Metabolic Engineering of Escherichia coli for High-Level Production of (R)-Acetoin from Low-Cost Raw Materials
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Strains and Plasmids
2.3. Culture Conditions
2.4. Screening of Acetoin Resistance Genes
2.5. Overexpression of Gene tsf
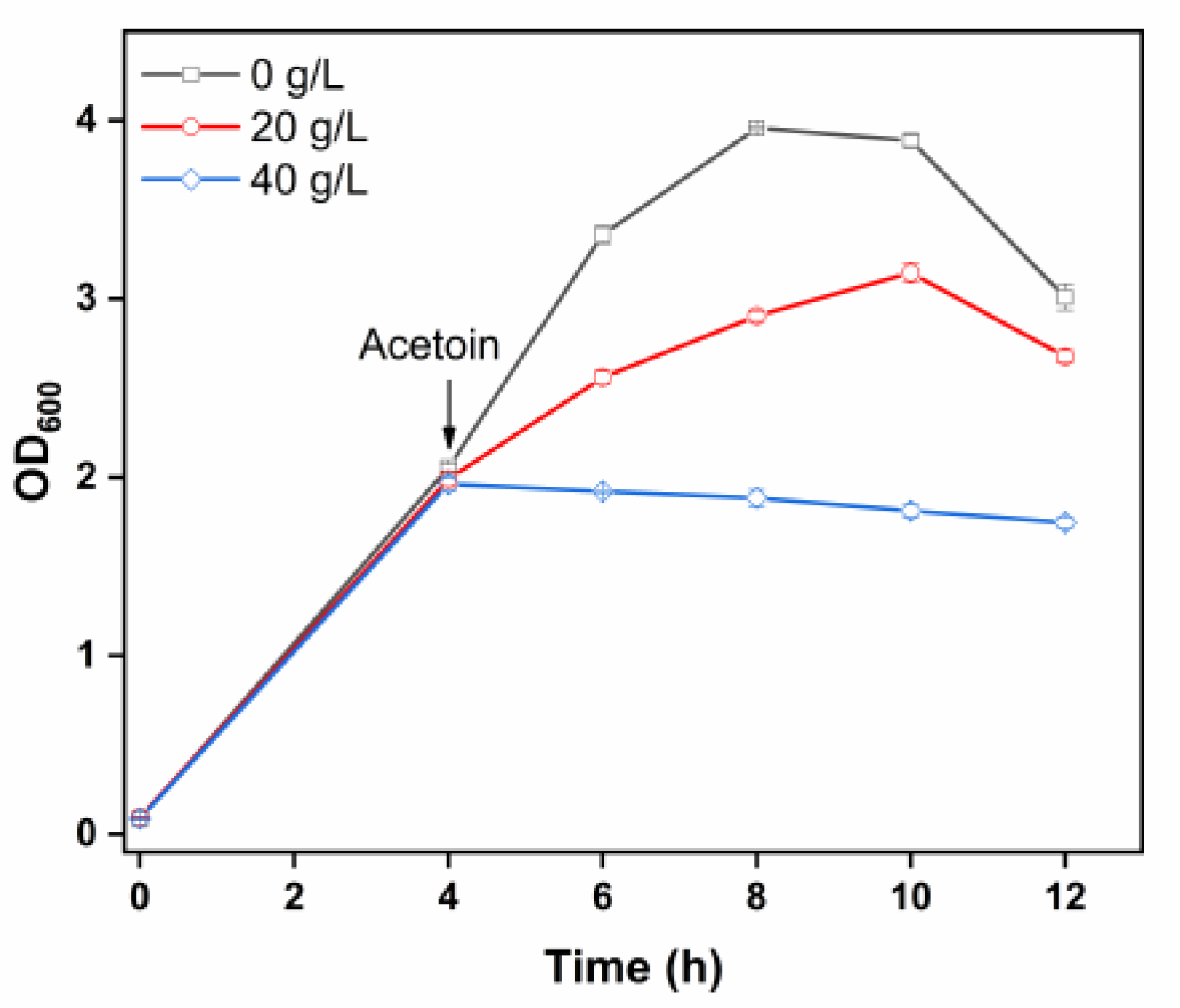
2.6. Stress Effects of Acetoin on Strain GXASR-48p
2.7. Production of Acetoin in Shake Flasks
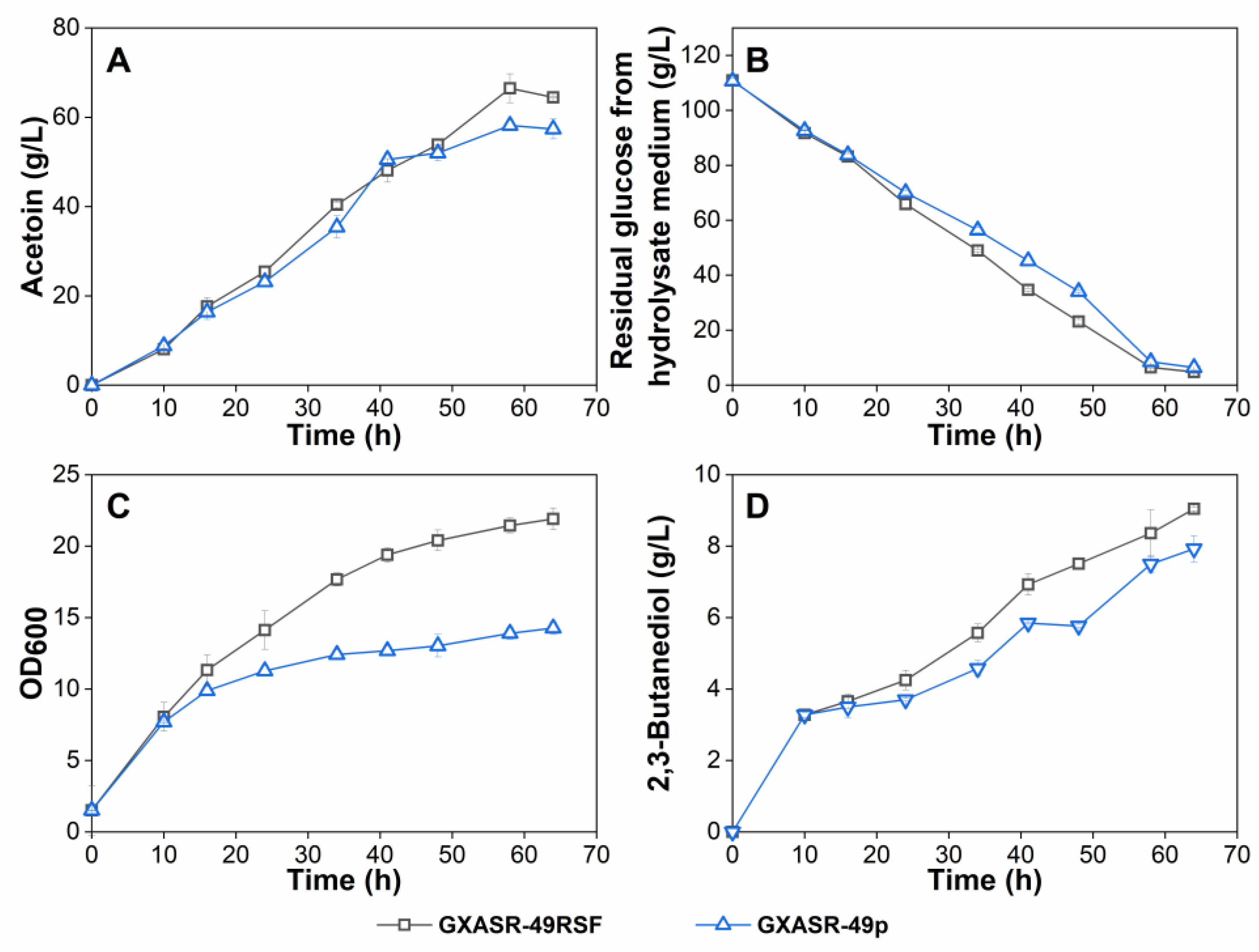
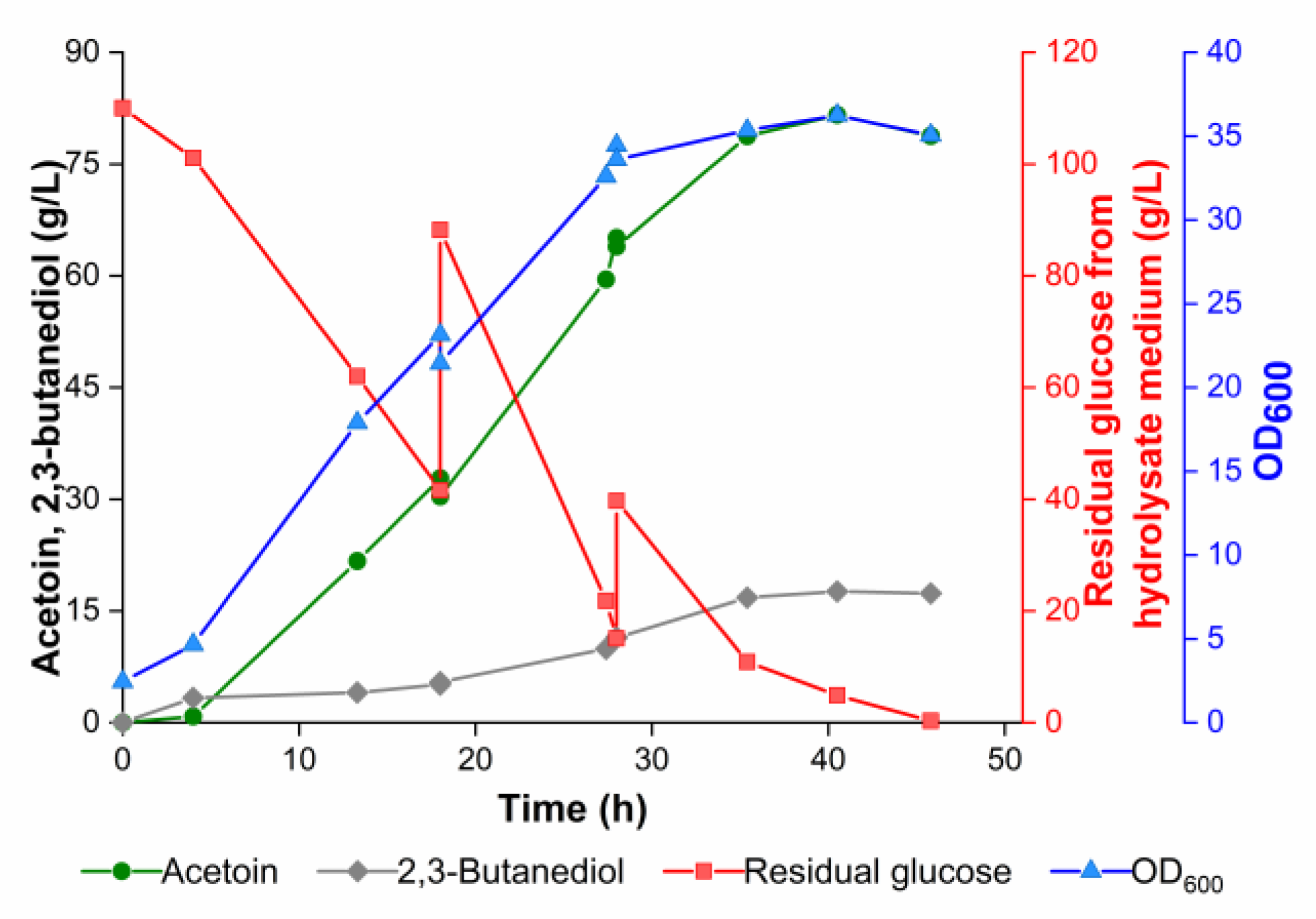
2.8. Fed-Batch Fermentation in a 3-L Fermenter
2.9. Analytical Methods
3. Results and Discussion
3.1. Optimization of Metabolic Pathways in Acetoin-Producting Strains
3.2. Evaluation of Acetoin Toxicity on Engineered Strain GXASR-48p
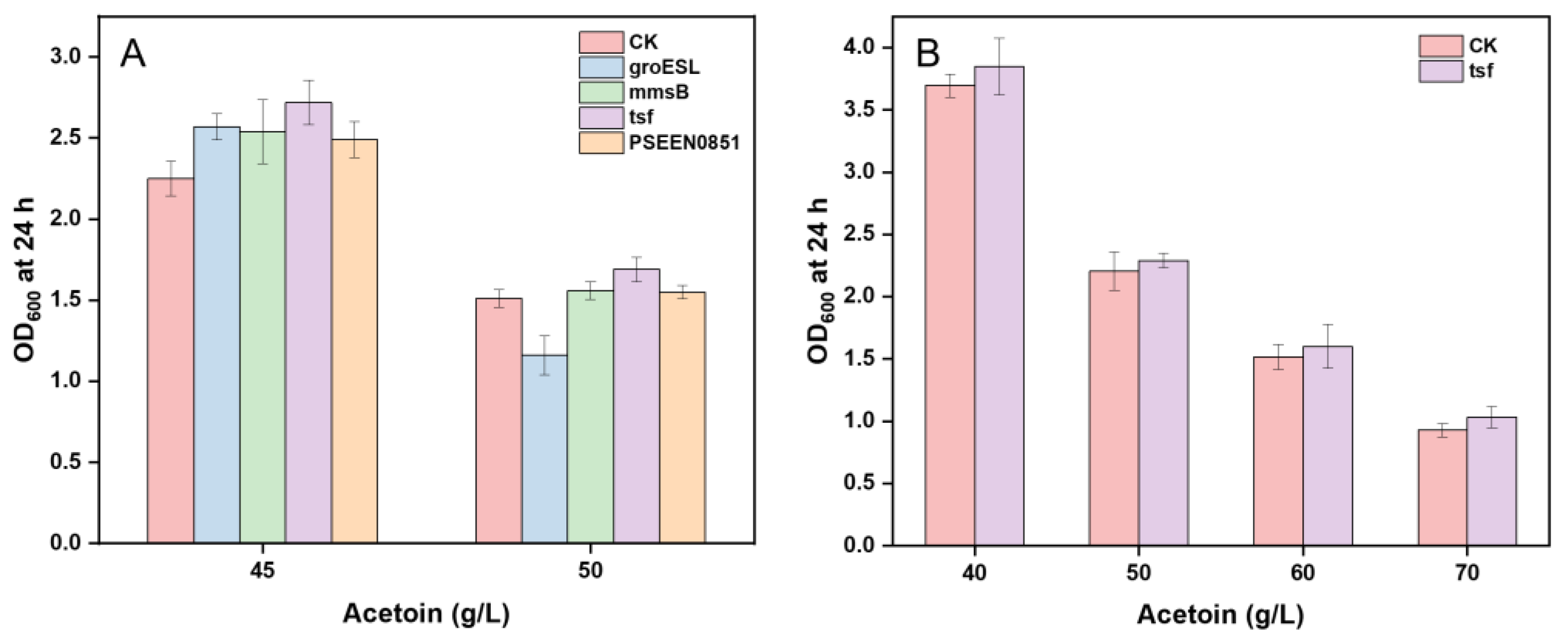
3.3. Screen of Acetoin Resistance Genes
3.4. Effect of Resistant Gene tsf on Acetoin Production
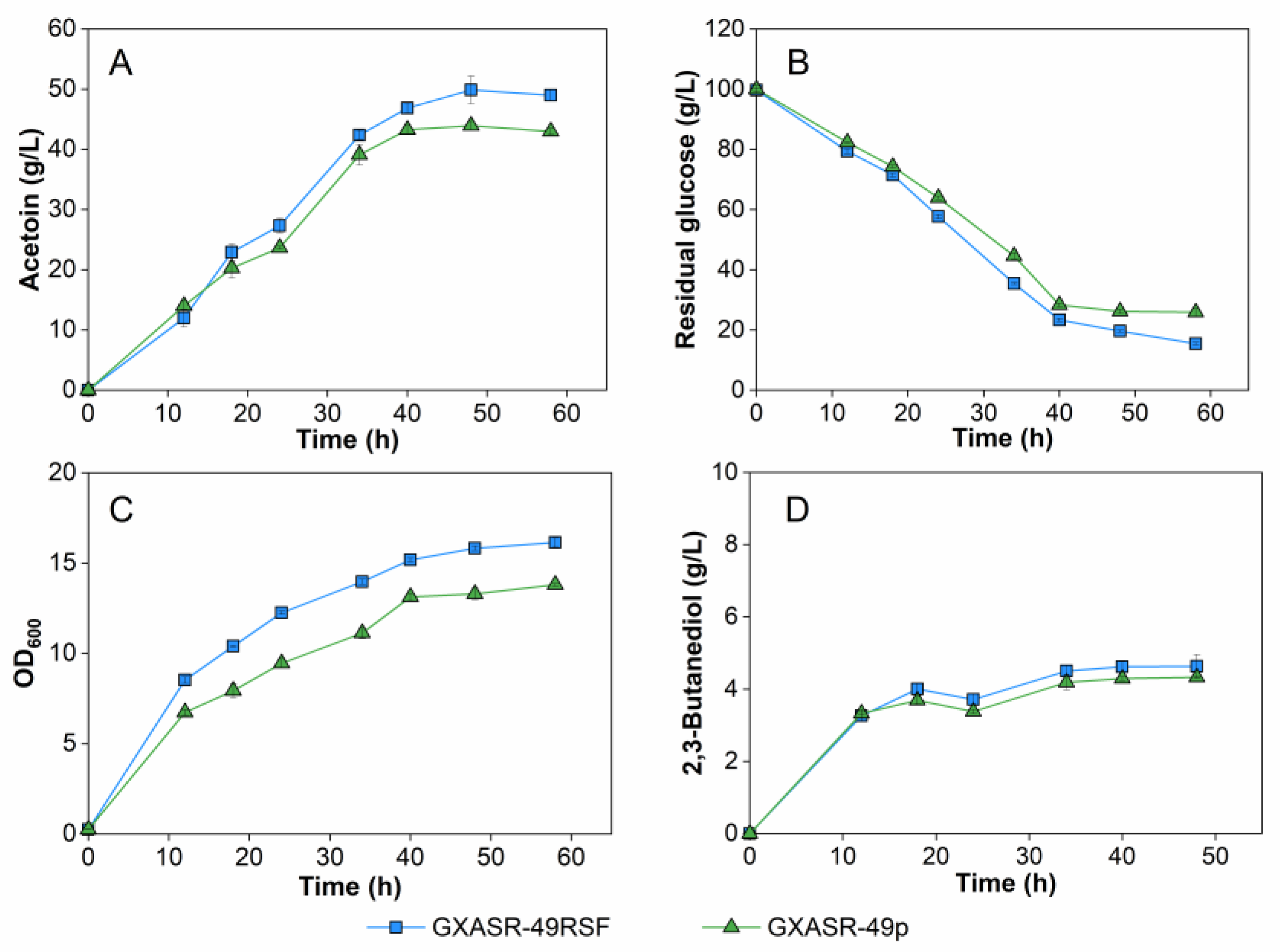
3.5. Effect of Altering the Expression Level of Pathway Genes on Acetoin Production
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yuan, H.; Xu, Y.; Chen, Y.; Zhan, Y.; Wei, X.; Li, L.; Wang, D.; He, P.; Li, S.; Chen, S. Metabolomics analysis reveals global acetoin stress response of Bacillus licheniformis. Metabolomics 2019, 15, 25. [Google Scholar] [CrossRef]
- Lu, L.; Mao, Y.; Kou, M.; Cui, Z.; Jin, B.; Chang, Z.; Wang, Z.; Ma, H.; Chen, T. Engineering central pathways for industrial-level (3R)-acetoin biosynthesis in Corynebacterium glutamicum. Microb. Cell Factories 2020, 19, 102. [Google Scholar] [CrossRef]
- Cui, Z.; Wang, Z.; Zheng, M.; Chen, T. Advances in biological production of acetoin: A comprehensive overview. Crit. Rev. Biotechnol. 2021, 42, 1135–1156. [Google Scholar] [CrossRef]
- Werpy, T.; Petersen, G. Top Value added Chemicals from Biomass: Volume I—Results of Screening for Potential Candidates from Sugars and Synthesis Gas; National Renewable Energy Lab.: Golden, CO, USA, 2004.
- Zhang, L.; Liu, Q.; Ge, Y.; Li, L.; Gao, C.; Xu, P.; Ma, C. Biotechnological production of acetoin, a bio-based platform chemical, from a lignocellulosic resource by metabolically engineered Enterobacter cloacae. Green Chem. 2016, 18, 1560–1570. [Google Scholar] [CrossRef]
- Kandasamy, V.; Liu, J.; Dantoft, S.H.; Solem, C.; Jensen, P.R. Synthesis of (3R)-acetoin and 2,3-butanediol isomers by metabolically engineered Lactococcus lactis. Sci. Rep. 2016, 6, 36769. [Google Scholar] [CrossRef]
- Li, J.; Lu, J.; Ma, Z.; Li, J.; Chen, X.; Diao, M.; Xie, N. A green route for high-yield production of tetramethylpyrazine from non-food raw materials. Front. Bioeng. Biotechnol. 2022, 9, 1350. [Google Scholar] [CrossRef]
- Xiao, Z.; Lu, J.R. Strategies for enhancing fermentative production of acetoin: A review. Biotechnol. Adv. 2014, 32, 492–503. [Google Scholar] [CrossRef]
- Meng, W.; Ma, C.; Xu, P.; Gao, C. Biotechnological production of chiral acetoin. Trends Biotechnol. 2022, 40, 958–973. [Google Scholar] [CrossRef]
- Jang, J.-W.; Jung, H.-M.; Kim, D.G.; Oh, M.-K. Acetoin production using metabolically engineered Klebsiella pneumoniae. Korean Chem. Eng. Res. 2017, 55, 237–241. [Google Scholar] [CrossRef]
- Bai, F.; Dai, L.; Fan, J.; Truong, N.; Rao, B.; Zhang, L.; Shen, Y. Engineered Serratia marcescens for efficient (3R)-acetoin and (2R,3R)-2,3-butanediol production. J. Ind. Microbiol. Biotechnol. 2015, 42, 779–786. [Google Scholar] [CrossRef]
- Yang, T.; Rao, Z.; Zhang, X.; Xu, M.; Xu, Z.; Yang, S.-T. Metabolic engineering strategies for acetoin and 2,3-butanediol production: Advances and prospects. Crit. Rev. Biotechnol. 2017, 37, 990–1005. [Google Scholar] [CrossRef]
- Bae, S.-J.; Kim, S.; Hahn, J.-S. Efficient production of acetoin in Saccharomyces cerevisiae by disruption of 2,3-butanediol dehydrogenase and expression of NADH oxidase. Sci. Rep. 2016, 6, 27667. [Google Scholar] [CrossRef]
- Sørensen, H.P.; Mortensen, K.K. Advanced genetic strategies for recombinant protein expression in Escherichia coli. J. Biotech. 2005, 115, 113–128. [Google Scholar] [CrossRef]
- Xu, Y.; Chu, H.; Gao, C.; Tao, F.; Zhou, Z.; Li, K.; Li, L.; Ma, C.; Xu, P. Systematic metabolic engineering of Escherichia coli for high-yield production of fuel bio-chemical 2,3-butanediol. Metab. Eng. 2014, 23, 22–33. [Google Scholar] [CrossRef]
- Maina, S.; Prabhu, A.A.; Vivek, N.; Vlysidis, A.; Koutinas, A.; Kumar, V. Prospects on bio-based 2,3-butanediol and acetoin production: Recent progress and advances. Biotechnol. Adv. 2022, 54, 107783. [Google Scholar] [CrossRef]
- Xu, Q.; Xie, L.; Li, Y.; Lin, H.; Sun, S.; Guan, X.; Hu, K.; Shen, Y.; Zhang, L. Metabolic engineering of Escherichia coli for efficient production of (3R)-acetoin. J. Chem. Technol. Biotechnol. 2015, 90, 93–100. [Google Scholar] [CrossRef]
- Xu, Y.; Xu, C.; Li, X.; Sun, B.; Eldin, A.A.; Jia, Y. A combinational optimization method for efficient synthesis of tetramethylpyrazine by the recombinant Escherichia coli. Biochem. Eng. J. 2018, 129, 33–43. [Google Scholar] [CrossRef]
- Guo, Z.; Zhao, X.; He, Y.; Yang, T.; Gao, H.; Li, G.; Chen, F.; Sun, M.; Lee, J.-K.; Zhang, L. Efficient (3R)-acetoin production from meso-2, 3-butanediol using a new whole-cell biocatalyst with co-expression of meso-2, 3-butanediol dehydrogenase, NADH oxidase, and Vitreoscilla hemoglobin. J. Microbiol. Biotechnol. 2017, 27, 92–100. [Google Scholar] [CrossRef]
- Peters, D. Raw Materials. In White Biotechnology; Ulber, R., Sell, D., Eds.; Springer: Berlin/Heidelberg, Germany, 2007; pp. 1–30. [Google Scholar] [CrossRef]
- Choi, G.-W.; Moon, S.-K.; Kang, H.-W.; Min, J.; Chung, B.-W. Simultaneous saccharification and fermentation of sludge-containing cassava mash for batch and repeated batch production of bioethanol by Saccharomyces cerevisiae CHFY0321. J. Chem. Technol. Biotechnol. 2009, 84, 547–553. [Google Scholar] [CrossRef]
- Papong, S.; Malakul, P. Life-cycle energy and environmental analysis of bioethanol production from cassava in Thailand. Bioresour. Technol. 2010, 101, S112–S118. [Google Scholar] [CrossRef]
- Wang, L.; Zhao, B.; Liu, B.; Yang, C.; Yu, B.; Li, Q.; Ma, C.; Xu, P.; Ma, Y. Efficient production of L-lactic acid from cassava powder by Lactobacillus rhamnosus. Bioresour. Technol. 2010, 101, 7895–7901. [Google Scholar] [CrossRef]
- Altaf, M.; Naveena, B.J.; Reddy, G. Use of inexpensive nitrogen sources and starch for L(+) lactic acid production in anaerobic submerged fermentation. Bioresour. Technol. 2007, 98, 498–503. [Google Scholar] [CrossRef]
- Li, Y.; Wang, L.; Ju, J.; Yu, B.; Ma, Y. Efficient production of polymer-grade d-lactate by Sporolactobacillus laevolacticus DSM442 with agricultural waste cottonseed as the sole nitrogen source. Bioresour. Technol. 2013, 142, 186–191. [Google Scholar] [CrossRef]
- Cesselin, B.; Henry, C.; Gruss, A.; Gloux, K.; Gaudu, P.; Björkroth, J. Mechanisms of acetoin toxicity and adaptive responses in an acetoin-producing species, Lactococcus lactis. Appl. Environ. Microbiol. 2021, 87, e01079-21. [Google Scholar] [CrossRef]
- Luo, Q.; Wu, J.; Wu, M. Enhanced acetoin production by Bacillus amyloliquefaciens through improved acetoin tolerance. Process Biochem. 2014, 49, 1223–1230. [Google Scholar] [CrossRef]
- Si, H.-M.; Zhang, F.; Wu, A.-N.; Han, R.-Z.; Xu, G.-C.; Ni, Y. DNA microarray of global transcription factor mutant reveals membrane-related proteins involved in n-butanol tolerance in Escherichia coli. Biotechnol. Biofuels 2016, 9, 114. [Google Scholar] [CrossRef]
- Yuan, Y.; Bi, C.; Nicolaou, S.A.; Zingaro, K.A.; Ralston, M.; Papoutsakis, E.T. Overexpression of the Lactobacillus plantarum peptidoglycan biosynthesis murA2 gene increases the tolerance of Escherichia coli to alcohols and enhances ethanol production. Appl. Microbiol. Biotechnol. 2014, 98, 8399–8411. [Google Scholar] [CrossRef]
- Kamthan, A.; Kamthan, M.; Datta, A. Expression of C-5 sterol desaturase from an edible mushroom in fisson yeast enhances its ethanol and thermotolerance. PLoS ONE 2017, 12, e0173381. [Google Scholar] [CrossRef]
- Fisher, M.A.; Boyarskiy, S.; Yamada, M.R.; Kong, N.; Bauer, S.; Tullman-Ercek, D. Enhancing tolerance to short-chain alcohols by engineering the Escherichia coli AcrB efflux pump to secrete the non-native substrate n-butanol. ACS Synth. Biol. 2014, 3, 30–40. [Google Scholar] [CrossRef]
- Blin, K.; Pedersen, L.E.; Weber, T.; Lee, S.Y. CRISPy-web: An online resource to design sgRNAs for CRISPR applications. Synth. Syst. Biotechnol. 2016, 1, 118–121. [Google Scholar] [CrossRef]
- Lv, X.; Dai, L.; Bai, F.; Wang, Z.; Zhang, L.; Shen, Y. Metabolic engineering of Serratia marcescens MG1 for enhanced production of (3R)-acetoin. Bioresour. Bioprocess. 2016, 3, 52. [Google Scholar] [CrossRef]
- Hao, Z.-k.; Li, P.-w.; Hao, Z.-c.; Chen, L.-f. Effect of knockouting frdB on anaerobic mixed acid fermentation for Escherichia coli. China Biotechnol. 2014, 34, 67–75. [Google Scholar] [CrossRef]
- Li, Z.-y.; Wang, J.; Tian, K.-m.; Dong, Z.-x.; Jin, P.; Liu, X.-g.; Wang, Z.-x. High-efficiency production of succinic acid from glycerol by metabolically engineered Escherichia coli. Sci. Technol. Food Ind. 2018, 39, 94. [Google Scholar] [CrossRef]
- Cao, J.; Zhou, L.; Zhang, L.; Wang, Z.; Shi, G. Construction and fermentation of succinate-producing recombinant Escherichia coli. Chin. J. Appl. Environ. Biol. 2010, 16, 851–857. [Google Scholar] [CrossRef]
- Zingaro, K.A.; Terry Papoutsakis, E. GroESL overexpression imparts Escherichia coli tolerance to i-, n-, and 2-butanol, 1,2,4-butanetriol and ethanol with complex and unpredictable patterns. Metab. Eng. 2013, 15, 196–205. [Google Scholar] [CrossRef] [PubMed]
- Ni, Y.; Song, L.; Qian, X.; Sun, Z. Proteomic analysis of Pseudomonas putida reveals an organic solvent tolerance-related gene mmsB. PLoS ONE 2013, 8, e55858. [Google Scholar] [CrossRef]
- Tan, Z.; Yoon, J.M.; Nielsen, D.R.; Shanks, J.V.; Jarboe, L.R. Membrane engineering via trans unsaturated fatty acids production improves Escherichia coli robustness and production of biorenewables. Metab. Eng. 2016, 35, 105–113. [Google Scholar] [CrossRef]
- Bui, L.M.; Lee, J.Y.; Geraldi, A.; Rahman, Z.; Lee, J.H.; Kim, S.C. Improved n-butanol tolerance in Escherichia coli by controlling membrane related functions. J. Biotech. 2015, 204, 33–44. [Google Scholar] [CrossRef]
- Werbowy, O.; Werbowy, S.; Kaczorowski, T. Plasmid stability analysis based on a new theoretical model employing stochastic simulations. PLoS ONE 2017, 12, e0183512. [Google Scholar] [CrossRef]
- Mesa-Pereira, B.; Rea, M.C.; Cotter, P.D.; Hill, C.; Ross, R.P. Heterologous expression of biopreservative bacteriocins with a view to low cost production. Front. Microbiol. 2018, 9, 1654. [Google Scholar] [CrossRef]



| Strains or Plasmids | Descriptions | Sources |
|---|---|---|
| Strains | ||
| E. coli MG1655 | Host strain for heterologous biosynthesis of acetoin | Lab collection |
| E. coli MG1655p | E. coli MG1655 harboring pTrc99A-budB-budA-noxE | Lab collection |
| E. coli DH5a | Host strain for cloning | Lab collection |
| GXASR-48 | E. coli MG1655 ΔgldA ΔfrdABCD ΔackA-pta ΔpoxB | This study |
| GXASR-48-CK | GXASR-48 harboring pTrc99A | This study |
| GXASR-48-groESL | GXASR-48 harboring pTrc99A-groESL | This study |
| GXASR-48-mmsB | GXASR-48 harboring pTrc99A-mmsB | This study |
| GXASR-48-tsf | GXASR-48 harboring pTrc99A-tsf | This study |
| GXASR-48-PSEEN0851 | GXASR-48 harboring pTrc99A-PSEEN0851 | This study |
| GXASR-48ΔyibT | E. coli MG1655 ΔgldA ΔfrdABCD ΔackA-pta ΔpoxB ΔyibT | This study |
| GXASR-48ΔyghW | E. coli MG1655 ΔgldA ΔfrdABCD ΔackA-pta ΔpoxB ΔyghW | This study |
| GXASR-49 | E. coli MG1655 ΔgldA ΔfrdABCD ΔackA-pta ΔpoxB yibT:: tsf | This study |
| GXASR-48p | GXASR-48 harboring pTrc99A-budB-budA-noxE | This study |
| GXASR-49p | GXASR-49 harboring pTrc99A-budB-budA-noxE | This study |
| GXASR-48RSF | GXASR-48 harboring pRSF-budB-budA-noxE | This study |
| GXASR-49RSF | GXASR-49 harboring pRSF-budB-budA-noxE | This study |
| Plasmids | ||
| pTrc99A | AmpR | Lab collection |
| pTrc99A-budB-budA-noxE | AmpR | Lab collection |
| pTrc99A-groESL | AmpR | This study |
| pTrc99A-mmsB | AmpR | This study |
| pTrc99A-tsf | AmpR | This study |
| pTrc99A-PSEEN0851 | AmpR | This study |
| pRSFDuet | KanR | Lab collection |
| pRSF-budB-budA-noxE | Replace the ori of pTrc99A with RSF replicon of pRSFDuet | This study |
| pTarget-tdcC | StrR (constitutive expression) | Kindly provided by Prof. Bo Yu, Chinese Academy of Sciences |
| pCas | KanR |
| Strains | Formate (g/L) | Acetate (g/L) | Succinate (g/L) |
|---|---|---|---|
| E. coli MG1655p | 0.41 ± 0.05 | 2.13 ± 0.08 | 1.44 ± 0.19 |
| GXASR-48p | ND a | 0.64 ± 0.10 | 0.15 ± 0.06 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Diao, M.; Chen, X.; Li, J.; Shi, Y.; Yu, B.; Ma, Z.; Li, J.; Xie, N. Metabolic Engineering of Escherichia coli for High-Level Production of (R)-Acetoin from Low-Cost Raw Materials. Microorganisms 2023, 11, 203. https://doi.org/10.3390/microorganisms11010203
Diao M, Chen X, Li J, Shi Y, Yu B, Ma Z, Li J, Xie N. Metabolic Engineering of Escherichia coli for High-Level Production of (R)-Acetoin from Low-Cost Raw Materials. Microorganisms. 2023; 11(1):203. https://doi.org/10.3390/microorganisms11010203
Chicago/Turabian StyleDiao, Mengxue, Xianrui Chen, Jing Li, Ya’nan Shi, Bo Yu, Zhilin Ma, Jianxiu Li, and Nengzhong Xie. 2023. "Metabolic Engineering of Escherichia coli for High-Level Production of (R)-Acetoin from Low-Cost Raw Materials" Microorganisms 11, no. 1: 203. https://doi.org/10.3390/microorganisms11010203
APA StyleDiao, M., Chen, X., Li, J., Shi, Y., Yu, B., Ma, Z., Li, J., & Xie, N. (2023). Metabolic Engineering of Escherichia coli for High-Level Production of (R)-Acetoin from Low-Cost Raw Materials. Microorganisms, 11(1), 203. https://doi.org/10.3390/microorganisms11010203