The Contribution of Actinobacteria to the Degradation of Chlorinated Compounds: Variations in the Activity of Key Degradation Enzymes
Abstract
:1. Introduction
2. Material and Methods
2.1. Bacterial Culture and Cultivation Conditions
2.2. PCR
2.3. Sequencing and Analysis of Nucleotide Sequences
2.4. Identification of Plasmid DNA
2.5. Preparation of Cell-Free Extract
2.6. Determination of Enzyme Activity and Protein Amount
2.7. Purification of Enzymes from the Biomass of R. opacus 6a, R.opacus 1CP, R.ruber P25, Microbacterium Foliorum B51 Strains Grown with 3-CBA as Substrate
2.8. Polarographic Measurement of Cell Respiration and BDO Activity in Bacterial Cells
2.9. Matrix Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry (MALDI-TOF MS) Analysis
3. Results
3.1. The Activity of Actinobacteria with Substituted Benzoate and Phenols
3.2. Enzymes of Strains R. opacus 6a, R. opacus 1CP, R. ruber P25, M. foliorum B51 Grown on 3-CBA
3.3. clcF Gene in Actinobacteria
3.4. MALDI-ToF Mass Spectrometric Analysis as a Method for Rapid Screening of Enzymes of Interest
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ghosal, D.; Ghosh, S.; Dutta, T.K.; Ahn, Y. Current State of Knowledge in Microbial Degradation of Polycyclic Aromatic Hydrocarbons (PAHs): A Review. Front. Microbiol. 2016, 7, 1369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Al-Limoun, M.O.; Khleifat, K.M.; Alsharafa, K.Y.; Qaralleh, H.N.; Alrawashdeh, S.A. Purification and characterization of a mesophilic organic solvent tolerant lipase produced by Acinetobacter sp. K5b4. Biocatal. Biotransform. 2018, 37, 139–151. [Google Scholar] [CrossRef]
- Yan, D.; Kang, J.; Liu, D.-Q. Genomic analysis of the aromatic catabolic pathways from Silicibacter pomeroyi DSS-3. Ann. Microbiol. 2009, 59, 789–800. [Google Scholar] [CrossRef]
- Pérez-Pantoja, D.; Leiva-Novoa, P.; Donoso, R.A.; Little, C.; Godoy, M.; Pieper, D.H.; González, B. Hierarchy of Carbon Source Utilization in Soil Bacteria: Hegemonic Preference for Benzoate in Complex Aromatic Compound Mixtures Degraded by Cupriavidus pinatubonensis Strain JMP134. Appl. Environ. Microbiol. 2015, 81, 3914–3924. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coleman, M.L.; Chisholm, S.W. Ecosystem-specific selection pressures revealed through comparative population genomics. Proc. Natl. Acad. Sci. USA 2010, 107, 18634–18639. [Google Scholar] [CrossRef] [Green Version]
- Crisp, A.; Boschetti, C.; Perry, M.; Tunnacliffe, A.; Micklem, G. Expression of multiple horizontally acquired genes is a hallmark of both vertebrate and invertebrate genomes. Genome Biol. 2015, 16, 50. [Google Scholar] [CrossRef] [Green Version]
- Hartmann, J.; Reineke, W.; Knackmuss, H.-J. Metabolism of 3-Chloro-, 4-Chloro-, and 3,5-Dichlorobenzoate by a Pseudomonad. Appl. Environ. Microbiol. 1979, 37, 421–428. [Google Scholar] [CrossRef] [Green Version]
- Jing, R.; Fusi, S.; Kjellerup, B.V. Remediation of Polychlorinated Biphenyls (PCBs) in Contaminated Soils and Sediment: State of Knowledge and Perspectives. Front. Environ. Sci. 2018, 6, 79. [Google Scholar] [CrossRef]
- Schmidt, E.; Knackmuss, H.-J. Production of cis,cis-muconate from benzoate and 2-fluoro-cis,cis-muconate from 3-fluorobenzoate by 3-chlorobenzoate degrading bacteria. Appl. Microbiol. Biotechnol. 1984, 20, 351–355. [Google Scholar] [CrossRef]
- Ajithkumar, P.V.; Kunhi, A.A.M. Pathways for 3-chloro- and 4-chlorobenzoate degradation in Pseudomonas aeruginosa 3mT. Biodegradation 2000, 11, 247–261. [Google Scholar] [CrossRef]
- Pieper, D.H.; Pollmann, K.; Nikodem, P.; Gonzalez, B.; Wray, V. Monitoring Key Reactions in Degradation of Chloroaromatics by In Situ 1 H Nuclear Magnetic Resonance: Solution Structures of Metabolites Formed from cis-Dienelactone. J. Bacteriol. 2002, 184, 1466–1470. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schell, U.; Seibert, V.; Vollmer, M.; Schlömann, M. TfdF—A second plasmid-encoded maleylacetate reductase of Alcaligenes eutrophus JMP134 (pJP4), abstr. P413. Bioengineering 1994, 10, 83. [Google Scholar]
- Zampolli, J.; Zeaiter, Z.; Di Canito, A.; Di Gennaro, P. Genome analysis and -omics approaches provide new insights into the biodegradation potential of Rhodococcus. Appl. Microbiol. Biotechnol. 2019, 103, 1069–1080. [Google Scholar] [CrossRef] [PubMed]
- Reineke, W.; Knackmuss, H.-J. Chemical structure and biodegradability of halogenated aromatic compounds substituent effects on 1,2-dioxygenation of benzoic acid. Biochim. Biophys. Acta (BBA)-Gen. Subj. 1978, 542, 412–423. [Google Scholar] [CrossRef]
- Solyanikova, I.P.; Emelyanova, E.V.; Shumkova, E.S.; Travkin, V.M. Pathways of 3-Chlorobenzoate Degradation by Rhodococcus opacus strains 1CP and 6a. Microbiology 2019, 88, 563–572. [Google Scholar] [CrossRef]
- Plotnikova, E.G.; Maltseva, O.V.; Tsoi, T.V.; Demakov, V.A.; Tiedje, J.M. Isolation and characterization of novel bacteria degrading polychlorinated biphenyls. In Proceedings of the Book of Abstracts, 4th Int. Conf. “Problems of Environment Pollution”, Moscow, Russia, 8–12 October 1998; p. 160. [Google Scholar]
- Plotnikova, E.G.; Solyanikova, I.P.; Egorova, D.O.; Shumkova, E.S.; Golovleva, L.A. Degradation of 4-chlorobiphenyl and chlorobenzoic acid by the strain Rhodococcus ruber P25. Microbiology 2012, 81, 143–153. [Google Scholar] [CrossRef]
- Plotnikova, E.G.; Rybkina, D.O.; Anan’ina, L.N.; Yastrebova, O.V.; Demakov, V.A. Characterization of microorganisms isolated from technogenic soils of the Kama region. Russ. J. Ecol. 2006, 4, 261–268. [Google Scholar]
- Gorlatov, S.N.; Maltseva, O.V.; Shevchenko, V.I.; Golovleva, L.A. Degradation of chlorophenols by a culture of Rhodococcus erythropolis. Mikrobiologiya 1989, 58, 647–651. [Google Scholar]
- Solyanikova, I.P.; Golovlev, E.L.; Lisnyak, O.V.; Golovleva, L.A. Isolation and characterization of catechol 1,2-dioxygenases from Rhodococcus rhodnii strain 135 and Rhodococcus rhodochrous strain 89: Comparison with analogous enzymes of the ordinary and modified ortho-cleavage pathways. Biochemistry 1999, 64, 824–831. [Google Scholar]
- Gröning, J.A.; Roth, C.; Kaschabek, S.R.; Sträter, N.; Schlömann, M. Recombinant expression of a unique chloromuconolactone dehalogenase ClcF from Rhodococcus opacus 1CP and identification of catalytically relevant residues by mutational analysis. Arch. Biochem. Biophys. 2012, 526, 69–77. [Google Scholar] [CrossRef]
- Hayaishi, O.; Katagiri, M.; Rothberg, S. Studies on oxygenases; pyrocatechase. J. Biol. Chem. 1957, 229, 905–920. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, E.; Knackmuss, H.-J. Chemical structure and biodegradability of halogenated aromatic compounds. Conversion of chlorinated muconic acids into maleoylacetic acid. Biochem. J. 1980, 192, 339–347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moiseeva, O.V.; Belova, O.V.; Solyanikova, I.P.; Schlömann, M.; Golovleva, L.A. Enzymes of a new modified ortho-pathway utilizing 2-chlorophenol in Rhodococcus opacus 1CP. Biochemistry 2001, 66, 548–555. [Google Scholar] [CrossRef] [PubMed]
- Dorn, E.; Knackmuss, H.J. Chemical structure and biodegradability of halogenated aromatic compounds. Substituent effects on 1,2-dioxygenation of catechol. Biochem. J. 1978, 174, 85–94. [Google Scholar] [CrossRef] [PubMed]
- Schlömann, M.; Schmidt, E.; Knackmuss, H.J. Different types of dienelactone hydrolase in 4-fluorobenzoate-utilizing bacteria. J. Bacteriol. 1990, 172, 5112–5118. [Google Scholar] [CrossRef] [Green Version]
- Emelyanova, E.V.; Solyanikova, I.P. Evaluation of 3-Chlorobenzoate 1,2-Dioxygenase Inhibition by 2- and 4-Chlorobenzoate with a Cell-Based Technique. Biosensors 2019, 9, 106. [Google Scholar] [CrossRef] [Green Version]
- Emelyanova, E.V.; Solyanikova, I.P. Evaluation of phenol-degradation activity of Rhodococcus opacus 1CP using immobilized and intact cells. Int. J. Environ. Sci. Technol. 2020, 17, 2279–2294. [Google Scholar] [CrossRef]
- Martínková, L.; Uhnáková, B.; Pátek, M.; Nešvera, J.; Křen, V. Biodegradation potential of the genus Rhodococcus. Environ. Int. 2009, 35, 162–177. [Google Scholar] [CrossRef]
- Emelyanova, E.V.; Solyanikova, I.P. Specificity of Rhodococcus opacus 1CP cells’ responses to benzoate and 3-chlorobenzoate. Front. Biosci. 2022, 14, 15. [Google Scholar] [CrossRef]
- Shumkova, E.S.; Solyanikova, I.P.; Plotnikova, E.G.; Golovleva, L.A. Degradation of para-toluate by the bacterium Rhodococcus ruber P25. Microbiology 2009, 78, 376–378. [Google Scholar] [CrossRef]
- König, C.; Eulberg, D.; Gröning, J.; Lakner, S.; Seibert, V.; Kaschabek, S.R.; Schlömann, M. A linear megaplasmid, p1CP, carrying the genes for chlorocatechol catabolism of Rhodococcus opacus 1CP. Microbiology 2004, 150, 3075–3087. [Google Scholar] [CrossRef] [PubMed]
- Solyanikova, I.P.; Plotnikova, E.G.; Shumkova, E.S.; Robota, I.V.; Prisyazhnaya, N.V.; Golovleva, L.A. Chloromuconolactone dehalogenase ClcF of actinobacteria. J. Environ. Sci. Health Part B 2014, 49, 422–431. [Google Scholar] [CrossRef] [PubMed]
- Retamal-Morales, G.; Heine, T.; Tischler, J.S.; Erler, B.; Gröning, J.A.; Kaschabek, S.R.; Schlömann, M.; Levicán, G.; Tischler, D. Draft genome sequence of Rhodococcus erythropolis B7g, a biosurfactant producing actinobacterium. J. Biotechnol. 2018, 280, 38–41. [Google Scholar] [CrossRef] [PubMed]
- Garrido-Sanz, D.; Redondo-Nieto, M.; Martín, M.; Rivilla, R. Comparative Genomics of the Rhodococcus Genus Shows Wide Distribution of Biodegradation Traits. Microorganisms 2020, 8, 774. [Google Scholar] [CrossRef]
- McLeod, M.P.; Warren, R.L.; Hsiao, W.W.; Araki, N.; Myhre, M.; Fernandes, C.; Miyazawa, D.; Wong, W.; Lillquist, A.L.; Wang, D.; et al. The complete genome of Rhodococcus sp. RHA1 provides insights into a catabolic powerhouse. Proc. Natl. Acad. Sci. USA 2006, 103, 15582–15587. [Google Scholar] [CrossRef] [Green Version]
- Wübbeler, J.H.; Bruland, N.; Wozniczka, M.; Steinbüchel, A. Biodegradation of the xenobiotic organic disulphide 4,4′-dithiodibutyric acid by Rhodococcus erythropolis strain MI2 and comparison with the microbial utilization of 3,3′-dithiodipropionic acid and 3,3′-thiodipropionic acid. Microbiology 2010, 156, 1221–1233. [Google Scholar] [CrossRef] [Green Version]
- de Carvalho, C.C.C.R. Adaptation of Rhodococcus erythropolis cells for growth and bioremediation under extreme conditions. Res. Microbiol. 2012, 163, 125–136. [Google Scholar] [CrossRef]
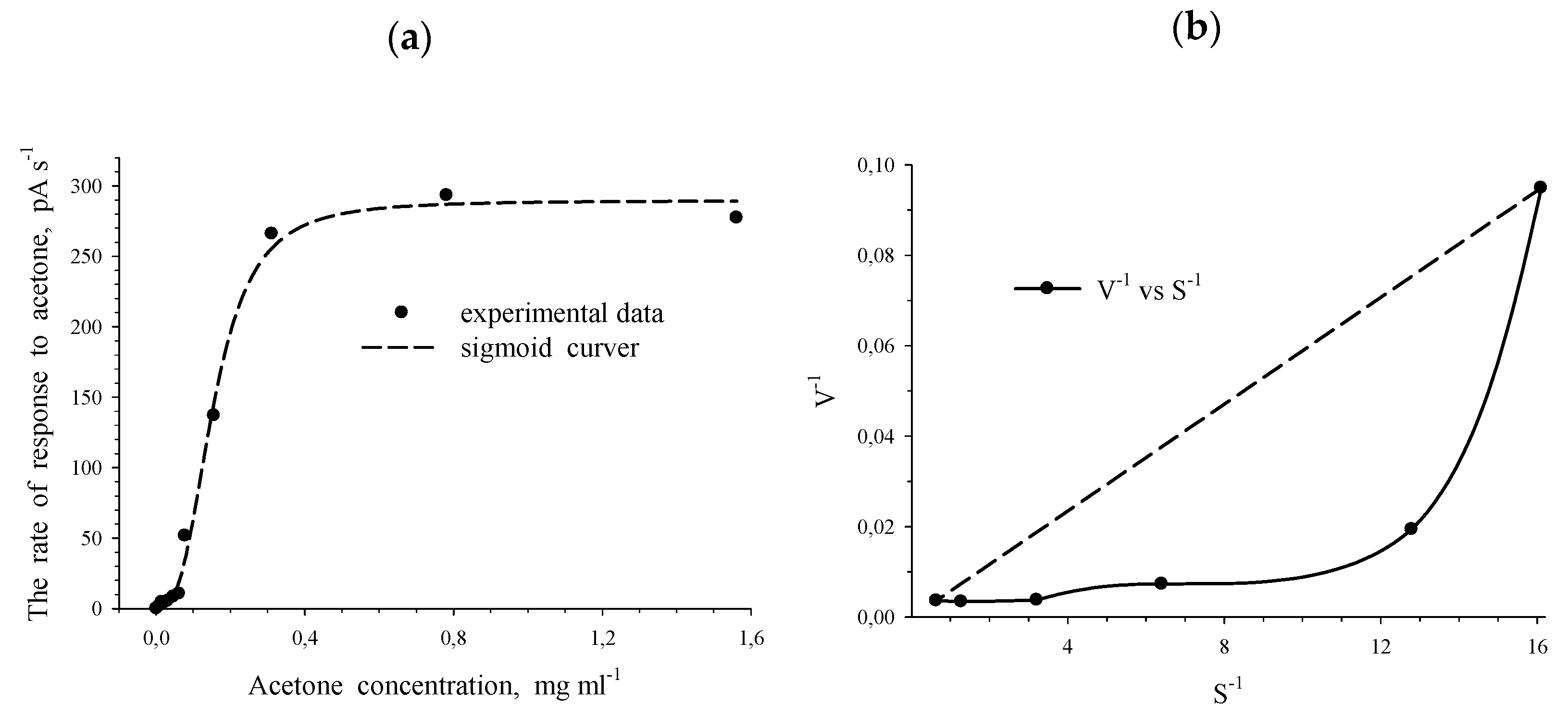
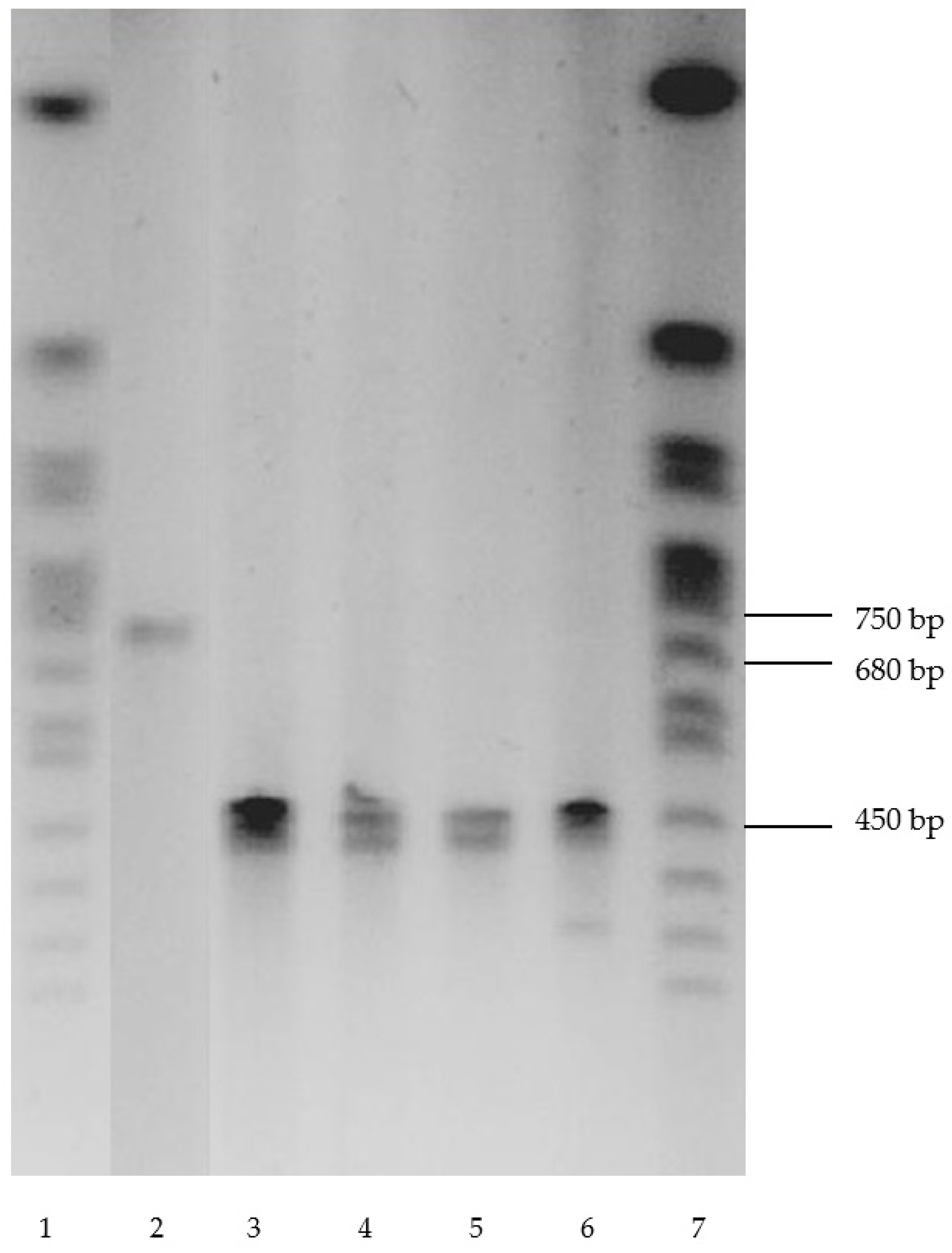
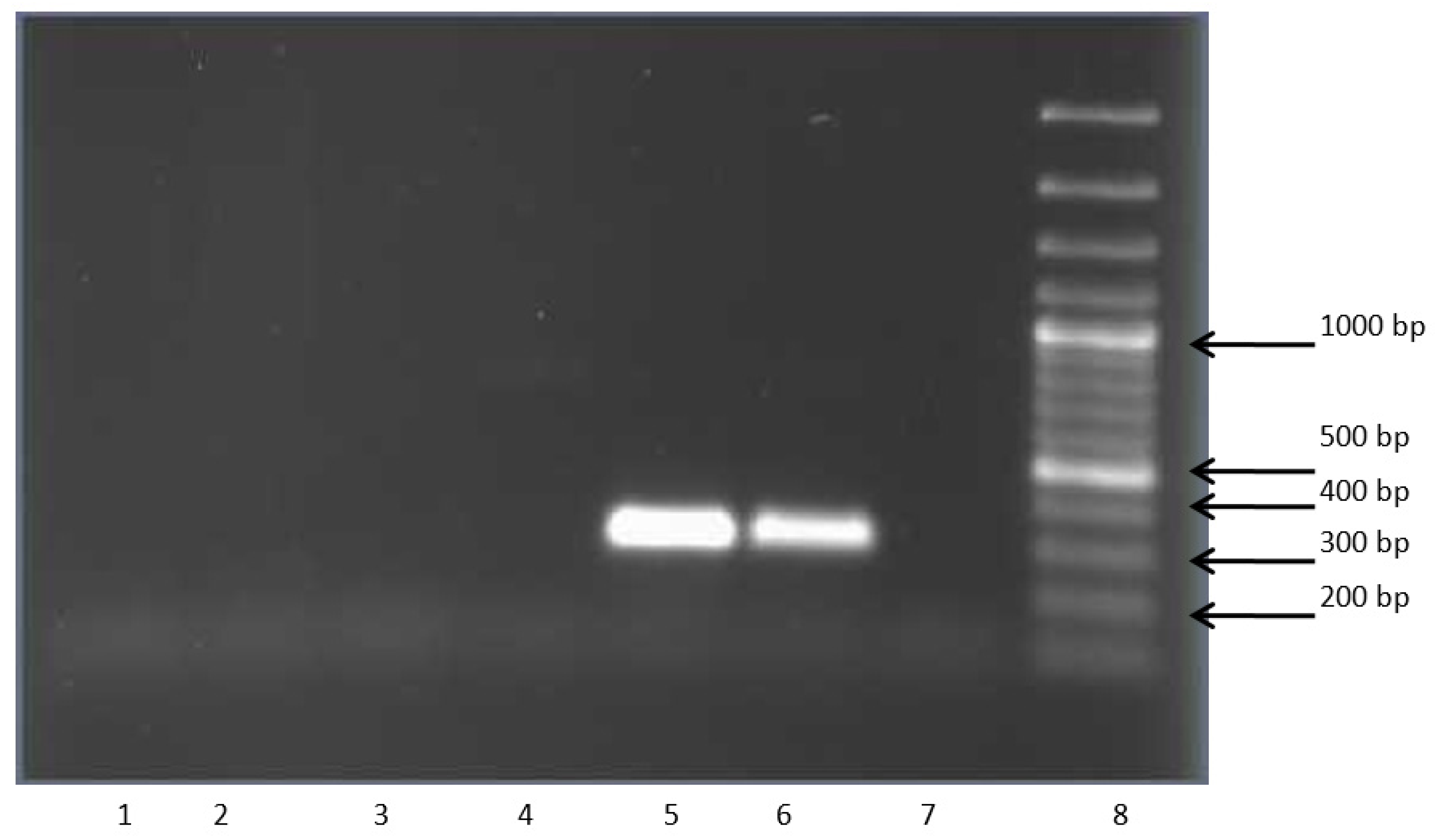
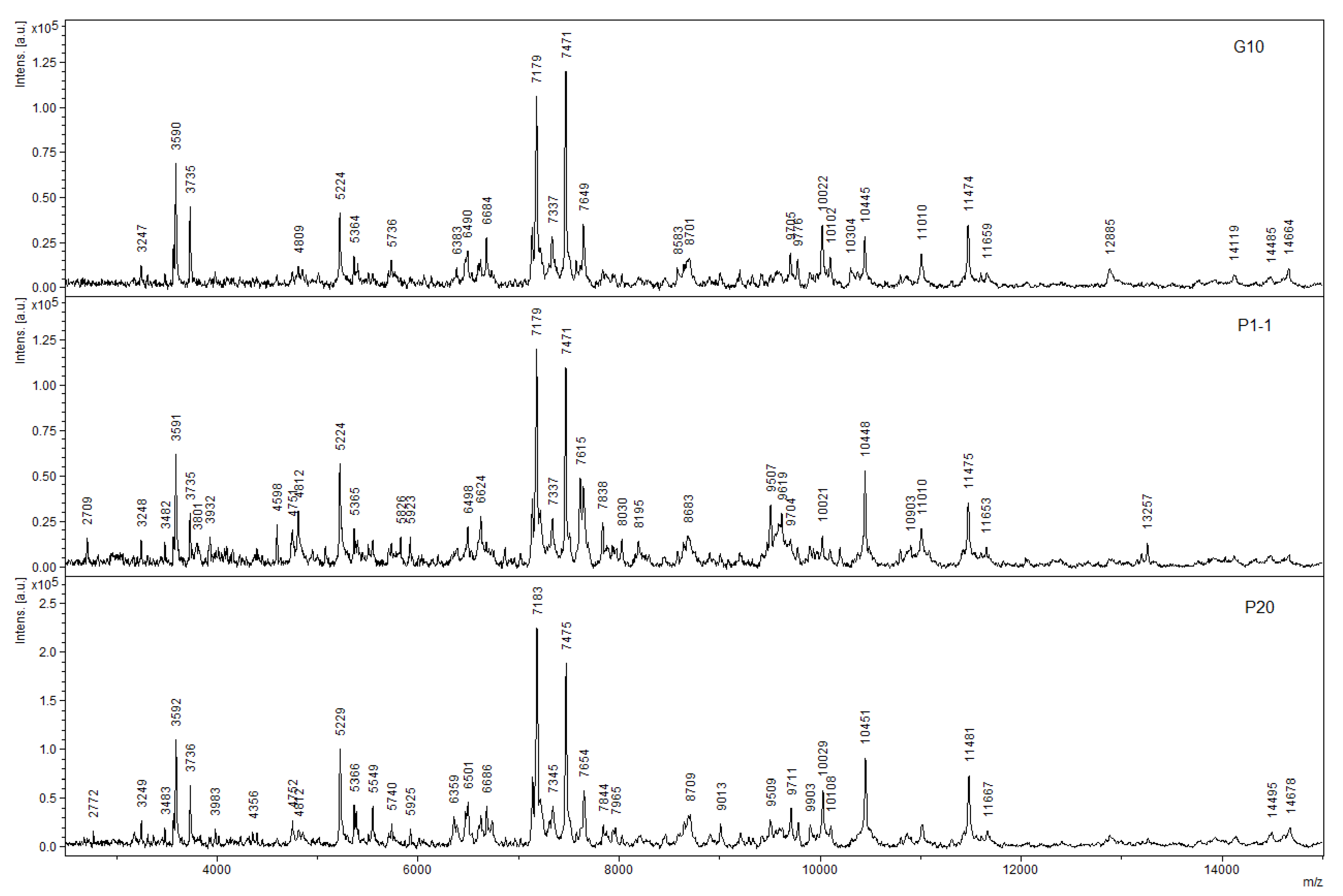
| Substrate | R. opacus 1CP | R. opacus 6a [15] | R. rhodochrous 89 | G. polyisopr. 135 | R. wratislaviensis G10 |
|---|---|---|---|---|---|
| Benzoate | 30 | 24 | 214 | 80 | 92 |
| 2-Chlorobenzoate | 102 | 0 | 0 | 0 | 431 |
| 3-Chlorobenzoate | 100 | 100 | 100 | 100 | 100 |
| 4-Chlorobenzoate | 57 | 0 | 20 | 0 | 0 |
| 2,4-Dichlorobenzoate | 0 | 47 | n.d. | 53 | 131 |
| 2,5-Dichlorobenzoate | 0 | 0 | n.d. | 0 | 0 |
| 2,6-Dichlorobenzoate | 124 | n.d. | n.d. | 40 | 469 |
| 3,5-Dichlorobenzoate | 0 | n.d. | n.d. | 0 | 0 |
| 2-Hydroxybenzoate | n.d. | 48 | n.d. | 0 | n.d. |
| 4-Hydroxybenzoate | 94 | 0 | n.d. | 0 | 162 |
| 3,4-Dihydroxybenzoate | 41 | n.d. | n.d. | 93 | 46 |
| 2,5-Dihydroxybenzoate | 80 | n.d. | n.d. | n.d. | 85 |
| Phenol | 15 | n.d. | 8 | 93 | 0 |
| 2Chlorophenol | 54 | n.d. | 10 | 120 | 0 |
| 3Chlorophenol | 61 | n.d. | 4 | 133 | 39 |
| 4Chlorophenol | 36 | n.d. | n.d. | 120 | 200 |
| Enzyme Substrate | Strain | |||
|---|---|---|---|---|
| R. opacus 6a | R.opacus 1CP | R.ruber P25 | M. foliorum B51 | |
| CCat 1,2-DO | ||||
| Catechol | 0.206 ± 0.008 | 0.007 ± 0.001 | 0.005 ± 0.0002 | 0.20 ± 0.005 |
| MCI | ||||
| Muconate | 0 | 0.003 ± 0.0001 | 0 | 0 |
| 2-Methylmuconate | 0 | 0 | 0.0003 ± 0.00002 | 0 |
| 3- Methylmuconate | 0.0042 ± 0.0002 | 0.043 ± 0.001 | 0.0003 ± 0.00001 | 0 |
| 2-Chloromuconate | 0 | 0.013 ± 0.001 | 0.0071 ± 0.0003 | 0.0221 ± 0.0008 |
| DLH | ||||
| cis-Dienelactone | 0.124 ± 0.005 | 0.054 ± 0.002 | 0.0033 ± 0.0001 | 0.095 ± 0.004 |
| Substrate | Strain | |||
|---|---|---|---|---|
| R. opacus 6a | R.opacus 1cp | R. ruber P25 | M. foliorum B51 | |
| Catechol | 100 | 100 | 100 | 100 |
| 3-Methylcatechol | 100 | 120 | 125 | 131 |
| 4-Methylcatechol | n.d. | 197 | 162 | 230 |
| 4-Chlorocatechol | 25 | 52 | 56 | 73 |
| 3-Chlorocatechol | 100 | 14 | n.d. | n.d. |
| 3,5-Dichlorocatechol | 4 | 9 | n.d. | n.d. |
| Substrate | Activity after Q-Sepharose Chromatography | |||
|---|---|---|---|---|
| R. opacus 1CP | R. ruber P25 | M. foliorum B 51 | ||
| Peak 1 | Peak 2 | |||
| Catechol | 100 | 100 | 100 | 100 |
| 3-Chlorocatechol | 94 | n.d. | n.d. | n.d. |
| 4-Chlorocatechol | 55 | 87 | 20 | 77 |
| 3-Methylcatechol | 78 | 225 | 98 | 115 |
| 4-Methylcatechol | 188 | 262 | 124 | 203 |
| 3,5-Dichlorocatechol | 233 | n.d. | 3.8 | 9.8 |
| 3,6-Dichlorocatechol | n.d. | 50 | n.d. | n.d. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Emelyanova, E.V.; Ramanaiah, S.V.; Prisyazhnaya, N.V.; Shumkova, E.S.; Plotnikova, E.G.; Wu, Y.; Solyanikova, I.P. The Contribution of Actinobacteria to the Degradation of Chlorinated Compounds: Variations in the Activity of Key Degradation Enzymes. Microorganisms 2023, 11, 141. https://doi.org/10.3390/microorganisms11010141
Emelyanova EV, Ramanaiah SV, Prisyazhnaya NV, Shumkova ES, Plotnikova EG, Wu Y, Solyanikova IP. The Contribution of Actinobacteria to the Degradation of Chlorinated Compounds: Variations in the Activity of Key Degradation Enzymes. Microorganisms. 2023; 11(1):141. https://doi.org/10.3390/microorganisms11010141
Chicago/Turabian StyleEmelyanova, Elena V., Sudarsu V. Ramanaiah, Nataliya V. Prisyazhnaya, Ekaterina S. Shumkova, Elena G. Plotnikova, Yonghong Wu, and Inna P. Solyanikova. 2023. "The Contribution of Actinobacteria to the Degradation of Chlorinated Compounds: Variations in the Activity of Key Degradation Enzymes" Microorganisms 11, no. 1: 141. https://doi.org/10.3390/microorganisms11010141







