green Version]
- Clark, S.; Hall, Y.; Williams, A. Animal Models of Tuberculosis: Guinea Pigs. Cold Spring Harb. Perspect. Med. 2015, 5, a018572. [Google Scholar] [CrossRef]
- Chambers, M.A.; Williams, A.; Gavier-Widén, D.; Whelan, A.; Hughes, C.; Hall, G.; Lever, M.S.; Marsh, P.D.; Hewinson, R.G. A Guinea Pig Model of Low-Dose Mycobacterium bovis Aerogenic Infection. Vet. Microbiol. 2001, 80, 213–226. [Google Scholar] [CrossRef]
- Chandran, A.; Williams, K.; Mendum, T.; Stewart, G.; Clark, S.; Zadi, S.; McLeod, N.; Williams, A.; Villarreal-Ramos, B.; Vordermeier, M.; et al. Development of a Diagnostic Compatible BCG Vaccine against Bovine Tuberculosis. Sci. Rep. 2019, 9, 17791. [Google Scholar] [CrossRef]
- Kashino, S.S.; Napolitano, D.R.; Skobe, Z.; Campos-Neto, A. Guinea Pig Model of Mycobacterium tuberculosis Dormant Infection. Microbes Infect. 2008, 10, 1469–1476. [Google Scholar] [CrossRef]
- Lenaerts, A.J.; Chapman, P.L.; Orme, I.M. Statistical Limitations to the Cornell Model of Latent Tuberculosis Infection for the Study of Relapse Rates. Tuberculosis 2004, 84, 361–364. [Google Scholar] [CrossRef] [PubMed]
- Zhan, L.; Tang, J.; Sun, M.; Qin, C. Animal Models for Tuberculosis in Translational and Precision Medicine. Front. Microbiol. 2017, 8, 717. [Google Scholar] [CrossRef] [PubMed]
- Bapat, P.R.; Husain, A.A.; Daginawala, H.F.; Agrawal, N.P.; Panchbhai, M.S.; Satav, A.R.; Taori, G.M.; Kashyap, R.S. The Assessment of Cytokines in Quantiferon Supernatants for the Diagnosis of Latent TB Infection in a Tribal Population of Melghat, India. J. Infect. Public Health 2015, 8, 329–340. [Google Scholar] [CrossRef] [PubMed]
- Sutherland, J.S.; Hill, P.C.; Adetifa, I.M.; de Jong, B.C.; Donkor, S.; Joosten, S.A.; Opmeer, L.; Haks, M.C.; Ottenhoff, T.H.M.; Adegbola, R.A.; et al. Identification of Probable Early-Onset Biomarkers for Tuberculosis Disease Progression. PLoS ONE 2011, 6, e25230. [Google Scholar] [CrossRef]
- Sloot, R.; Schim van der Loeff, M.F.; van Zwet, E.W.; Haks, M.C.; Keizer, S.T.; Scholing, M.; Ottenhoff, T.H.M.; Borgdorff, M.W.; Joosten, S.A. Biomarkers Can Identify Pulmonary Tuberculosis in HIV-Infected Drug Users Months Prior to Clinical Diagnosis. EbioMedicine 2015, 2, 172–179. [Google Scholar] [CrossRef]
- Chegou, N.N.; Black, G.F.; Kidd, M.; van Helden, P.D.; Walzl, G. Host Markers in Quantiferon Supernatants Differentiate Active TB from Latent TB Infection: Preliminary Report. BMC Pulm. Med. 2009, 9, 21. [Google Scholar] [CrossRef]
- Kim, S.Y.; Park, M.S.; Kim, Y.S.; Kim, S.K.; Chang, J.; Lee, H.J.; Cho, S.N.; Kang, Y.A. The Responses of Multiple Cytokines Following Incubation of Whole Blood from TB Patients, Latently Infected Individuals and Controls with the TB Antigens ESAT-6, CFP-10 and TB7.7. Scand. J. Immunol. 2012, 76, 580–586. [Google Scholar] [CrossRef]
- Suzukawa, M.; Akashi, S.; Nagai, H.; Nagase, H.; Nakamura, H.; Matsui, H.; Hebisawa, A.; Ohta, K. Combined Analysis of IFN-γ, IL-2, IL-5, IL-10, IL-1RA and MCP-1 in QFT Supernatant Is Useful for Distinguishing Active Tuberculosis from Latent Infection. PLoS ONE 2016, 11, e0152483. [Google Scholar] [CrossRef]
- Singh, G.; Yadav, M.; Ghosh, C.; Rathore, J.S. Bacterial Toxin-Antitoxin Modules: Classification, Functions, and Association with Persistence. Curr. Res. Microb. Sci. 2021, 2, 100047. [Google Scholar] [CrossRef]
- Boon, C.; Li, R.; Qi, R.; Dick, T. Proteins of Mycobacterium bovis BCG Induced in the Wayne Dormancy Model. J. Bacteriol. 2001, 183, 2672–2676. [Google Scholar] [CrossRef] [PubMed]
- Leistikow, R.L.; Morton, R.A.; Bartek, I.L.; Frimpong, I.; Wagner, K.; Voskuil, M.I. The Mycobacterium tuberculosis DosR Regulon Assists in Metabolic Homeostasis and Enables Rapid Recovery from Nonrespiring Dormancy. J. Bacteriol. 2010, 192, 1662–1670. [Google Scholar] [CrossRef] [PubMed]
- Arroyo, L.; Marín, D.; Franken, K.L.M.C.; Ottenhoff, T.H.M.; Barrera, L.F. Potential of DosR and Rpf Antigens from Mycobacterium tuberculosis to Discriminate between Latent and Active Tuberculosis in a Tuberculosis Endemic Population of Medellin Colombia. BMC Infect. Dis. 2018, 18, 26. [Google Scholar] [CrossRef]
- Black, G.F.; Thiel, B.A.; Ota, M.O.; Parida, S.K.; Adegbola, R.; Boom, W.H.; Dockrell, H.M.; Franken, K.L.M.C.; Friggen, A.H.; Hill, P.C.; et al. Immunogenicity of Novel DosR Regulon-Encoded Candidate Antigens of Mycobacterium tuberculosis in Three High-Burden Populations in Africa. Clin. Vaccine Immunol. 2009, 16, 1203–1212. [Google Scholar] [CrossRef]
- Keren, I.; Minami, S.; Rubin, E.; Lewis, K. Characterization and Transcriptome Analysis of Mycobacterium tuberculosis Persisters. mBio 2011, 2, 11. [Google Scholar] [CrossRef] [PubMed]
- O’ Toole, R.; Smeulders, M.J.; Blokpoel, M.C.; Kay, E.J.; Lougheed, K.; Williams, H.D. A Two-Component Regulator of Universal Stress Protein Expression and Adaptation to Oxygen Starvation in Mycobacterium smegmatis. J. Bacteriol. 2003, 185, 1543–1554. [Google Scholar] [CrossRef] [PubMed]
- Stewart, G.R.; Robertson, B.D.; Young, D.B. Tuberculosis: A Problem with Persistence. Nat. Rev. Microbiol. 2003, 1, 97–105. [Google Scholar] [CrossRef]
- Flores-Valdez, M.A.; Aceves-Sánchez, M.D.J.; Peterson, E.J.R.; Baliga, N.; Bravo-Madrigal, J.; De la Cruz-Villegas, M.Á.; Ares, M.A.; Born, S.; Voskuil, M.; Pérez-Padilla, N.A.; et al. Transcriptional Portrait of M. Bovis BCG during Biofilm Production Shows Genes Differentially Expressed during Intercellular Aggregation and Substrate Attachment. Sci. Rep. 2020, 10, 12578. [Google Scholar] [CrossRef]
- Hozumi, H.; Tsujimura, K.; Yamamura, Y.; Seto, S.; Uchijima, M.; Nagata, T.; Miwa, S.; Hayakawa, H.; Fujisawa, T.; Hashimoto, D.; et al. Immunogenicity of Dormancy-Related Antigens in Individuals Infected with Mycobacterium tuberculosis in Japan. Int. J. Tuberc. Lung Dis. 2013, 17, 818–824. [Google Scholar] [CrossRef]
- Demaio, J.; Zhang, Y.; Ko, C.; Youngt, D.B.; Bishai, W.R. A Stationary-Phase Stress-Response Sigma Factor from Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 1996, 93, 2790–2794. [Google Scholar] [CrossRef]
- Michele, T.M.; Ko, C. Exposure to Antibiotics Induces Expression of the Mycobacterium tuberculosis SigF Gene: Implications for Chemotherapy against Mycobacterial Persistors. Society 2010, 43, 218–225. [Google Scholar] [CrossRef] [PubMed]
- Williams, E.P.; Lee, J.-H.; Bishai, W.R.; Colantuoni, C.; Karakousis, P.C. Mycobacterium tuberculosis SigF Regulates Genes Encoding Cell Wall-Associated Proteins and Directly Regulates the Transcriptional Regulatory Gene PhoY1. J. Bacteriol. 2007, 189, 4234–4242. [Google Scholar] [CrossRef] [PubMed]
- Sherman, R.A.; Shimoda, K.J. Tuberculosis Tracking: Determining the Frequency of the Booster Effect in Patients and Staff. Am. J. Infect. Control 2001, 29, 7–12. [Google Scholar] [CrossRef] [PubMed]
- Jee, B.; Singh, Y.; Yadav, R.; Lang, F. Small Heat Shock Protein16.3 of Mycobacterium tuberculosis: After Two Decades of Functional Characterization. Cell. Physiol. Biochem. 2018, 49, 368–380. [Google Scholar] [CrossRef]
- Rodrigue, S.; Provvedi, R.; Jacques, P.É.; Gaudreau, L.; Manganelli, R. The σ Factors of Mycobacterium tuberculosis. FEMS Microbiol. Rev. 2006, 30, 926–941. [Google Scholar] [CrossRef] [PubMed]
- Sachdeva, P.; Misra, R.; Tyagi, A.K.; Singh, Y. The Sigma Factors of Mycobacterium tuberculosis: Regulation of the Regulators. FEBS J. 2010, 277, 605–626. [Google Scholar] [CrossRef]
- Manganelli, R.; Dubnau, E.; Tyagi, S.; Kramer, F.R.; Smith, I. Differential Expression of 10 Sigma Factor Genes in Mycobacterium tuberculosis. Mol. Microbiol. 1999, 30, 715–724. [Google Scholar] [CrossRef]
- Cuerda, M.X.; Colombatti, M.A.; Gravisaco, M.J.; Marfil, M.J.; Barandiaran, S.; Sevilla, I.A.; Garrido, J.M.; Moyano, R.D.; Zumárraga, M.J.; Romano, M.I.; et al. Pathogenesis of Domestic Pigs Submitted to Mycobacterial Sensitizations Previous to Experimental Infection with Mycobacterium bovis. Span. J. Agric. Res. 2022, 20, e0502. [Google Scholar] [CrossRef]
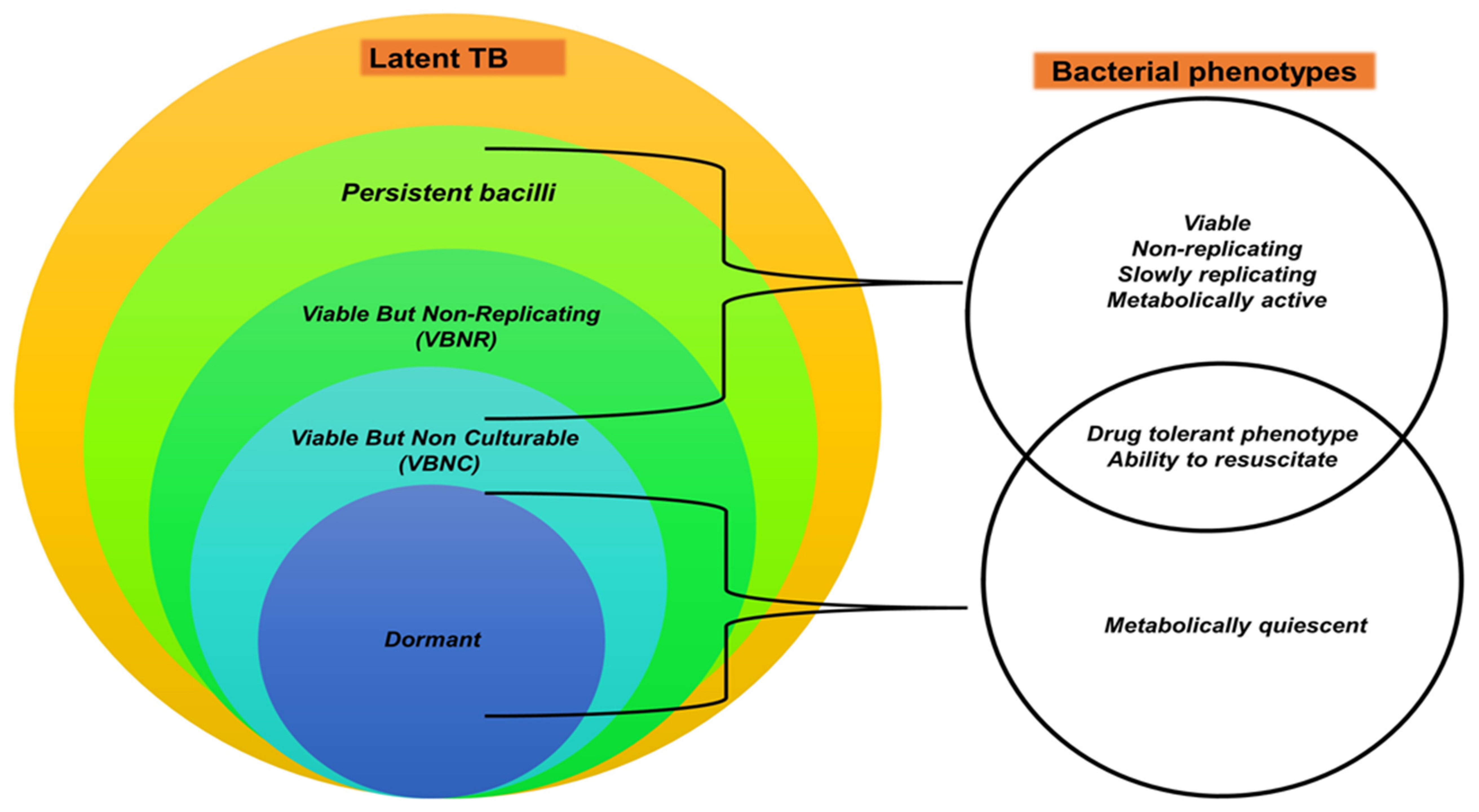
| MTBC Pathogens | Primary Hosts | Secondary/Sporadic Hosts |
|---|---|---|
| M. bovis | African buffaloes (Syncerus caffer) [44,55,56], Asian elephants (Elephas maximus) [57,58,59,60,61,62], bushbucks (Tragelaphus scriptus) [28,63,64], bush pigs [40,65,66] (Potamochoerus porcus), cattle [18,67,68,69], chacma baboons (Papio ursinus) [28,70,71], cheetahs (Acinonyx jubatus) [70,72], common duiker (Syvicapra grimmia) [12,73], African elephants (Loxodonta africana) [28,59,74], eland (Taurotragus oryx) [28,40,66], giraffe (Giraffa camelopardalis) [75], european badger (Meles meles) [76,77,78,79], honey badger (Mellivora capensis) [12,28,80], greater kudu (Tragelaphus strepsiceros) [72,81], hippopotamus (Hippopotamus amphibius) [82,83], impala (Aepyceros melampus) [28,40,84], African leopards (Panthera pardus) [28,35,85], African lions (Panthera leo) [6,11,28,86,87,88,89], banded mongoose (Mungos mungo) [90,91], nyala (Tragelaphus angasii) [28,92] black rhinoceros (Diceros bicornis) [6,93,94,95], red fox (Vulpes vulpes) [96], spotted genet (Genetta tigrine) [28,65,81], springbok (Antidorcas marsupialis) [12,28], common warthog (Phacochoerus africanus) [28,97,98,99], wild boars (Sus scrofa) [96], African wild dog (Lycaon pictus) [100,101,102,103], blue wildebeest (Connochaetes taurinus) [104,105], wild deer (Capreolus capreolus, Cervus elaphus) [96], white rhinoceros (Ceratotherium simum) [35,95,104,106] | Humans [17,19,43], cats [107] |
| M. tuberculosis | Humans [1,19,28] | Cattle [108,109], non-human primates [110,111], dogs [112,113,114], cats [115], zoo animals e.g., black and white rhinos [116], African elephants, and Asian elephants [117,118,119], goats [120] |
| Mycobacterium africanum | Humans [121,122,123] | Cattle [124], rock hyraxes (Procavia capensis) [29,125] |
| Mycobacterium pinnipedii | Seals (pinnipedts) [126] | Humans [127,128], cattle [129] |
| Mycobacterium caprae | Goats, sheep, and pigs [130,131] | Humans [132] |
| Mycobacterium microti | Cats, pigs [133], bank vole, wood mouse, shrew [134] | Humans [135] |
| Mycobacterium orygis | Antelope, deer, waterbuck [136], oryxes, gazelles [137], | Humans [138] |
| Mycobacterium canettii | Humans [29,139,140,141] | - |
| Mycobacterium mungi | Banded mongooses [142] | - |
| Mycobacterium suricattae | Meerkats (Suricata suricatta) [143] | - |
| Chimpanzee bacillus | Chimpanzee (Pan troglodytes) [16] | - |
| Dassie bacillus | Rock hyraxes [14,144], meerkats [143,145] | - |
| bTB Stage | Clinical Signs | Host–Pathogen Interactions | |
|---|---|---|---|
| Uninfected | Absent |
| |
| Infected | Cleared infection | Absent |
|
| Latent infection | Absent |
| |
| Subclinical infection | Absent |
| |
| Active TB disease | Present |
| |
| Animal TB Stage (a) | Antemortem Tests (b,c) | Postmortem Tests (d) | |||
|---|---|---|---|---|---|
| TST | IGRA | Bacterial Culture | Tissue Histopathology Consistent with bTB (Yes/No) | ||
| Uninfected | Negative | Negative | Negative | No | |
| Infected | Cleared infection | Negative (innate immune system clearance) Positive (adaptive system immune clearance) | Positive for a short period before waning (adaptive immune clearance) | Negative | No |
| Latent infection | Positive | Positive | Negative | No | |
| Subclinical infection | Positive | Positive | Positive | Yes (early-stage granulomas) | |
| Active disease | Positive | Positive | Positive | Yes (multifocal and/or confluent late-stage granulomas | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ncube, P.; Bagheri, B.; Goosen, W.J.; Miller, M.A.; Sampson, S.L. Evidence, Challenges, and Knowledge Gaps Regarding Latent Tuberculosis in Animals. Microorganisms 2022, 10, 1845. https://doi.org/10.3390/microorganisms10091845
Ncube P, Bagheri B, Goosen WJ, Miller MA, Sampson SL. Evidence, Challenges, and Knowledge Gaps Regarding Latent Tuberculosis in Animals. Microorganisms. 2022; 10(9):1845. https://doi.org/10.3390/microorganisms10091845
Chicago/Turabian StyleNcube, Pamela, Bahareh Bagheri, Wynand Johan Goosen, Michele Ann Miller, and Samantha Leigh Sampson. 2022. "Evidence, Challenges, and Knowledge Gaps Regarding Latent Tuberculosis in Animals" Microorganisms 10, no. 9: 1845. https://doi.org/10.3390/microorganisms10091845
APA StyleNcube, P., Bagheri, B., Goosen, W. J., Miller, M. A., & Sampson, S. L. (2022). Evidence, Challenges, and Knowledge Gaps Regarding Latent Tuberculosis in Animals. Microorganisms, 10(9), 1845. https://doi.org/10.3390/microorganisms10091845






