Optimization of Transposon Mutagenesis Methods in Pseudomonas antarctica
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains, Plasmids, Media, and Growth Conditions
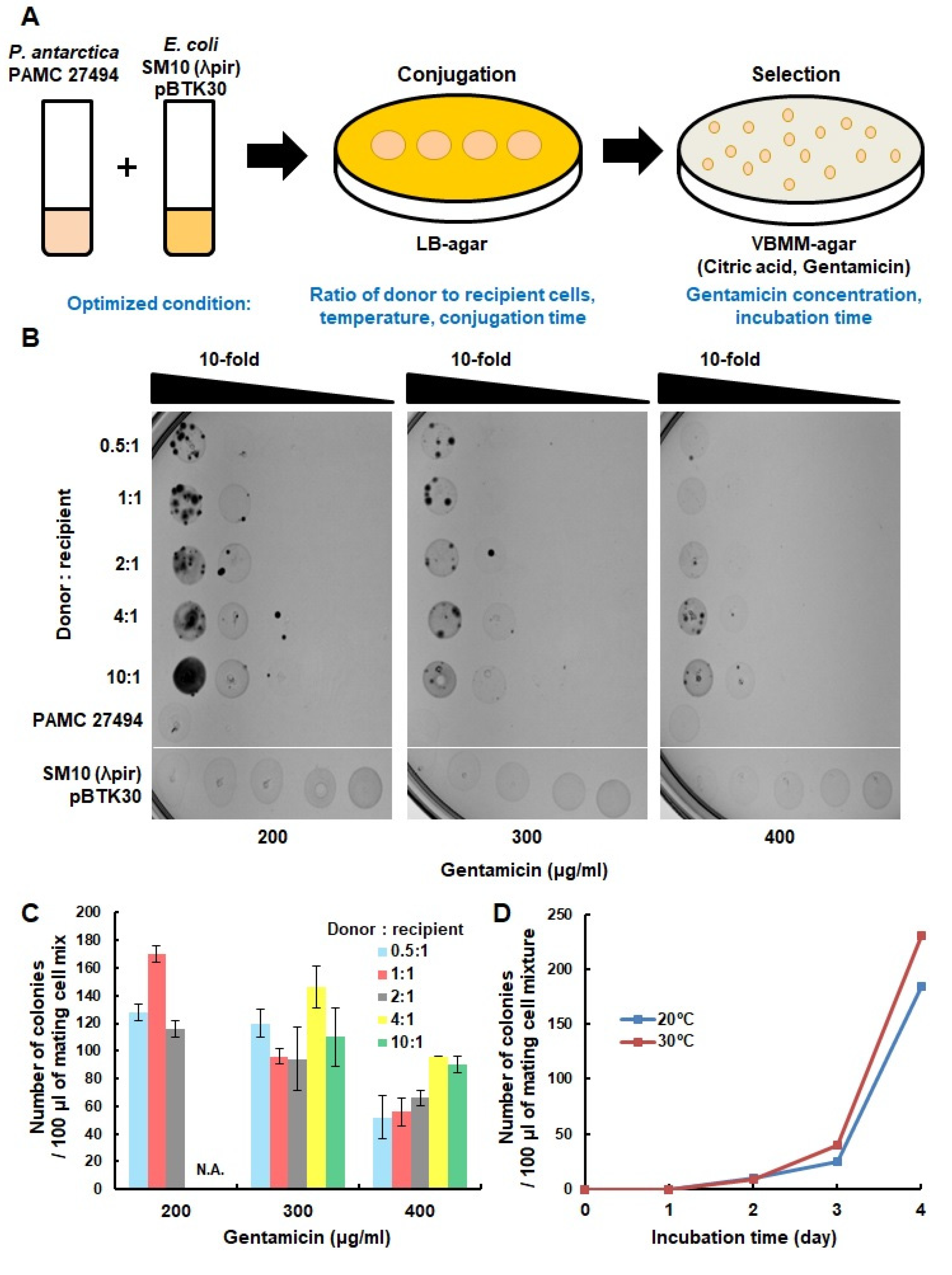
2.2. Transposon Insertion
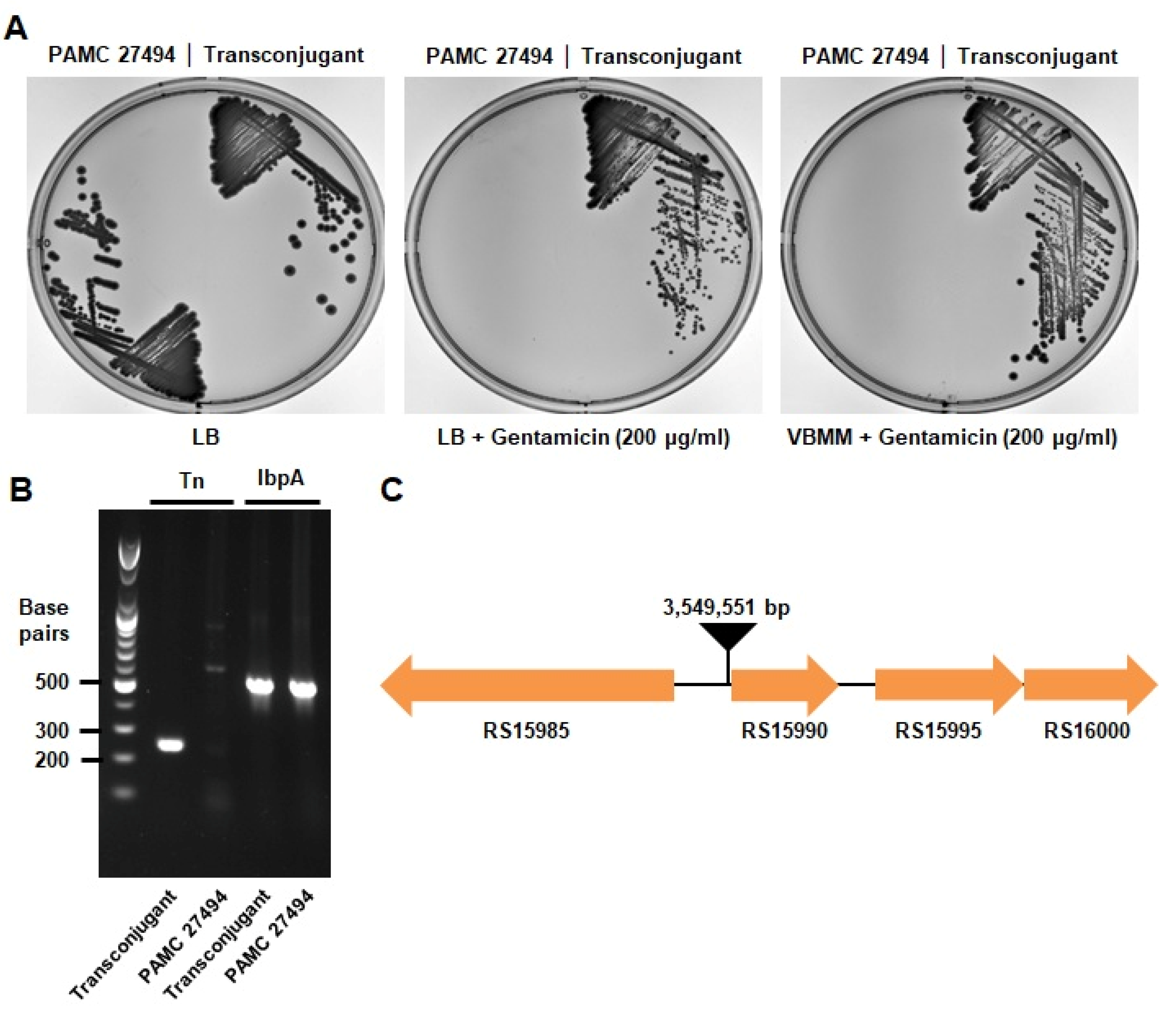
2.3. Spot Titer Assay
2.4. PCR for the Verification of Transposon Insertion
2.5. Inverse PCR and Sequencing
3. Results and Discussions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Diggle, S.P.; Whiteley, M. Microbe Profile: Pseudomonas aeruginosa: Opportunistic pathogen and lab rat. Microbiology 2020, 166, 30. [Google Scholar] [CrossRef] [PubMed]
- Xin, X.-F.; Kvitko, B.; He, S.Y. Pseudomonas syringae: What it takes to be a pathogen. Nat. Rev. Microbiol. 2018, 16, 316–328. [Google Scholar] [CrossRef] [PubMed]
- Spiers, A.J.; Buckling, A.; Rainey, P.B. The causes of Pseudomonas diversity. Microbiology 2000, 146, 2345–2350. [Google Scholar] [CrossRef] [PubMed]
- Römling, U.; Wingender, J.; Müller, H.; Tümmler, B. A major Pseudomonas aeruginosa clone common to patients and aquatic habitats. Appl. Environ. Microbiol. 1994, 60, 1734–1738. [Google Scholar] [CrossRef] [PubMed]
- Reddy, G.S.; Matsumoto, G.I.; Schumann, P.; Stackebrandt, E.; Shivaji, S. Psychrophilic pseudomonads from Antarctica: Pseudomonas antarctica sp. nov., Pseudomonas meridiana sp. nov. and Pseudomonas proteolytica sp. nov. Int. J. Syst. Evol. Microbiol. 2004, 54, 713–719. [Google Scholar] [CrossRef]
- Kriss, A.; Mitskevich, I.; Rozanova, E.; Osnitskaia, L. Microbiological studies of the Wanda Lake (Antarctica). Mikrobiologiia 1976, 45, 1075–1081. [Google Scholar]
- Yarzábal, L.A.; Monserrate, L.; Buela, L.; Chica, E. Antarctic Pseudomonas spp. promote wheat germination and growth at low temperatures. Polar Biol. 2018, 41, 2343–2354. [Google Scholar] [CrossRef]
- Shukor, M.; Hassan, N.; Jusoh, A.; Perumal, N.; Shamaan, N.; MacCormack, W.; Syed, M. Isolation and characterization of a Pseudomonas diesel-degrading strain from Antarctica. J. Environ. Biol. 2009, 30, 1–6. [Google Scholar]
- Ma, Y.; Wang, L.; Shao, Z. Pseudomonas, the dominant polycyclic aromatic hydrocarbon-degrading bacteria isolated from Antarctic soils and the role of large plasmids in horizontal gene transfer. Environ. Microbiol. 2006, 8, 455–465. [Google Scholar] [CrossRef]
- Stallwood, B.; Shears, J.; Williams, P.; Hughes, K. Low temperature bioremediation of oil-contaminated soil using biostimulation and bioaugmentation with a Pseudomonas sp. from maritime Antarctica. J. Appl. Microbiol. 2005, 99, 794–802. [Google Scholar] [CrossRef]
- López, N.I.; Pettinari, M.J.; Stackebrandt, E.; Tribelli, P.M.; Põtter, M.; Steinbüchel, A.; Méndez, B.S. Pseudomonas extremaustralis sp. nov., a poly (3-hydroxybutyrate) producer isolated from an Antarctic environment. Curr. Microbiol. 2009, 59, 514–519. [Google Scholar] [CrossRef]
- Ayub, N.D.; Pettinari, M.J.; Ruiz, J.A.; López, N.I. A polyhydroxybutyrate-producing Pseudomonas sp. isolated from Antarctic environments with high stress resistance. Curr. Microbiol. 2004, 49, 170–174. [Google Scholar] [CrossRef]
- Tan, I.K.P.; Foong, C.P.; Tan, H.T.; Lim, H.; Zain, N.-A.A.; Tan, Y.C.; Hoh, C.C.; Sudesh, K. Polyhydroxyalkanoate (PHA) synthase genes and PHA-associated gene clusters in Pseudomonas spp. and Janthinobacterium spp. isolated from Antarctica. J. Biotechnol. 2020, 313, 18–28. [Google Scholar] [CrossRef]
- Lee, C.; Klockgether, J.; Fischer, S.; Trcek, J.; Tümmler, B.; Römling, U. Why?–Successful Pseudomonas aeruginosa clones with a focus on clone C. FEMS Microbiol. Rev. 2020, 44, 740–762. [Google Scholar] [CrossRef]
- Lee, J.; Cho, Y.-J.; Yang, J.Y.; Jung, Y.-J.; Hong, S.G.; Kim, O.-S. Complete genome sequence of Pseudomonas antarctica PAMC 27494, a bacteriocin-producing psychrophile isolated from Antarctica. J. Biotechnol. 2017, 259, 15–18. [Google Scholar] [CrossRef]
- Muryoi, N.; Kawahara, H.; Obata, H. Properties of a novel extracellular cell-free ice nuclei from ice-nucleating Pseudomonas antarctica IN-74. Biosci. Biotechnol. Biochem. 2003, 67, 1950–1958. [Google Scholar] [CrossRef]
- Dupeyron, M.; Baril, T.; Bass, C.; Hayward, A. Phylogenetic analysis of the Tc1/mariner superfamily reveals the unexplored diversity of pogo-like elements. Mob. DNA 2020, 11, 21. [Google Scholar] [CrossRef]
- Trubitsyna, M.; Morris, E.R.; Finnegan, D.J.; Richardson, J.M. Biochemical characterization and comparison of two closely related active mariner transposases. Biochemistry 2014, 53, 682–689. [Google Scholar] [CrossRef]
- Crénès, G.; Moundras, C.; Demattei, M.-V.; Bigot, Y.; Petit, A.; Renault, S. Target site selection by the mariner-like element, Mos1. Genetica 2010, 138, 509–517. [Google Scholar] [CrossRef]
- Sonnabend, M.S.; Klein, K.; Beier, S.; Angelov, A.; Kluj, R.; Mayer, C.; Groß, C.; Hofmeister, K.; Beuttner, A.; Willmann, M. Identification of drug resistance determinants in a clinical isolate of Pseudomonas aeruginosa by high-density transposon mutagenesis. Antimicrob. Agents Chemother. 2020, 64, e01771-19. [Google Scholar] [CrossRef]
- Harayama, S.; Lehrbach, P.R.; Timmis, K.N. Transposon mutagenesis analysis of meta-cleavage pathway operon genes of the TOL plasmid of Pseudomonas putida mt-2. J. Bacteriol. 1984, 160, 251–255. [Google Scholar] [CrossRef] [PubMed]
- Andreani, N.A.; Carraro, L.; Zhang, L.; Vos, M.; Cardazzo, B. Transposon mutagenesis in Pseudomonas fluorescens reveals genes involved in blue pigment production and antioxidant protection. Food Microbiol. 2019, 82, 497–503. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.K.; Pindi, P.K.; Dube, S.; Sundareswaran, V.; Shivaji, S. Importance of trmE for growth of the psychrophile Pseudomonas syringae at low temperatures. Appl. Environ. Microbiol. 2009, 75, 4419–4426. [Google Scholar] [CrossRef] [PubMed]
- Kiran, M.; Prakash, J.; Annapoorni, S.; Dube, S.; Kusano, T.; Okuyama, H.; Murata, N.; Shivaji, S. Psychrophilic Pseudomonas syringae requires trans-monounsaturated fatty acid for growth at higher temperature. Extremophiles 2004, 8, 401–410. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.K.; Jeong, H.-J.; Lee, H.; Hong, S.G.; Cho, J.-C.; Lee, H.K. Transposon mutagenesis of Psychrobacter cryohalolentis PAMC 21807 by tri-parental conjugation. Adv. Polar Sci. 2013, 24, 223–230. [Google Scholar]
- Cho, H. Transposon insertion site sequencing (TIS) of Pseudomonas aeruginosa. J. Microbiol. 2021, 59, 1067–1074. [Google Scholar] [CrossRef]
- Choi, K.-H.; Schweizer, H.P. mini-Tn7 insertion in bacteria with single attTn7 sites: Example Pseudomonas aeruginosa. Nat. Protoc. 2006, 1, 153–161. [Google Scholar] [CrossRef]
- Green, M.R.; Sambrook, J. Inverse polymerase chain reaction (PCR). Cold Spring Harb. Protoc. 2019, 2019, pdb.prot095166. [Google Scholar] [CrossRef]
- Lütgens, M.; Gottschalk, G. Why a co-substrate is required for anaerobic growth of Escherichia coli on citrate. Microbiology 1980, 119, 63–70. [Google Scholar] [CrossRef]
- Van Opijnen, T.; Bodi, K.L.; Camilli, A. Tn-seq: High-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nat. Methods 2009, 6, 767–772. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, S.; Lee, C. Optimization of Transposon Mutagenesis Methods in Pseudomonas antarctica. Microorganisms 2023, 11, 118. https://doi.org/10.3390/microorganisms11010118
Kim S, Lee C. Optimization of Transposon Mutagenesis Methods in Pseudomonas antarctica. Microorganisms. 2023; 11(1):118. https://doi.org/10.3390/microorganisms11010118
Chicago/Turabian StyleKim, Sangha, and Changhan Lee. 2023. "Optimization of Transposon Mutagenesis Methods in Pseudomonas antarctica" Microorganisms 11, no. 1: 118. https://doi.org/10.3390/microorganisms11010118
APA StyleKim, S., & Lee, C. (2023). Optimization of Transposon Mutagenesis Methods in Pseudomonas antarctica. Microorganisms, 11(1), 118. https://doi.org/10.3390/microorganisms11010118




