Lactiplantibacillus plantarum Strains Modulate Intestinal Innate Immune Response and Increase Resistance to Enterotoxigenic Escherichia coli Infection
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Culture Conditions
2.2. Porcine Intestinal Epitheliocyte Cells
2.3. Immunomodulatory Assay in PIE Cells
2.4. Quantitative Expression Analysis by RT-PCR
2.5. ETEC Challenge in Mice
2.6. Ex Vivo Peritoneal Macrophage Phagocytosis Assay
2.7. Bactericidal Activity of Peritoneal Macrophages
2.8. Cytokine Concentrations
2.9. Statistical Analysis
3. Results
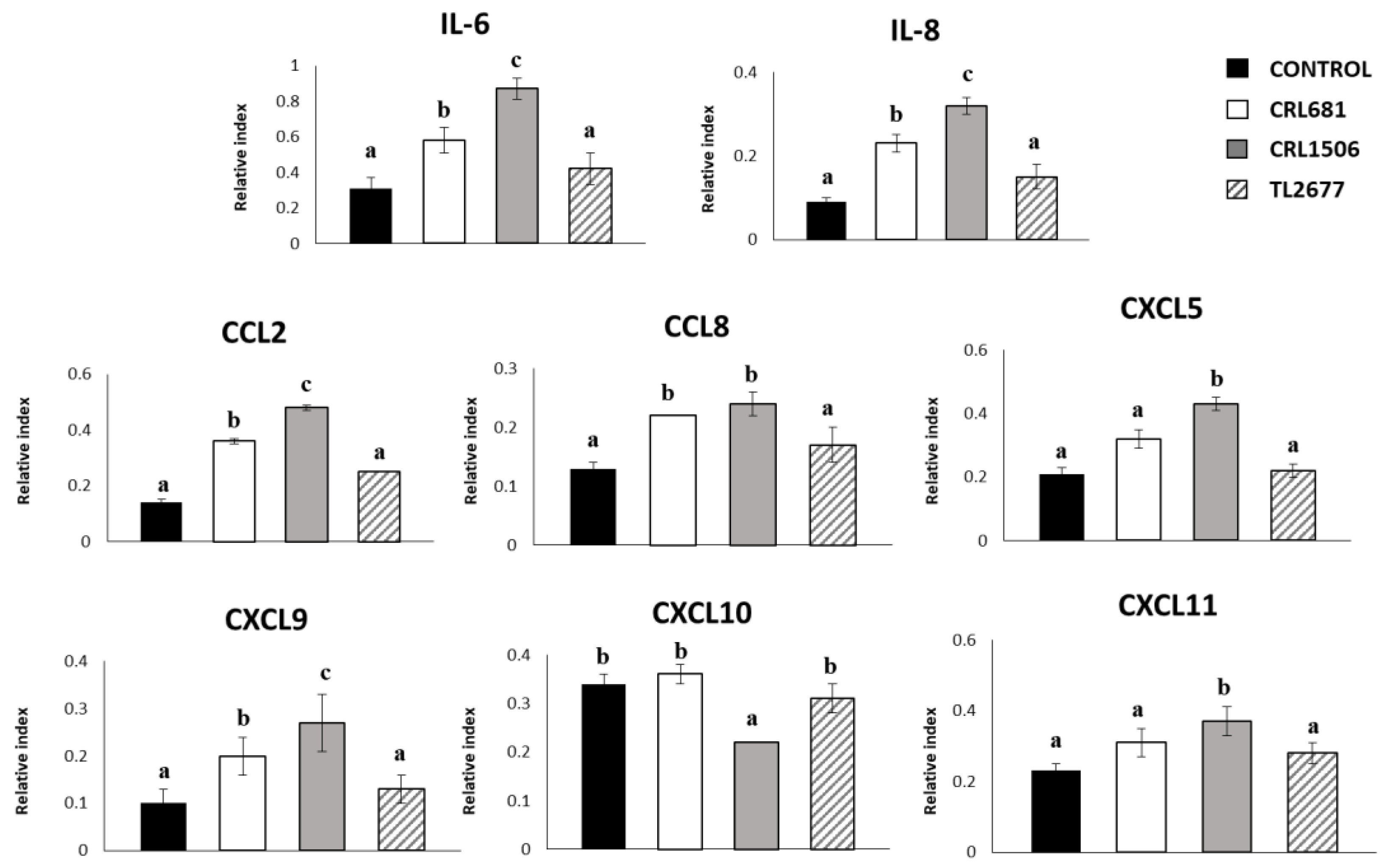
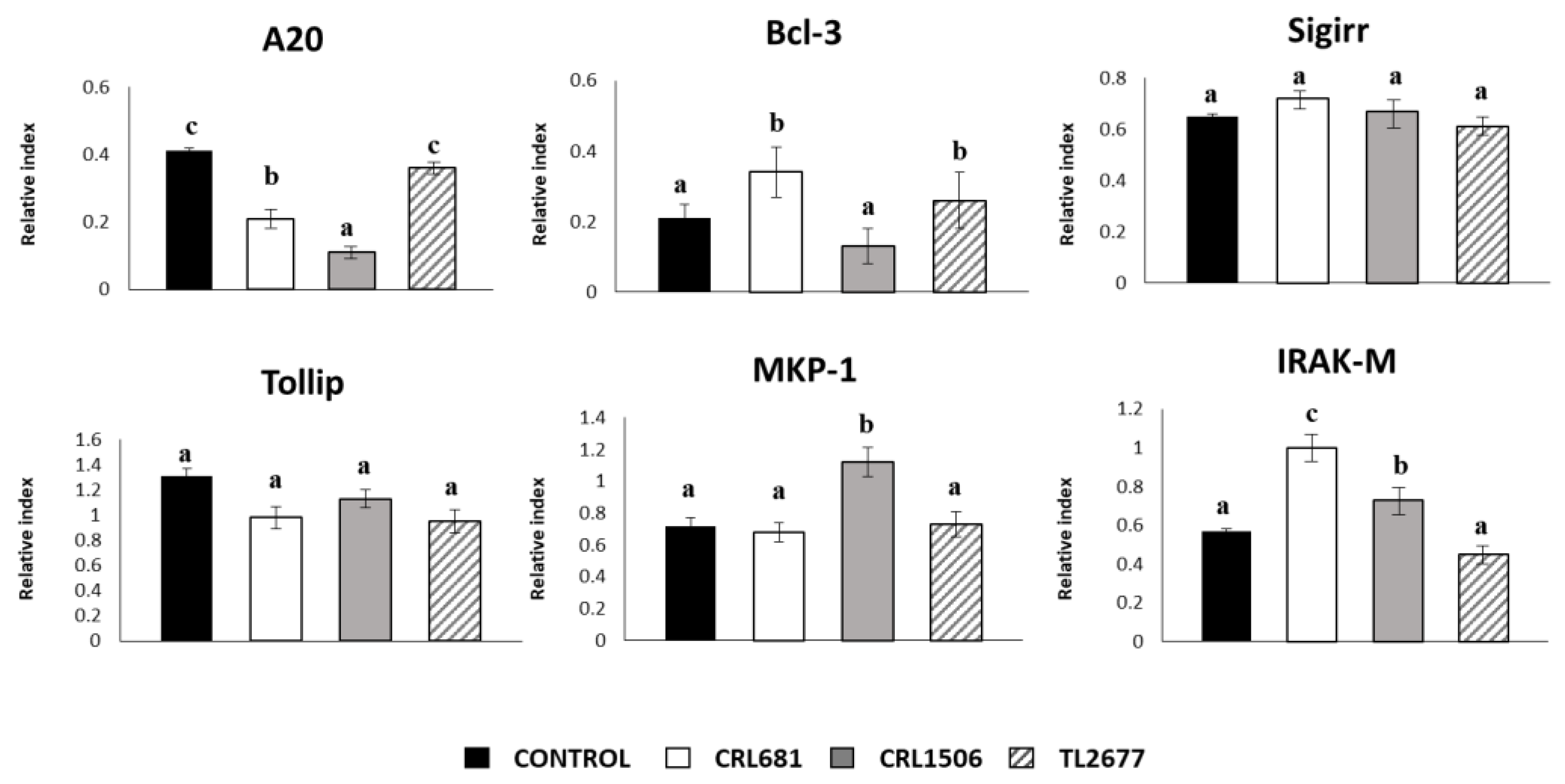
3.1. Effect of L. plantarum Strains on the Expression of Cytokines and the Negative Regulators of the TLR4 Signaling in PIE Cells
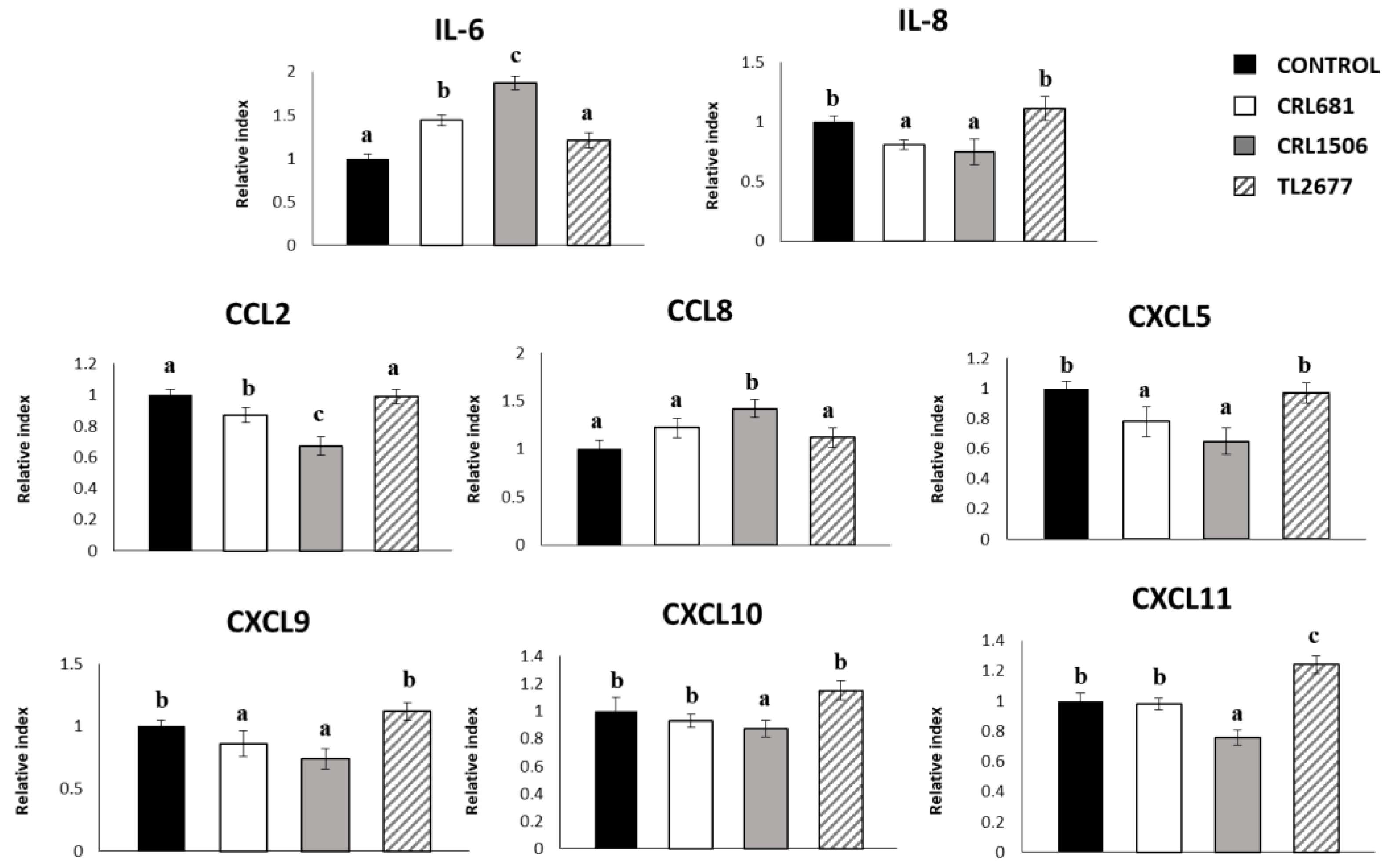
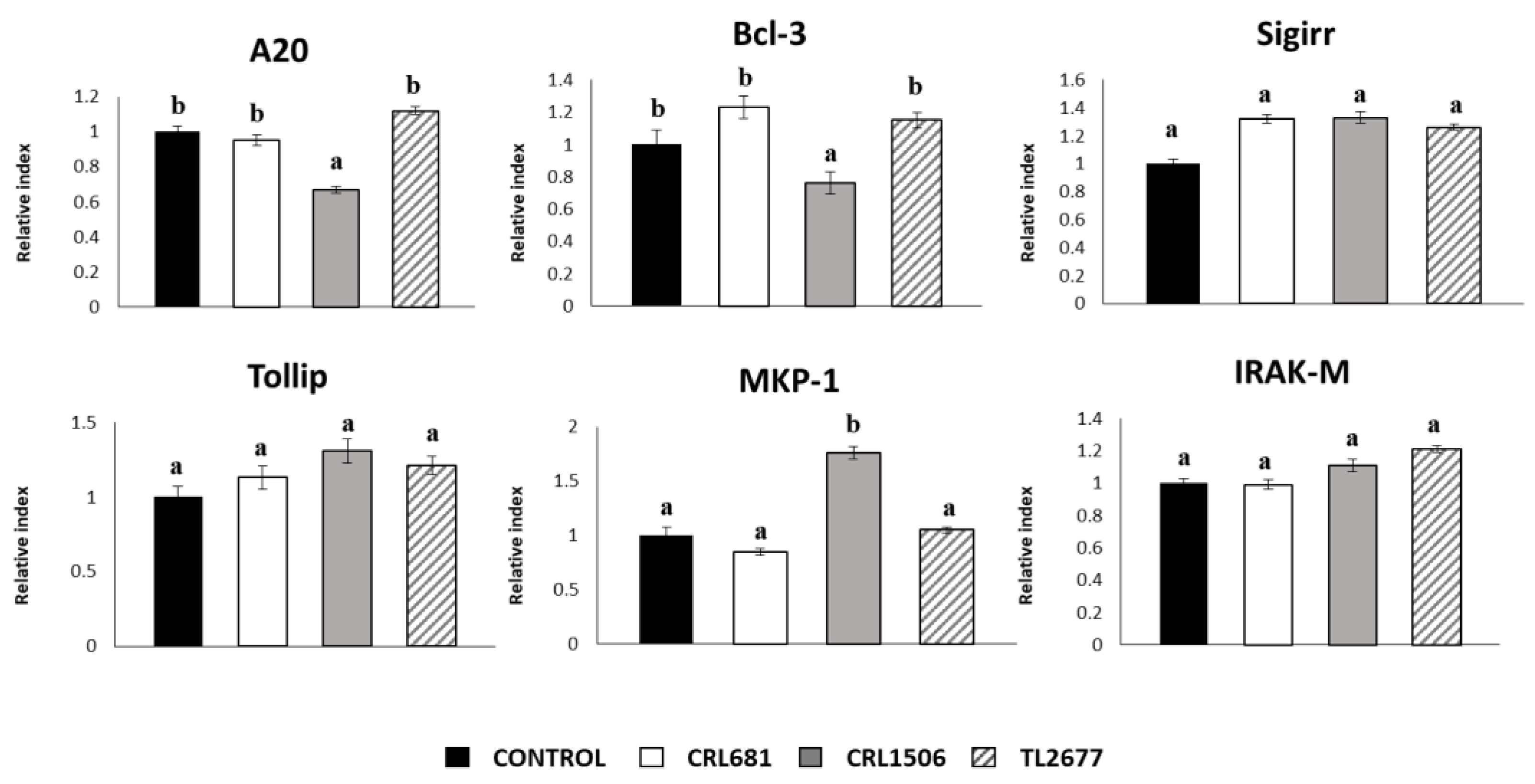
3.2. Effect of L. plantarum Strains on ETEC-Activated Innate Immune Response in PIE Cells
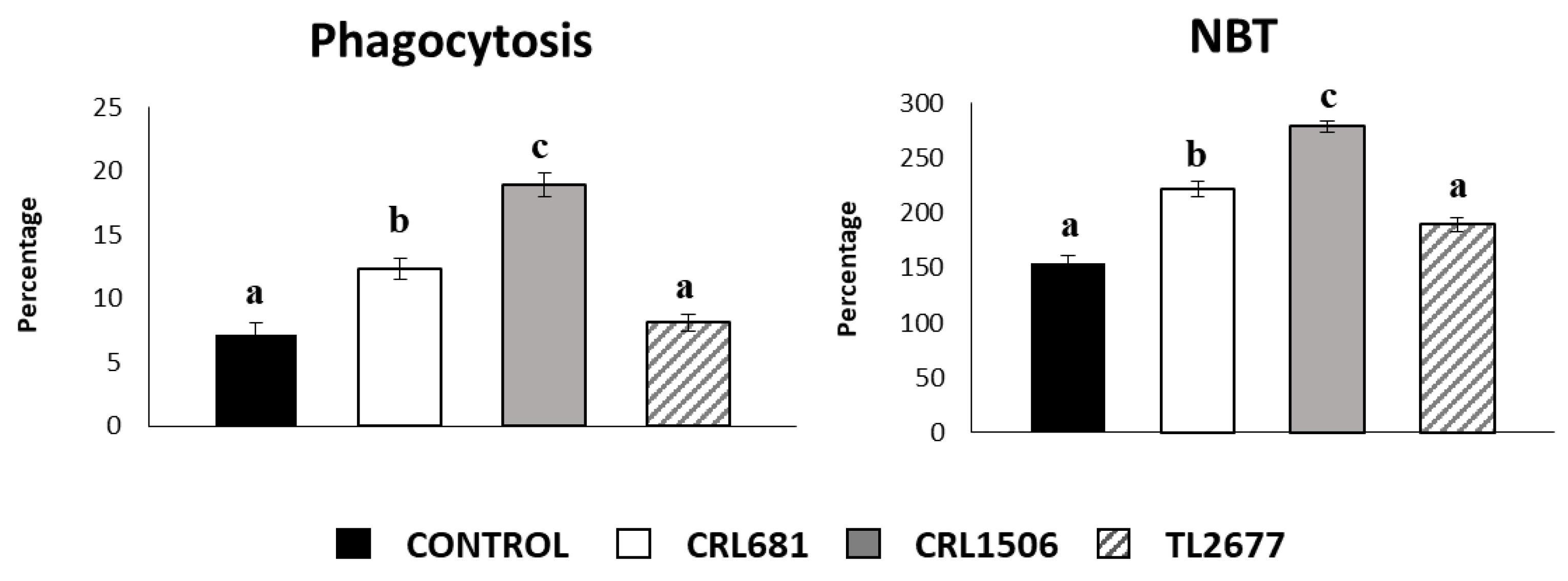
3.3. L. plantarum CRL681 and CRL1506 Modulate Intestinal Immunity In Vivo
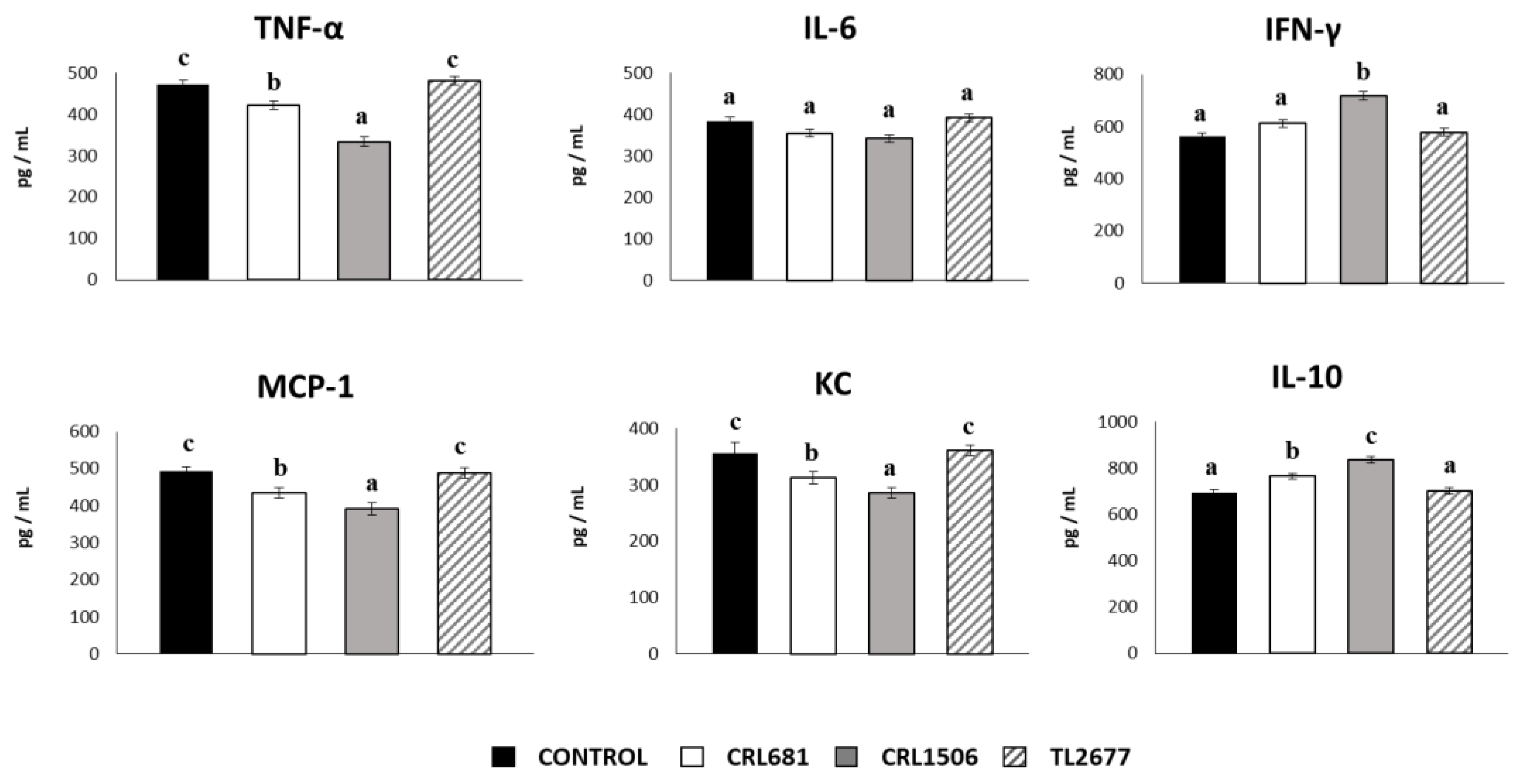
3.4. L. plantarum CRL681 and CRL1506 prevent the Spread of ETEC and Modulate the Expression of Intestinal Cytokines after Bacterial Challenge
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Khalil, I.A.; Troeger, C.; Blacker, B.F.; Rao, P.C.; Brown, A.; Atherly, D.E.; Brewer, T.G.; Engmann, C.M.; Houpt, E.R.; Kang, G.; et al. Morbidity and Mortality Due to Shigella and Enterotoxigenic Escherichia coli Diarrhoea: The Global Burden of Disease Study 1990–2016. Lancet Infect. Dis. 2018, 18, 1229–1240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laird, T.J.; Abraham, S.; Jordan, D.; Pluske, J.R.; Hampson, D.J.; Trott, D.J.; O’Dea, M. Porcine Enterotoxigenic Escherichia coli: Antimicrobial Resistance and Development of Microbial-Based Alternative Control Strategies. Vet. Microbiol. 2021, 258, 109117. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Cui, D.; Chao, X.; Chen, P.; Liu, J.; Wang, Y.; Su, T.; Li, M.; Xu, R.; Zhu, Y.; et al. Transcriptome Analysis Identifies Strategies Targeting Immune Response-Related Pathways to Control Enterotoxigenic Escherichia coli Infection in Porcine Intestinal Epithelial Cells. Front. Vet. Sci. 2021, 8, 677897. [Google Scholar] [CrossRef] [PubMed]
- Dubreuil, J.D.; Isaacson, R.E.; Schifferli, D.M. Animal Enterotoxigenic Escherichia coli. EcoSal Plus 2016, 7, 10. [Google Scholar] [CrossRef] [Green Version]
- Kobayashi, H.; Albarracin, L.; Sato, N.; Kanmani, P.; Kober, A.; Ikeda-Ohtsubo, W.; Suda, Y.; Nochi, T.; Aso, H.; Makino, S.; et al. Modulation of Porcine Intestinal Epitheliocytes Immunetranscriptome Response by Lactobacillus jensenii TL2937. Benef. Microbes 2016, 7, 769–782. [Google Scholar] [CrossRef] [Green Version]
- Hu, M.M.; Shu, H.B. Cytoplasmic Mechanisms of Recognition and Defense of Microbial Nucleic Acids. Annu. Rev. Cell Dev. Biol. 2018, 34, 357–379. [Google Scholar] [CrossRef]
- Afonina, I.S.; Zhong, Z.; Karin, M.; Beyaert, R. Limiting Inflammation-the Negative Regulation of NF-ΚB and the NLRP3 Inflammasome. Nat. Immunol. 2017, 18, 861–869. [Google Scholar] [CrossRef]
- Zhou, M.; Xu, W.; Wang, J.; Yan, J.; Shi, Y.; Zhang, C.; Ge, W.; Wu, J.; Du, P.; Chen, Y. Boosting MTOR-Dependent Autophagy via Upstream TLR4-MyD88-MAPK Signalling and Downstream NF-ΚB Pathway Quenches Intestinal Inflammation and Oxidative Stress Injury. EBioMedicine 2018, 35, 345–360. [Google Scholar] [CrossRef] [Green Version]
- Brubaker, J.; Zhang, X.; Bourgeois, A.L.; Harro, C.; Sack, D.A.; Chakraborty, S. Intestinal and Systemic Inflammation Induced by Symptomatic and Asymptomatic Enterotoxigenic E. coli Infection and Impact on Intestinal Colonization and ETEC Specific Immune Responses in an Experimental Human Challenge Model. Gut Microbes 2021, 13, 1–13. [Google Scholar] [CrossRef]
- Jiang, F.; Wu, Z.; Zheng, Y.; Frana, T.S.; Sahin, O.; Zhang, Q.; Li, G. Genotypes and Antimicrobial Susceptibility Profiles of Hemolytic Escherichia coli from Diarrheic Piglets. Foodborne Pathog. Dis. 2019, 16, 94–103. [Google Scholar] [CrossRef]
- de Lagarde, M.; Vanier, G.; Arsenault, J.; Fairbrother, J.M. High Risk Clone: A Proposal of Criteria Adapted to the One Health Context with Application to Enterotoxigenic Escherichia coli in the Pig Population. Antibiotics 2021, 10, 244. [Google Scholar] [CrossRef] [PubMed]
- Dubreuil, J.D. Enterotoxigenic Escherichia coli and Probiotics in Swine: What the Bleep Do We Know? Biosci. Microbiota Food Health 2017, 36, 75–90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shu, Q.; Gill, H.S. A dietary probiotic (Bifidobacterium lactis HN019) reduces the severity of Escherichia coli O157:H7 infection in mice. Med. Microbiol. Immunol. 2001, 189, 147–152. [Google Scholar] [CrossRef] [PubMed]
- Shu, Q.; Gill, H.S. Immune protection mediated by the probiotic Lactobacillus rhamnosus HN001 (DR20) against Escherichia coli O157:H7 infection in mice. FEMS Immunol. Med. Microbiol. 2002, 34, 59–64. [Google Scholar] [CrossRef]
- Shu, Q.; Qu, F.; Gill, H.S. Probiotic treatment using Bifidobacterium lactis HN019 reduces weanling diarrhea associated with rotavirus and Escherichia coli infection in a piglet model. J. Pediatr. Gastroenterol. Nutr. 2001, 33, 171–177. [Google Scholar] [CrossRef]
- Sun, Y.; Duarte, M.E.; Kim, S.W. Dietary inclusion of multispecies probiotics to reduce the severity of post-weaning diarrhea caused by Escherichia coli F18+ in pigs. Anim. Nutr. 2021, 7, 326–333. [Google Scholar] [CrossRef]
- Daudelin, J.F.; Lessard, M.; Beaudoin, F.; Nadeau, E.; Bissonnette, N.; Boutin, Y.; Brousseau, J.P.; Lauzon, K.; Fairbrother, J.M. Administration of probiotics influences F4 (K88)-positive enterotoxigenic Escherichia coli attachment and intestinal cytokine expression in weaned pigs. Veter Res. 2011, 42, 69. [Google Scholar] [CrossRef] [Green Version]
- Garcia-Castillo, V.; Komatsu, R.; Clua, P.; Indo, Y.; Takagi, M.; Salva, S.; Islam, M.A.; Alvarez, S.; Takahashi, H.; Garcia-Cancino, A.; et al. Evaluation of the Immunomodulatory Activities of the Probiotic Strain Lactobacillus fermentum UCO-979C. Front. Immunol. 2019, 10, 1376. [Google Scholar] [CrossRef] [Green Version]
- Villena, J.; Kitazawa, H. Modulation of Intestinal TLR4-Inflammatory Signaling Pathways by Probiotic Microorganisms: Lessons Learned from Lactobacillus jensenii TL2937. Front. Immunol. 2014, 4, 512. [Google Scholar] [CrossRef] [Green Version]
- Moue, M.; Tohno, M.; Shimazu, T.; Kido, T.; Aso, H.; Saito, T.; Kitazawa, H. Toll-like Receptor 4 and Cytokine Expression Involved in Functional Immune Response in an Originally Established Porcine Intestinal Epitheliocyte Cell Line. Biochim. Biophys. Acta Gen. Subj. 2008, 1780, 134–144. [Google Scholar] [CrossRef]
- Shimazu, T.; Villena, J.; Tohno, M.; Fujie, H.; Hosoya, S.; Shimosato, T.; Aso, H.; Suda, Y.; Kawai, Y.; Saito, T.; et al. Immunobiotic Lactobacillus jensenii Elicits Anti-Inflammatory Activity in Porcine Intestinal Epithelial Cells by Modulating Negative Regulators of the Toll-like Receptor Signaling Pathway. Infect. Immun. 2012, 80, 276–288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Vries, M.C.; Vaughan, E.E.; Kleerebezem, M.; de Vos, W.M. Lactobacillus plantarum-Survival, Functional and Potential Probiotic Properties in the Human Intestinal Tract. Int. Dairy J. 2006, 16, 1018–1028. [Google Scholar] [CrossRef]
- Siezen, R.J.; Tzeneva, V.A.; Castioni, A.; Wels, M.; Phan, H.T.K.; Rademaker, J.L.W.; Starrenburg, M.J.C.; Kleerebezem, M.; van Hylckama Vlieg, J.E.T. Phenotypic and Genomic Diversity of Lactobacillus plantarum Strains Isolated from Various Environmental Niches. Environ. Microbiol. 2010, 12, 758–773. [Google Scholar] [CrossRef] [PubMed]
- Villena, J.; Li, C.; Vizoso-Pinto, M.G.; Sacur, J.; Ren, L.; Kitazawa, H. Lactiplantibacillus plantarum as a Potential Adjuvant and Delivery System for the Development of SARS-CoV-2 Oral Vaccines. Microorganisms 2021, 9, 683. [Google Scholar] [CrossRef] [PubMed]
- Behera, S.S.; Ray, R.C.; Zdolec, N. Lactobacillus plantarum with Functional Properties: An Approach to Increase Safety and Shelf-Life of Fermented Foods. Biomed Res. Int 2018, 2018, 9361614. [Google Scholar] [CrossRef] [Green Version]
- Albarracin, L.; Garcia-Castillo, V.; Masumizu, Y.; Indo, Y.; Islam, M.A.; Suda, Y.; Garcia-Cancino, A.; Aso, H.; Takahashi, H.; Kitazawa, H.; et al. Efficient Selection of New Immunobiotic Strains With Antiviral Effects in Local and Distal Mucosal Sites by Using Porcine Intestinal Epitheliocytes. Front. Immunol. 2020, 11, 543. [Google Scholar] [CrossRef]
- Mizuno, H.; Arce, L.; Tomotsune, K.; Albarracin, L.; Funabashi, R.; Vera, D.; Islam, M.A.; Vizoso-Pinto, M.G.; Takahashi, H.; Sasaki, Y.; et al. Lipoteichoic Acid Is Involved in the Ability of the Immunobiotic Strain Lactobacillus plantarum CRL1506 to Modulate the Intestinal Antiviral Innate Immunity Triggered by TLR3 Activation. Front. Immunol. 2020, 11, 571. [Google Scholar] [CrossRef]
- Islam, M.A.; Albarracin, L.; Tomokiyo, M.; Valdez, J.C.; Sacur, J.; Vizoso-Pinto, M.G.; Andrade, B.G.N.; Cuadrat, R.R.C.; Kitazawa, H.; Villena, J. Immunobiotic Lactobacilli Improve Resistance of Respiratory Epithelial Cells to SARS-CoV-2 Infection. Pathogens 2021, 10, 1197. [Google Scholar] [CrossRef]
- Tada, A.; Zelaya, H.; Clua, P.; Salva, S.; Alvarez, S.; Kitazawa, H.; Villena, J. Immunobiotic Lactobacillus Strains Reduce Small Intestinal Injury Induced by Intraepithelial Lymphocytes after Toll-like Receptor 3 Activation. Inflamm. Res. 2016, 65, 771–783. [Google Scholar] [CrossRef]
- Albarracin, L.; Tonetti, F.R.; Fukuyama, K.; Suda, Y.; Zhou, B.; Baillo, A.A.; Fadda, S.; Saavedra, L.; Kurata, S.; Hebert, E.M.; et al. Genomic Characterization of Lactiplantibacillus plantarum Strains Possessing Differential Antiviral Immunomodulatory Activities. Bacteria 2022, 1, 136–160. [Google Scholar] [CrossRef]
- Fadda, S.; Vildoza, M.J.; Vignolo, G. The acidogenic metabolism of Lactobacillus plantarum CRL 681 improves sarcoplasmic protein hydrolysis during meat fermentation. J. Muscle Foods 2010, 21, 545–556. [Google Scholar] [CrossRef]
- Fadda, S.; Chambon, C.; Champomier-Vergès, M.C.; Talon, R.; Vignolo, G. Lactobacillus Role during Conditioning of Refrigerated and Vacuum-Packaged Argentinean Meat. Meat Sci. 2008, 79, 603–610. [Google Scholar] [CrossRef] [PubMed]
- de Almeida, M.A.; Saldaña, E.; da Silva Pinto, J.S.; Palacios, J.; Contreras-Castillo, C.J.; Sentandreu, M.A.; Fadda, S.G. A Peptidomic Approach of Meat Protein Degradation in a Low-Sodium Fermented Sausage Model Using Autochthonous Starter Cultures. Food Res. Int. 2018, 109, 368–379. [Google Scholar] [CrossRef] [PubMed]
- Fadda, S.; López, C.; Vignolo, G. Role of Lactic Acid Bacteria during Meat Conditioning and Fermentation: Peptides Generated as Sensorial and Hygienic Biomarkers. Meat Sci. 2010, 86, 66–79. [Google Scholar] [CrossRef] [PubMed]
- Orihuel, A.; Terán, L.; Renaut, J.; Vignolo, G.M.; de Almeida, A.M.; Saavedra, M.L.; Fadda, S. Differential Proteomic Analysis of Lactic Acid Bacteria- Escherichia coli O157:H7 Interaction and Its Contribution to Bioprotection Strategies in Meat. Front. Microbiol. 2018, 9, 1083. [Google Scholar] [CrossRef]
- Tomosada, Y.; Villena, J.; Murata, K.; Chiba, E.; Shimazu, T.; Aso, H.; Iwabuchi, N.; Xiao, J.Z.; Saito, T.; Kitazawa, H. Immunoregulatory Effect of Bifidobacteria Strains in Porcine Intestinal Epithelial Cells through Modulation of Ubiquitin-Editing Enzyme A20 Expression. PLoS One 2013, 8, e59259. [Google Scholar] [CrossRef] [PubMed]
- Indo, Y.; Kitahara, S.; Tomokiyo, M.; Araki, S.; Islam, M.A.; Zhou, B.; Albarracin, L.; Miyazaki, A.; Ikeda-Ohtsubo, W.; Nochi, T.; et al. Ligilactobacillus Salivarius Strains Isolated From the Porcine Gut Modulate Innate Immune Responses in Epithelial Cells and Improve Protection Against Intestinal Viral-Bacterial Superinfection. Front. Immunol. 2021, 12, 1. [Google Scholar] [CrossRef]
- Marranzino, G.; Villena, J.; Salva, S.; Alvarez, S. Stimulation of Macrophages by Immunobiotic Lactobacillus Strains: Influence beyond the Intestinal Tract. Microbiol. Immunol. 2012, 56, 771–781. [Google Scholar] [CrossRef]
- Adetoye, A.; Pinloche, E.; Adeniyi, B.A.; Ayeni, F.A. Characterization and Anti-Salmonella Activities of Lactic Acid Bacteria Isolated from Cattle Faeces. BMC Microbiol. 2018, 18, 96. [Google Scholar] [CrossRef] [Green Version]
- Markowiak, P.; Ślizewska, K. The Role of Probiotics, Prebiotics and Synbiotics in Animal Nutrition. Gut Pathog. 2018, 10, 21. [Google Scholar] [CrossRef]
- Chaillou, S.; Champomier-Vergès, M.C.; Cornet, M.; Crutz-Le Coq, A.M.; Dudez, A.M.; Martin, V.; Beaufils, S.; Darbon-Rongère, E.; Bossy, R.; Loux, V.; et al. The Complete Genome Sequence of the Meat-Borne Lactic Acid Bacterium Lactobacillus sakei 23K. Nat. Biotechnol. 2005, 23, 1527–1533. [Google Scholar] [CrossRef] [Green Version]
- Hebert, E.M.; Saavedra, L.; Taranto, M.P.; Mozzi, F.; Magni, C.; Nader, M.E.F.; de Valdez, G.F.; Sesma, F.; Vignolo, G.; Raya, R.R. Genome Sequence of the Bacteriocin-Producing Lactobacillus curvatus Strain CRL705. J. Bacteriol. 2012, 194, 538–539. [Google Scholar] [CrossRef] [Green Version]
- Corsetti, A.; Valmorri, S. Lactic Acid Bacteria|Lactobacillus spp.: Lactobacillus plantarum. Encycl. Dairy Sci. Second. Ed. 2011, 3, 111–118. [Google Scholar] [CrossRef]
- Vignolo, G.M.; de Ruiz Holgado, A.P.; Oliver, G. Acid Production and Proteolytic Activity of Lactobacillus Strains Isolated from Dry Sausages. J. Food Prot. 1988, 51, 481–484. [Google Scholar] [CrossRef]
- Fadda, S.; Vignolo, G.; Oliver, G. Tyramine Degradation and Tyramine/Histamine Production by Lactic Acid Bacteria and Kocuria Strains. Biotechnol. Lett. 2001, 23, 2015–2019. [Google Scholar] [CrossRef]
- Albarracin, L.; Kobayashi, H.; Iida, H.; Sato, N.; Nochi, T.; Aso, H.; Salva, S.; Alvarez, S.; Kitazawa, H.; Villena, J. Transcriptomic Analysis of the Innate Antiviral Immune Response in Porcine Intestinal Epithelial Cells: Influence of Immunobiotic Lactobacilli. Front. Immunol. 2017, 8, 57. [Google Scholar] [CrossRef] [Green Version]
- Maldonado Galdeano, C.; de Moreno De Leblanc, A.; Vinderola, G.; Bibas Bonet, M.E.; Perdigón, G. Proposed Model: Mechanisms of Immunomodulation Induced by Probiotic Bacteria. Clin. Vaccine Immunol. 2007, 14, 485–492. [Google Scholar] [CrossRef] [Green Version]
- Salva, S.; Villena, J.; Alvarez, S. Immunomodulatory Activity of Lactobacillus rhamnosus Strains Isolated from Goat Milk: Impact on Intestinal and Respiratory Infections. Int. J. Food Microbiol. 2010, 141, 82–89. [Google Scholar] [CrossRef]
- Villena, J.; Chiba, E.; Vizoso-Pinto, M.G.; Tomosada, Y.; Takahashi, T.; Ishizuka, T.; Aso, H.; Salva, S.; Alvarez, S.; Kitazawa, H. Immunobiotic Lactobacillus rhamnosus Strains Differentially Modulate Antiviral Immune Response in Porcine Intestinal Epithelial and Antigen Presenting Cells. BMC Microbiol. 2014, 14, 126. [Google Scholar] [CrossRef] [Green Version]
- Grivennikov, S.; Karin, E.; Terzic, J.; Mucida, D.; Yu, G.Y.; Vallabhapurapu, S.; Scheller, J.; Rose-John, S.; Cheroutre, H.; Eckmann, L.; et al. IL-6 and Stat3 Are Required for Survival of Intestinal Epithelial Cells and Development of Colitis-Associated Cancer. Cancer Cell 2009, 15, 103–113. [Google Scholar] [CrossRef]
- Li, Y.Y.; Ishihara, S.; Aziz, M.M.; Oka, A.; Kusunoki, R.; Tada, Y.; Yuki, T.; Amano, Y.; Ansary, M.U.; Kinoshita, Y. Autophagy Is Required for Toll-like Receptor-Mediated Interleukin-8 Production in Intestinal Epithelial Cells. Int. J. Mol. Med. 2011, 27, 337–344. [Google Scholar] [CrossRef]
- Elean, M.; Albarracin, L.; Fukuyama, K.; Zhou, B.; Tomokiyo, M.; Kitahara, S.; Araki, S.; Suda, Y.; Saavedra, L.; Villena, J.; et al. Lactobacillus delbrueckii CRL 581 Differentially Modulates TLR3-Triggered Antiviral Innate Immune Response in Intestinal Epithelial Cells and Macrophages. Microorganisms 2021, 9, 2449. [Google Scholar] [CrossRef] [PubMed]
- Roselli, M.; Finamore, A.; Britti, M.S.; Mengheri, E. Probiotic Bacteria Bifidobacterium animalis MB5 and Lactobacillus rhamnosus GG Protect Intestinal Caco-2 Cells from the Inflammation-Associated Response Induced by Enterotoxigenic Escherichia coli K88. Br. J. Nutr. 2006, 95, 1177–1184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, Z.; Liu, X.; Dai, R.; Xiao, Y.; Wang, X.; Bi, D.; Shi, D. Enterococcus faecium HDRsEf1 Protects the Intestinal Epithelium and Attenuates ETEC-Induced IL-8 Secretion in Enterocytes. Mediat. Inflamm. 2016, 6, 7474306. [Google Scholar] [CrossRef] [Green Version]
- Roselli, M.; Finamore, A.; Britti, M.S.; Konstantinov, S.R.; Smidt, H.; de Vos, W.M.; Mengheri, E. The Novel Porcine Lactobacillus sobrius Strain Protects Intestinal Cells from Enterotoxigenic Escherichia coli K88 Infection and Prevents Membrane Barrier Damage. J. Nutr. 2007, 137, 2709–2716. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J.; Qiu, Y.; Hu, S.; Zhu, C.; Wang, L.; Wen, X.; Yang, X.; Jiang, Z. Lactobacillus plantarum Inhibited the Inflammatory Response Induced by Enterotoxigenic Escherichia coli K88 via Modulating MAPK and NF-ΚB Signalling in Intestinal Porcine Epithelial Cells. J. Appl. Microbiol. 2021, 130, 1684–1694. [Google Scholar] [CrossRef] [PubMed]
- Kessel, A.; Toubi, E.; Pavlotzky, E.; Mogilner, J.; Coran, A.G.; Lurie, M.; Karry, R.; Sukhotnik, I. Treatment with Glutamine Is Associated with Down-Regulation of Toll-like Receptor-4 and Myeloid Differentiation Factor 88 Expression and Decrease in Intestinal Mucosal Injury Caused by Lipopolysaccharide Endotoxaemia in a Rat. Clin. Exp. Immunol. 2008, 151, 341–347. [Google Scholar] [CrossRef] [Green Version]
- Yue, Y.; He, Z.; Zhou, Y.; Ross, R.P.; Stanton, C.; Zhao, J.; Zhang, H.; Yang, B.; Chen, W. Lactobacillus plantarum Relieves Diarrhea Caused by Enterotoxin-Producing Escherichia coli through Inflammation Modulation and Gut Microbiota Regulation. Food Funct. 2020, 11, 10362–10374. [Google Scholar] [CrossRef]
- Hubbard, L.L.N.; Moore, B.B. IRAK-M Regulation and Function in Host Defense and Immune Homeostasis. Infect. Dis. Rep. 2010, 2, 22–29. [Google Scholar] [CrossRef] [Green Version]
- Escoll, P.; del Fresno, C.; García, L.; Vallés, G.; Lendínez, M.J.; Arnalich, F.; López-Collazo, E. Rapid up-Regulation of IRAK-M Expression Following a Second Endotoxin Challenge in Human Monocytes and in Monocytes Isolated from Septic Patients. Biochem. Biophys. Res. Commun. 2003, 311, 465–472. [Google Scholar] [CrossRef]
- Ning, S.; Pagano, J.S. The A20 Deubiquitinase Activity Negatively Regulates LMP1 Activation of IRF7. J. Virol. 2010, 84, 6130–6138. [Google Scholar] [CrossRef] [Green Version]
- Vereecke, L.; Sze, M.; Mc Guire, C.; Rogiers, B.; Chu, Y.; Schmidt-Supprian, M.; Pasparakis, M.; Beyaert, R.; van Loo, G. Enterocyte-Specific A20 Deficiency Sensitizes to Tumor Necrosis Factor-Induced Toxicity and Experimental Colitis. J. Exp. Med. 2010, 207, 1513–1523. [Google Scholar] [CrossRef] [Green Version]
- Wessells, J.; Baer, M.; Young, H.A.; Claudio, E.; Brown, K.; Siebenlist, U.; Johnson, P.F. BCL-3 and NF-KappaB P50 Attenuate Lipopolysaccharide-Induced Inflammatory Responses in Macrophages. J. Biol. Chem. 2004, 279, 49995–50003. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Ford, H.R.; Grishin, A.V. NF-KappaB-Mediated Expression of MAPK Phosphatase-1 Is an Early Step in Desensitization to TLR Ligands in Enterocytes. Mucosal Immunol. 2010, 3, 523–534. [Google Scholar] [CrossRef] [Green Version]
- Stephens, M.; von der Weid, P.Y. Lipopolysaccharides Modulate Intestinal Epithelial Permeability and Inflammation in a Species-Specific Manner. Gut Microbes 2020, 11, 421–432. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baillo, A.; Villena, J.; Albarracín, L.; Tomokiyo, M.; Elean, M.; Fukuyama, K.; Quilodrán-Vega, S.; Fadda, S.; Kitazawa, H. Lactiplantibacillus plantarum Strains Modulate Intestinal Innate Immune Response and Increase Resistance to Enterotoxigenic Escherichia coli Infection. Microorganisms 2023, 11, 63. https://doi.org/10.3390/microorganisms11010063
Baillo A, Villena J, Albarracín L, Tomokiyo M, Elean M, Fukuyama K, Quilodrán-Vega S, Fadda S, Kitazawa H. Lactiplantibacillus plantarum Strains Modulate Intestinal Innate Immune Response and Increase Resistance to Enterotoxigenic Escherichia coli Infection. Microorganisms. 2023; 11(1):63. https://doi.org/10.3390/microorganisms11010063
Chicago/Turabian StyleBaillo, Ayelen, Julio Villena, Leonardo Albarracín, Mikado Tomokiyo, Mariano Elean, Kohtaro Fukuyama, Sandra Quilodrán-Vega, Silvina Fadda, and Haruki Kitazawa. 2023. "Lactiplantibacillus plantarum Strains Modulate Intestinal Innate Immune Response and Increase Resistance to Enterotoxigenic Escherichia coli Infection" Microorganisms 11, no. 1: 63. https://doi.org/10.3390/microorganisms11010063
APA StyleBaillo, A., Villena, J., Albarracín, L., Tomokiyo, M., Elean, M., Fukuyama, K., Quilodrán-Vega, S., Fadda, S., & Kitazawa, H. (2023). Lactiplantibacillus plantarum Strains Modulate Intestinal Innate Immune Response and Increase Resistance to Enterotoxigenic Escherichia coli Infection. Microorganisms, 11(1), 63. https://doi.org/10.3390/microorganisms11010063








