Identification of a Phylogenetically Divergent Vanillate O-Demethylase from Rhodococcus ruber R1 Supporting Growth on Meta-Methoxylated Aromatic Acids
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Plasmids, and Growth Conditions
2.2. Construction of a Plasmid Expressing vanAB Genes and Growth Tests of Strain Derivatives
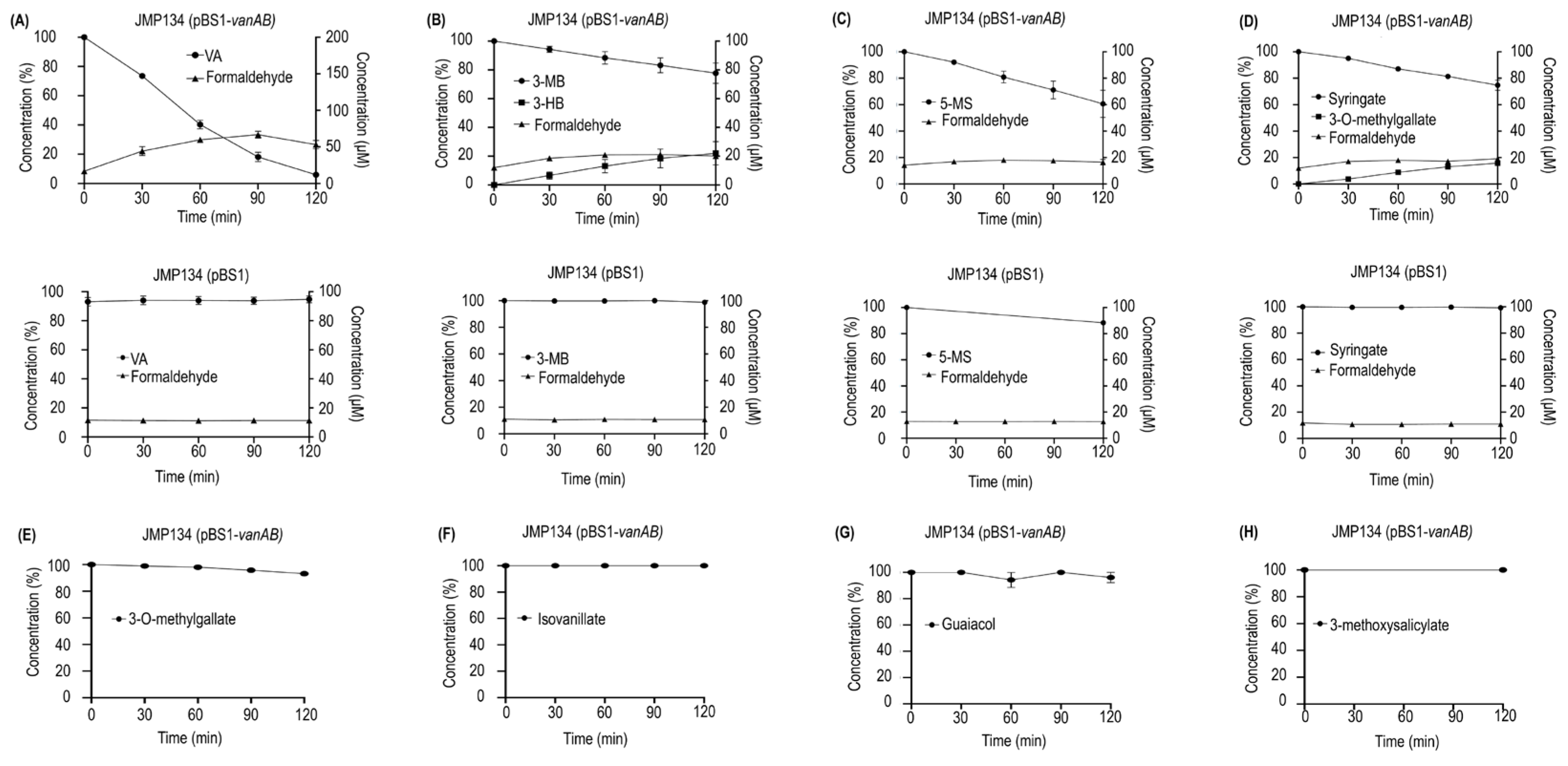
2.3. Resting Cell Assays
2.4. Analytical Methods
2.5. Bioinformatic Tools
2.6. Chemicals
3. Results and Discussion
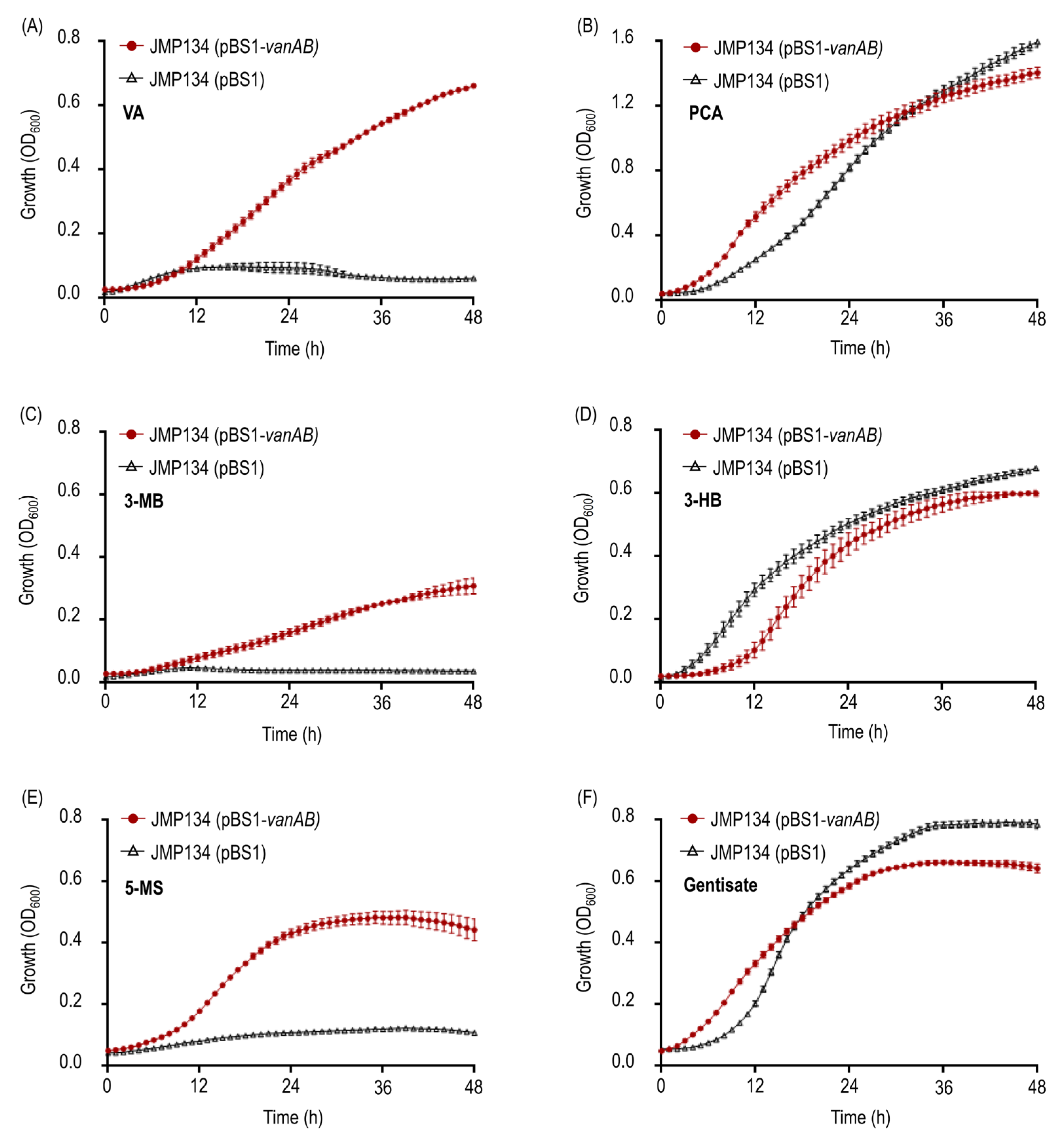
3.1. Heterologous Expression and Resting Cell Assays Suggest a Key Role of VanOD from Rhodococcus ruber R1 in meta-Methoxylated Aromatic Acids Degradation
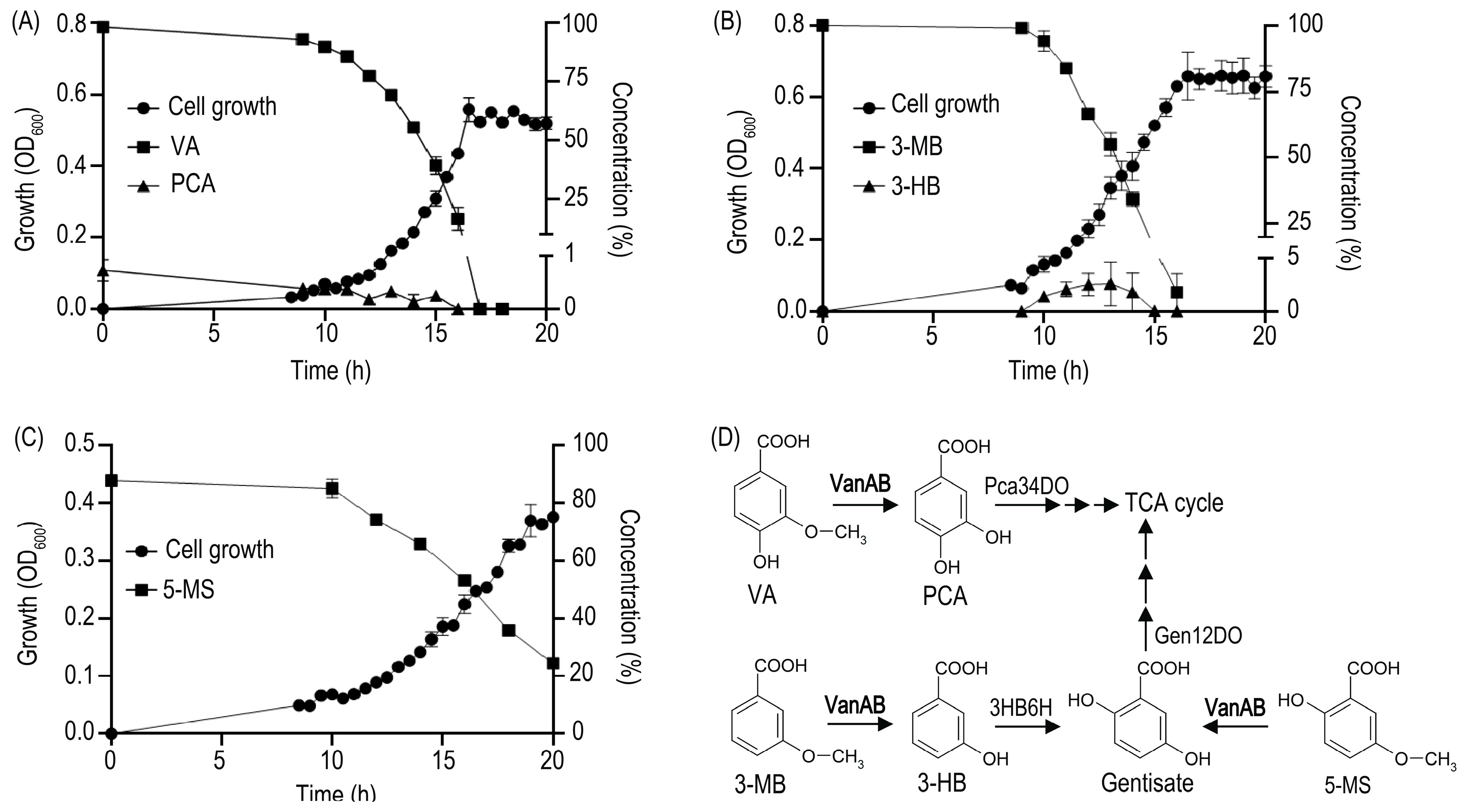
3.2. Rhodococcus Strains Carrying VanAB Homologues Closely Related to VanOD of R. ruber R1 Strain Are Able to Grow on VA, 3-MB, and 5-MS as a Sole Carbon and Energy Sources

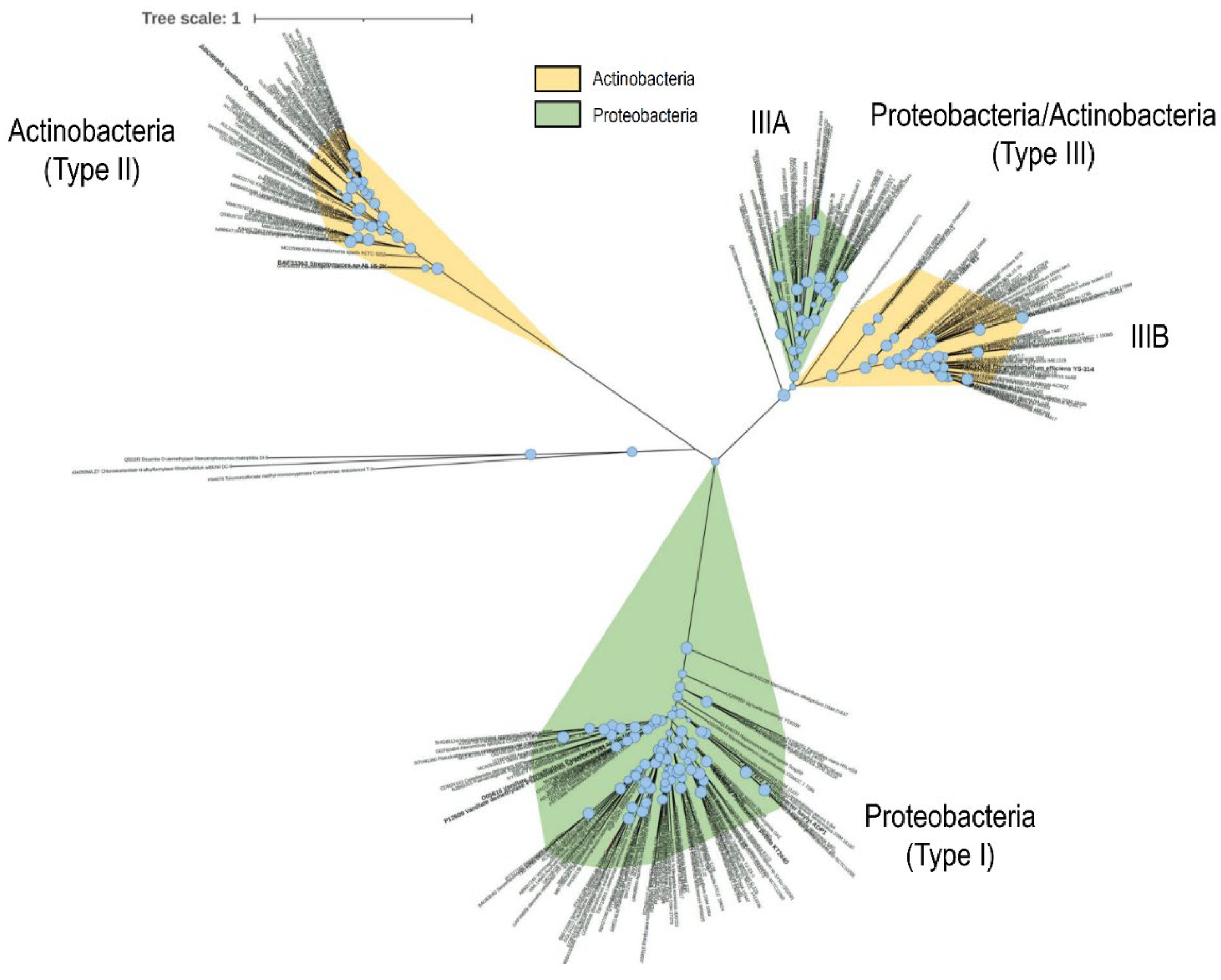
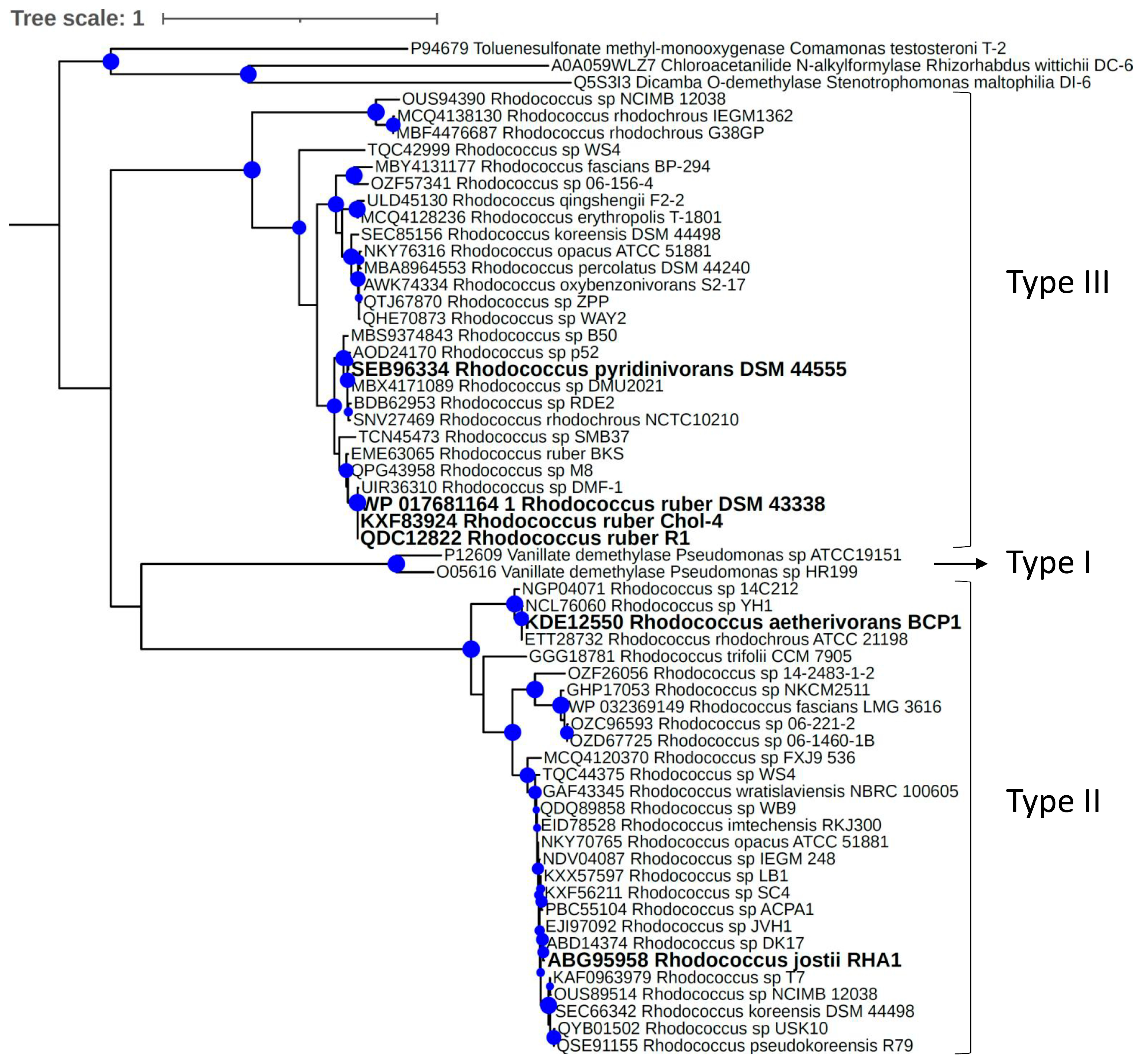
3.3. Two-Component Rieske-Type VanOD of Rhodococcus Species Are Allocated in Two Divergent Phylogenetic Clades
4. Conclusions
| Strain or Plasmid | Relevant Phenotype and/or Genotype | Reference or Source |
|---|---|---|
| Rhodococcus strains | ||
| R. aetherivorans BCP1 | VA+, 3-MB-, 5-MS- | [47] |
| R. jostii RHA1 | VA+, 3-MB-, 5-MS- | [46] |
| R. ruber R1 | VA+, 3-MB+, 5-MS+ | [8] |
| R. ruber Chol-4 | VA+, 3-MB+, 5-MS+ | [45] |
| R. ruber DSM 43338 T | VA+, 3-MB+, 5-MS+ | DSMZ a |
| R. pyridinivorans JCM 10940 T | VA+, 3-MB-, 5-MS- | [44] |
| Rhodococcus sp. H-CA8f | VA-, 3-MB−, 5-MS- | [48] |
| Rhodococcus sp. MS13 | VA-, 3-MB-, 5-MS- | [49] |
| Other strains | ||
| E. coli Mach1 | ∆recA1398 endA1 tonA Φ80∆lacM15 ∆lacX74 hsdR (rK- mK+) | Invitrogen, Carlsbad, CA, USA |
| C. pinatubonensis JMP134 | PCA+, Gentisate+, 3-HB+, VA-, 3-MB-, 5-MS- | [40] |
| Plasmids | ||
| pBS1 | Broad host range vector, araC-PBAD, GmR | [55] |
| pBS1-vanAB | pBS1 derivative expressing vanAB genes, GmR | This study |
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Warhurst, A.M.; Fewson, C.A. Biotransformations catalyzed by the genus Rhodococcus. Crit. Rev. Biotechnol. 1994, 14, 29–73. [Google Scholar] [CrossRef] [PubMed]
- Martínková, L.; Uhnáková, B.; Pátek, M.; Nesvera, J.; Kren, V. Biodegradation potential of the genus Rhodococcus. Environ. Int. 2009, 35, 162–177. [Google Scholar] [CrossRef] [PubMed]
- Yam, K.C.; Okamoto, S.; Roberts, J.N.; Eltis, L.D. Adventures in Rhodococcus—From steroids to explosives. Can. J. Microbiol. 2011, 57, 155–168. [Google Scholar] [CrossRef] [PubMed]
- Zampolli, J.; Zeaiter, Z.; Di Canito, A.; Di Gennaro, P. Genome analysis and -omics approaches provide new insights into the biodegradation potential of Rhodococcus. Appl. Microbiol. Biotechnol. 2019, 103, 1069–1080. [Google Scholar] [CrossRef] [PubMed]
- Busch, H.; Hagedoorn, P.L.; Hanefeld, U. Rhodococcus as a versatile biocatalyst in organic synthesis. Int. J. Mol. Sci. 2019, 20, 4787. [Google Scholar] [CrossRef] [Green Version]
- Ivshina, I.; Bazhutin, G.; Tyumina, E. Rhodococcus strains as a good biotool for neutralizing pharmaceutical pollutants and obtaining therapeutically valuable products: Through the past into the future. Front. Microbiol. 2022, 13, 967127. [Google Scholar] [CrossRef]
- Nazari, M.T.; Simon, V.; Machado, B.S.; Crestani, L.; Marchezi, G.; Concolato, G.; Ferrari, V.; Colla, L.M.; Piccin, J.S. Rhodococcus: A promising genus of actinomycetes for the bioremediation of organic and inorganic contaminants. J. Environ. Manag. 2022, 323, 116220. [Google Scholar] [CrossRef]
- Farkas, C.; Donoso, R.A.; Melis-Arcos, F.; Gárate-Castro, C.; Pérez-Pantoja, D. Complete Genome Sequence of Rhodococcus ruber R1, a Novel Strain Showing a Broad Catabolic Potential toward Lignin-Derived Aromatics. Microbiol. Resour. Announc. 2020, 9, e00905.19. [Google Scholar] [CrossRef] [Green Version]
- Jadan, A.P.; van Berkel, W.J.; Golovleva, L.A.; Golovlev, E.L. Purification and properties of p-hydroxybenzoate hydroxylases from Rhodococcus strains. Biochemistry 2001, 66, 898–903. [Google Scholar]
- Otani, H.; Lee, Y.E.; Casabon, I.; Eltis, L.D. Characterization of p-hydroxycinnamate catabolism in a soil Actinobacterium. J. Bacteriol. 2014, 196, 4293–4303. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.P.; Chow, M.; Liu, C.C.; Lau, A.; Liu, J.; Eltis, L.D. Vanillin catabolism in Rhodococcus jostii RHA1. Appl. Environ. Microbiol. 2012, 78, 586–588. [Google Scholar] [CrossRef] [PubMed]
- Correll, C.C.; Batie, C.J.; Ballou, D.P.; Ludwig, M.L. Phthalate dioxygenase reductase: A modular structure for electron transfer from pyridine nucleotides to [2Fe-2S]. Science 1992, 258, 1604–1610. [Google Scholar] [CrossRef] [PubMed]
- Dehmel, U.; Engesser, K.H.; Timmis, K.N.; Dwyer, D.F. Cloning, nucleotide sequence, and expression of the gene encoding a novel dioxygenase involved in metabolism of carboxydiphenyl ethers in Pseudomonas pseudoalcaligenes POB310. Arch. Microbiol. 1995, 163, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Priefert, H.; Rabenhorst, J.; Steinbüchel, A. Molecular characterization of genes of Pseudomonas sp. strain HR199 involved in bioconversion of vanillin to protocatechuate. J. Bacteriol. 1997, 179, 2595–2607. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Venturi, V.; Zennaro, F.; Degrassi, G.; Okeke, B.C.; Bruschi, C.V. Genetics of ferulic acid bioconversion to protocatechuic acid in plant-growth-promoting Pseudomonas putida WCS358. Microbiology 1998, 144, 965–973. [Google Scholar] [CrossRef] [Green Version]
- Morawski, B.; Segura, A.; Ornston, L.N. Substrate range and genetic analysis of Acinetobacter vanillate demethylase. J. Bacteriol. 2000, 182, 1383–1389. [Google Scholar] [CrossRef] [Green Version]
- Merkens, H.; Beckers, G.; Wirtz, A.; Burkovski, A. Vanillate metabolism in Corynebacterium glutamicum. Curr. Microbiol. 2005, 51, 59–65. [Google Scholar] [CrossRef]
- Providenti, M.A.; O’Brien, J.M.; Ruff, J.; Cook, A.M.; Lambert, I.B. Metabolism of isovanillate, vanillate, and veratrate by Comamonas testosteroni strain BR6020. J. Bacteriol. 2006, 188, 3862–3869. [Google Scholar] [CrossRef] [Green Version]
- Nishimura, M.; Nishimura, Y.; Abe, C.; Kohhata, M. Expression and substrate range of Streptomyces vanillate demethylase. Biol. Pharm. Bull. 2014, 37, 1564–1568. [Google Scholar] [CrossRef] [Green Version]
- Notonier, S.; Werner, A.Z.; Kuatsjah, E.; Dumalo, L.; Abraham, P.E.; Hatmaker, E.A.; Hoyt, C.B.; Amore, A.; Ramirez, K.J.; Woodworth, S.P.; et al. Metabolism of syringyl lignin-derived compounds in Pseudomonas putida enables convergent production of 2-pyrone-4,6-dicarboxylic acid. Metab. Eng. 2021, 65, 111–122. [Google Scholar] [CrossRef] [PubMed]
- Mueller, J.; Willett, H.; Feist, A.M.; Niu, W. Engineering Pseudomonas putida for improved utilization of syringyl aromatics. Biotechnol. Bioeng. 2022, 119, 2541–2550. [Google Scholar] [CrossRef] [PubMed]
- Berman, M.H.; Frazer, A.C. Importance of tetrahydrofolate and ATP in the anaerobic O-demethylation reaction for phenylmethylethers. Appl. Environ. Microbiol. 1992, 58, 925–931. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaufmann, F.; Wohlfarth, G.; Diekert, G. O-demethylase from Acetobacterium dehalogenans—Substrate specificity and function of the participating proteins. Eur. J. Biochem. 1998, 253, 706–711. [Google Scholar] [CrossRef] [PubMed]
- Naidu, D.; Ragsdale, S.W. Characterization of a three-component vanillate O-demethylase from Moorella thermoacetica. J. Bacteriol. 2001, 183, 3276–3281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abe, T.; Masai, E.; Miyauchi, K.; Katayama, Y.; Fukuda, M. A tetrahydrofolate-dependent O-demethylase, LigM, is crucial for catabolism of vanillate and syringate in Sphingomonas paucimobilis SYK-6. J. Bacteriol. 2005, 187, 2030–2037. [Google Scholar] [CrossRef] [Green Version]
- Hibi, M.; Sonoki, T.; Mori, H. Functional coupling between vanillate-O-demethylase and formaldehyde detoxification pathway. FEMS Microbiol. Lett. 2005, 253, 237–242. [Google Scholar] [CrossRef] [Green Version]
- Kohler, A.C.; Mills, M.J.L.; Adams, P.D.; Simmons, B.A.; Sale, K.L. Structure of aryl O-demethylase offers molecular insight into a catalytic tyrosine-dependent mechanism. Proc. Natl. Acad. Sci. USA 2017, 114, E3205–E3214. [Google Scholar] [CrossRef] [Green Version]
- Lanfranchi, E.; Trajković, M.; Barta, K.; de Vries, J.G.; Janssen, D.B. Exploring the Selective Demethylation of Aryl Methyl Ethers with a Pseudomonas Rieske Monooxygenase. ChemBioChem 2019, 20, 118–125. [Google Scholar] [CrossRef] [Green Version]
- Donoso, R.A.; González-Toro, F.; Pérez-Pantoja, D. Widespread distribution of hmf genes in Proteobacteria reveals key enzymes for 5-hydroxymethylfurfural conversion. Comput. Struct. Biotechnol. J. 2021, 19, 2160–2169. [Google Scholar] [CrossRef]
- Donoso, R.A.; Ruiz, D.; Gárate-Castro, C.; Villegas, P.; González-Pastor, J.E.; de Lorenzo, V.; González, B.; Pérez-Pantoja, D. Identification of a self-sufficient cytochrome P450 monooxygenase from Cupriavidus pinatubonensis JMP134 involved in 2-hydroxyphenylacetic acid catabolism, via homogentisate pathway. Microb. Biotechnol. 2021, 14, 1944–1960. [Google Scholar] [CrossRef]
- NCBI. Resource Coordinators. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2018, 46, D8–D13. [Google Scholar] [CrossRef] [PubMed]
- Johnson, M.; Zaretskaya, I.; Raytselis, Y.; Merezhuk, Y.; McGinnis, S.; Madden, T.L. NCBI BLAST: A better web interface. Nucleic Acids Res. 2008, 36, W5–W9. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoang, D.T.; Chernomor, O.; von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the Ultrafast Bootstrap Approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef] [Green Version]
- Montersino, S.; van Berkel, W.J. Functional annotation and characterization of 3-hydroxybenzoate 6-hydroxylase from Rhodococcus jostii RHA1. Biochim. Biophys. Acta 2012, 1824, 433–442. [Google Scholar] [CrossRef]
- Sucharitakul, J.; Tongsook, C.; Pakotiprapha, D.; van Berkel, W.J.; Chaiyen, P. The reaction kinetics of 3-hydroxybenzoate 6-hydroxylase from Rhodococcus jostii RHA1 provide an understanding of the para-hydroxylation enzyme catalytic cycle. J. Biol. Chem. 2013, 288, 35210–35221. [Google Scholar] [CrossRef] [Green Version]
- Pérez-Pantoja, D.; De la Iglesia, R.; Pieper, D.H.; González, B. Metabolic reconstruction of aromatic compounds degradation from the genome of the amazing pollutant-degrading bacterium Cupriavidus necator JMP134. FEMS Microbiol. Rev. 2008, 32, 736–794. [Google Scholar] [CrossRef] [Green Version]
- Pérez-Pantoja, D.; Donoso, R.A.; Sánchez, M.A.; González, B. Genuine genetic redundancy in maleylacetate-reductase-encoding genes involved in degradation of haloaromatic compounds by Cupriavidus necator JMP134. Microbiology 2009, 155, 3641–3651. [Google Scholar] [CrossRef] [PubMed]
- Nogales, J.; Canales, A.; Jiménez-Barbero, J.; García, J.L.; Díaz, E. Molecular characterization of the gallate dioxygenase from Pseudomonas putida KT2440. The prototype of a new subgroup of extradiol dioxygenases. J. Biol. Chem. 2005, 280, 35382–35390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nogales, J.; Canales, A.; Jiménez-Barbero, J.; Serra, B.; Pingarrón, J.M.; García, J.L.; Díaz, E. Unravelling the gallic acid degradation pathway in bacteria: The gal cluster from Pseudomonas putida. Mol. Microbiol. 2011, 79, 359–374. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.H.; Kang, S.S.; Cho, Y.G.; Lee, S.T.; Kho, Y.H.; Kim, C.J.; Park, Y.H. Rhodococcus pyridinivorans sp. nov., a pyridine-degrading bacterium. Int. J. Syst. Evol. Microbiol. 2000, 50, 2173–2180. [Google Scholar] [CrossRef] [PubMed]
- Heras, L.F.D.L.; Fernández, E.G.; Llorens, J.M.N.; Perera, J.; Drzyzga, O. Morphological, physiological, and molecular characterization of a newly isolated steroid-degrading actinomycete, identified as Rhodococcus ruber strain Chol-4. Curr. Microbiol. 2009, 59, 548–553. [Google Scholar] [CrossRef]
- McLeod, M.P.; Warren, R.L.; Hsiao, W.W.L.; Araki, N.; Myhre, M.; Fernandes, C.; Miyazawa, D.; Wong, W.; Lillquist, A.L.; Wang, D.; et al. The complete genome of Rhodococcus sp. RHA1 provides insights into a catabolic powerhouse. Proc. Natl. Acad. Sci. USA 2006, 103, 15582–15587. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cappelletti, M.; Di Gennaro, P.; D’Ursi, P.; Orro, A.; Mezzelani, A.; Landini, M.; Fedi, S.; Frascari, D.; Presentato, A.; Zannoni, D.; et al. Genome Sequence of Rhodococcus sp. Strain BCP1, a Biodegrader of Alkanes and Chlorinated Compounds. Genome Announc. 2013, 1, e00657.13. [Google Scholar] [CrossRef] [Green Version]
- Undabarrena, A.; Salvà-Serra, F.; Jaén-Luchoro, D.; Castro-Nallar, E.; Mendez, K.N.; Valencia, R.; Ugalde, J.A.; Moore, E.R.B.; Seeger, M.; Cámara, B. Complete genome sequence of the marine Rhodococcus sp. H-CA8f isolated from Comau fjord in Northern Patagonia, Chile. Mar. Genom. 2018, 40, 13–17. [Google Scholar] [CrossRef] [PubMed]
- Aguila-Torres, P.; Maldonado, J.; Gaete, A.; Figueroa, J.; González, A.; Miranda, R.; González-Stegmaier, R.; Martin, C.; González, M. Biochemical and Genomic Characterization of the Cypermethrin-Degrading and Biosurfactant-Producing Bacterial Strains Isolated from Marine Sediments of the Chilean Northern Patagonia. Mar. Drugs 2020, 18, 252. [Google Scholar] [CrossRef]
- Yamanashi, T.; Kim, S.Y.; Hara, H.; Funa, N. In vitro reconstitution of the catabolic reactions catalyzed by PcaHG, PcaB, and PcaL: The protocatechuate branch of the β-ketoadipate pathway in Rhodococcus jostii RHA1. Biosci. Biotechnol. Biochem. 2015, 79, 830–835. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.; Monnappa, A.K.; Mitchell, R.J. Biological activities of lignin hydrolysate-related compounds. BMB Rep. 2012, 45, 265–274. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wang, P.; Salam, N.; Li, X.; Ahmad, M.; Tian, Y.; Duan, L.; Huang, L.; Xiao, M.; Mou, X.; et al. Unraveling bacteria-mediated degradation of lignin-derived aromatic compounds in a freshwater environment. Sci. Total Environ. 2020, 749, 141236. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, Y.; Otsuka, Y.; Araki, T.; Kamimura, N.; Masai, E.; Nakamura, M.; Katayama, Y. Lignin valorization through efficient microbial production of β-ketoadipate from industrial black liquor. Bioresour. Technol. 2021, 337, 125489. [Google Scholar] [CrossRef] [PubMed]
- Venkatesagowda, B.; Dekker, R.F.H. Microbial demethylation of lignin: Evidence of enzymes participating in the removal of methyl/methoxyl groups. Enzym. Microb. Technol. 2021, 147, 109780. [Google Scholar] [CrossRef]
- Bronstein, P.A.; Marrichi, M.; Cartinhour, S.; Schneider, D.J.; DeLisa, M.P. Identification of a twin-arginine translocation system in Pseudomonas syringae pv. tomato DC3000 and its contribution to pathogenicity and fitness. J. Bacteriol. 2005, 187, 8450–8461. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Donoso, R.A.; Corbinaud, R.; Gárate-Castro, C.; Galaz, S.; Pérez-Pantoja, D. Identification of a Phylogenetically Divergent Vanillate O-Demethylase from Rhodococcus ruber R1 Supporting Growth on Meta-Methoxylated Aromatic Acids. Microorganisms 2023, 11, 78. https://doi.org/10.3390/microorganisms11010078
Donoso RA, Corbinaud R, Gárate-Castro C, Galaz S, Pérez-Pantoja D. Identification of a Phylogenetically Divergent Vanillate O-Demethylase from Rhodococcus ruber R1 Supporting Growth on Meta-Methoxylated Aromatic Acids. Microorganisms. 2023; 11(1):78. https://doi.org/10.3390/microorganisms11010078
Chicago/Turabian StyleDonoso, Raúl A., Ricardo Corbinaud, Carla Gárate-Castro, Sandra Galaz, and Danilo Pérez-Pantoja. 2023. "Identification of a Phylogenetically Divergent Vanillate O-Demethylase from Rhodococcus ruber R1 Supporting Growth on Meta-Methoxylated Aromatic Acids" Microorganisms 11, no. 1: 78. https://doi.org/10.3390/microorganisms11010078
APA StyleDonoso, R. A., Corbinaud, R., Gárate-Castro, C., Galaz, S., & Pérez-Pantoja, D. (2023). Identification of a Phylogenetically Divergent Vanillate O-Demethylase from Rhodococcus ruber R1 Supporting Growth on Meta-Methoxylated Aromatic Acids. Microorganisms, 11(1), 78. https://doi.org/10.3390/microorganisms11010078







