Abstract
Streptococcus (S.) suis presents a serious threat to the pig industry as well as food safety and public health. Although several LAMP assays have been developed for the identification of S. suis, no universal assay is so far available for the field-suitable examination of clinical pig specimens. Based on the thrA housekeeping gene, a new loop-mediated isothermal amplification (LAMP) assay was developed and validated for the detection of S. suis in the brain and joints of pigs. For this LAMP assay, two different methods for the extraction of DNA from brain and joint swabs were compared. Using the LPTV boiling method, the detection limit of LAMP was 1.08 CFU/reaction, while the detection limit was 53.8 CFU/reaction using a commercial DNA extraction kit. The detection limits of thrA-LAMP in combination with the LPTV boiling method were 104–105 CFU/swab in the presence of brain tissue and 103–104 CFU/swab in the presence of joint tissue. The diagnostic quality criteria of LAMP were determined by the examination of 49 brain swabs and 34 joint swabs obtained during routine diagnostic necropsies. Applying the LPTV boiling method to brain swabs, the sensitivity, specificity, and positive and negative predictive values of thrA-LAMP were 88.0, 95.8, 95.7, and 88.5% using cultural investigation as a reference method, and 76.7, 100, 100, and 73.1% using real-time PCR as a reference method. Based on these results, the thrA-LAMP assay combined with the LPTV boiling method is suitable for rapid detection of S. suis from brain swabs.
1. Introduction
Streptococcus (S.) suis is a gram-positive bacterium that inhabits the upper respiratory tract, specifically the tonsils and nasal cavities, of pigs [1]. Based on the capsular polysaccharides (CPS), S. suis strains have been serologically classified into 35 serotypes (1–34 and 1/2) until the end of the last century [2]. However, due to new phylogenetic analyses, serotypes 20, 22, 26, and 32–34 have been recently allocated to other species [3,4]. Furthermore, serotypes 20, 22, and 26 were proposed as a novel species called Streptococcus parasuis sp. nov. [5]. A recent study suggested that S. parasuis and S. suis still share genetic characteristics, and even horizontal gene transfer is considered possible [6]. Serotype 33 was reclassified as the new species Streptococcus ruminantium sp. nov. [7]. S. suis serotypes 32 and 34 reference strains were assigned to the species S. orisratti [8]. Investigation of the epidemiological situation of S. suis-serotypes in German pig herds in 2019 by Prüfer et al. revealed serotypes 2 and 1/2, 9, 7, 4, 1, and 14 to be the most common within the more recent collection of invasive isolates [9]. Data on global distribution showed that serotypes 2, 9, 3, 1/2, and 7 were predominant in clinically infected pigs [10]. Despite enormous progress in research focusing on virulence factor genes, the pathogenesis of a systemic S. suis infection is still not fully understood [11]. S. suis might cause disease when different biotic or abiotic predisposing factors, such as virus infections or stress, impact the host’s immunity [1,12]. In case of an outbreak, typical clinical signs are meningitis, arthritis, endocarditis, pneumonia, or peracute death [1,12]. Outbreaks most frequently occur in nursery piglets, but also suckling piglets can be affected [1]. Without antibiotic treatment, mortality can rise up to 20–30% [1,13]. Mortality during the nursery phase can lead to relevant economic losses [14]. S. suis is also a zoonotic pathogen, with most cases reported in China and Southeast Asia, mainly by serotype 2 [10,15]. Sporadic cases of S. suis infections in humans have also been reported in other regions of the world [10,15]. Humans in close contact with pigs or pork can get infected through minor skin lesions [12]. Consumption of raw pork products is also considered a possible route of infection [16]. Similar to pigs, human infections often result in meningitis or other manifestations like streptococcal toxic shock-like syndrome, septicaemia, arthritis, pneumonia, and peritonitis [12,17]. Therefore, S. suis represents a serious threat, not only to livestock and the economy but also in terms of food safety and public health [14,15,16]. Rapid and reliable diagnosis of this pathogen and its diversity of serotypes in various different samples is crucial. Detection of S. suis by standard culture methods is laborious, and biochemical characteristics may be inconclusive [18,19]. PCR-based assays show high sensitivity and specificity but require expensive equipment and are not suitable for on-site operations [20,21]. In contrast, loop-mediated isothermal amplification (LAMP) proved to be a suitable technique to overcome these shortcomings [21,22,23].
Since the LAMP reaction is performed under isothermal conditions, a simple water bath or heating block can be used [24]. On this basis, portable devices have been developed, allowing the method to be performed directly on-site [25]. In previous studies, it was shown that LAMP is less impacted by the effects of the biological matrix than PCR, suggesting that the step of DNA purification could be omitted for a LAMP assay [25,26,27,28]. This would facilitate the use of LAMP for diagnostics outside of laboratories [26]. Previously developed serotype-specific LAMP assays enabled the detection of highly pathogenic S. suis serotype 2 and 14 strains in samples of infected human patients [21,29]. The recN-based LAMP by Arai et al., could correctly identify all serotypes, except those recently excluded from the species S. suis [23]. Using these LAMP assays, the prevalence of S. suis in raw pig samples in Thailand was investigated [30]. Results revealed deficiencies in food safety due to cross-contamination with S. suis during meat handling, especially in local retail stores and markets [30]. Although several LAMP assays have been developed focusing on S. suis with regard to human public health and food safety, no systems have been established to detect infections in pigs. An appropriate diagnostic LAMP assay would facilitate early detection of S. suis outbreaks in pig herds. Consequently, antibiotic treatment could be initiated quickly, reducing mortality and economic loss as well as preventing further infection of humans in close contact [12,14]. The present study aimed at developing and validating a field-suitable LAMP assay for detecting the broad spectrum of S. suis variants with high specificity and sensitivity in the brain and joints of infected pigs.
2. Materials and Methods
2.1. Bacterial Strains
A total of 220 bacterial strains were examined in this study. Reference strains originated from varying culture collections such as the American Type Culture Collection (Manassas, VA, USA), the Culture Collection University of Gothenburg (Göteborg, Sweden), the DSMZ-German Collection of Microorganisms and Cell Cultures GmbH (Leibniz Institute, Braunschweig, Germany), and the National Collection of Type Cultures (Salisbury, UK). Field strains were provided by the Field Station for Epidemiology in Bakum (University of Veterinary Medicine Hannover, Foundation, Hannover, Germany) and the Institute for Food Quality and Food Safety (University of Veterinary Medicine Hannover, Foundation, Hannover, Germany). The S. suis field strains used in this study were obtained during internal routine diagnostics from various pig tissues. The S. suis isolates were recovered by standard culture methods and subsequently confirmed and serotyped by PCR (see Section 2.10). Detailed biochemical identification using the API® 20 STREP system (bioMérieux, Marcy-I’Etoile, France) was performed and interpreted according to the manufacturer’s manual. Field strains of non-target species were isolated in the respective institutes by standard culture methods during routine diagnostics, and confirmed via biochemical characterization or MALDI-TOF MS analysis. All bacterial strains were stored in cryobanks (Mast Diagnostica GmbH, Reinfeld, Germany) at −80 °C until further use.
2.2. Extraction of Genomic DNA from Bacterial Cultures
Glaesserella parasuis, Histophilus somni, and Actinobacillus spp. were plated out on Chocolate Agar (Blood Agar No. 2 Base, Becton Dickinson GmbH, Heidelberg, Germany) and incubated for 24 h at 36 °C under microaerobic (≥2.5% CO2) and aerobic conditions, respectively. Actinomyces hyovaginalis (36 °C), Brachyspira spp. (36 °C), and Clostridium pefringens (45 °C) isolates were cultured on Columbia Agar containing 5% sheep blood (Becton Dickinson GmbH, Heidelberg, Germany) at anaerobic conditions (≥13% CO2) for 24–48 h. For cultivation of Campylobacter jejuni and Campylobacter coli, strains were incubated on Columbia agar with sheep blood under microaerobic conditions at 42 °C for 24–48 h. All other strains, including S. suis, were cultured on Columbia sheep blood agar (Becton Dickinson GmbH, Heidelberg, Germany) under aerobic conditions at 36 °C for 24–48 h. Genomic DNA of five to ten colonies of each strain was extracted using the DNeasy® Blood & Tissue Kit (Qiagen GmbH, Hilden, Germany) according to the manufacturer‘s manual. The DNA concentration of each eluate was measured in duplicate using the Thermo ScientificTM MultiskanTM GO spectrophotometer together with the Thermo ScientificTM μDropTM Plate and calculated using the SkanItTM Software Version 4.1 (Thermo Fisher Scientific Corporation, Waltham, MA, USA). For standardization of running conditions in analytical specificity testing, each eluate was adjusted to a DNA concentration of 0.1 ng/μL.
2.3. Design of LAMP Primers
LAMP primer design was based on the nucleotide sequence of the thrA gene (Acc No.: DQ205250.1). Corresponding sequence data was obtained from the database of the National Center for Biotechnology Information (NCBI) (Bethesda, MD, USA). A set of six primers, including forward and backward outer primers (F3 and B3), forward and backward inner primers (FIP and BIP), as well as forward and backward loop primers (LF and LB), for accelerating amplification, was created using LAMP Designer software (OptiGene Ltd., Horsham, UK) [31]. Primer sequences are shown in Table 1. All LAMP primers were ordered from Eurofins Genomics Germany GmbH (Ebersberg, Germany).

Table 1.
LAMP Primers used in this study targeting the thrA gene (Acc No.: DQ205250.1) of Streptococcus (S.) suis.
2.4. LAMP Reaction
LAMP reactions were carried out as suggested in the OptiGene Ltd. LAMP User Guides (Version 1.1, OptiGene Ltd., Horsham, UK). For each LAMP reaction, a 25-µL reaction mixture containing 15 μL of GspSSD isothermal mastermix (ISO-001) (OptiGene Ltd.), 2.5 μL primer mix, 2.5 μL PCR-grade water (Qiagen GmbH, Hilden, Germany), as well as 5 μL template, was prepared. LAMP reactions were performed using the real-time fluorometer Genie® II (OptiGene Ltd., Horsham, UK).
2.5. Optimization of the LAMP Assay
Optimization of the LAMP assay was performed in accordance with OptiGene Ltd. LAMP User Guides (Version 1.1, OptiGene Ltd., Horsham, UK). At first, a standard and a concentrated primer mix version were prepared. Single primer concentrations per reaction were 0.2 µM F3/B3, 0.8 µM FIP/BIP, and 0.4 µM LF/LB using the standard primer mix, and 0.2 µM F3/B3, 2.0 µM FIP/BIP, and 1.0 µM LF/LB using the concentrated primer mix. The optimum reaction temperature of both primer mixes was determined by testing S. suis reference strain (DSM 9682) DNA (0.1 ng/µL) at different temperatures ranging from 62 to 69 °C (∆T = 1 °C). Each run consisted of a 40-min amplification phase followed by an annealing period to generate a melting curve in the range from 98 to 80 °C (ramp rate 0.05 °C/s). Considering the standard deviation of all measurements, the shortest mean detection time determined the individual optimal reaction temperature of the standard and concentrated primer mixes. Subsequently, the most suitable primer mix version was selected based on analytical sensitivity and the corresponding detection times. For this purpose, a ten-fold series dilution of S. suis reference strain DNA was prepared with concentrations ranging from 10 ng/µL to 10 fg/µL. Each dilution stage served as a template for the LAMP reaction using the standard and concentrated primer mix at the previously determined optimal reaction temperature. The amplification and annealing phases of each run were set as described above. The lowest DNA concentration that could be successfully detected in triplicate determined the analytical sensitivity of the primer mix. All optimization experiments were performed three times in a row.
For all subsequent runs, the instrument settings were chosen as described above. Furthermore, each run included one reaction with a 5 µL template of S. suis DSM 9682 DNA (0.1 ng/µL) as a positive control and one reaction using 5 µL PCR-grade water instead of the DNA template as a negative control.
2.6. Analytical Specificity Testing
To investigate the analytical specificity of the thrA-LAMP assay, DNA from a total of 220 bacterial reference strains and field isolates was tested. Inclusivity testing was performed using 104 S. suis isolates. In order to examine as many genetically distinct S. suis strains as possible, isolates of every available serotype were chosen. Within the same serotype, isolates were selected to represent every available pattern of virulence-associated factor genes. Isolates from as many different tissues and herds as possible were selected within a combination. Exclusivity was verified by testing 116 non-S. suis isolates. Non-target isolates were selected based on their close genetic relationship with S. suis sp. or similar clinical and pathomorphological characteristics. Species that share the pig as a host or occur in its environment were also of interest. To confirm the reaction specificity, a melting curve was generated for each reaction as described above.
2.7. Determination of the Bacterial Cell-Based Detection Limit Using Different DNA Extraction Methods
The cell-based detection limit of the assay was determined in triplicate using two different methods for extracting DNA. For this purpose, the S. suis reference strain (DSM 9682) was cultured on Columbia sheep blood agar (Becton Dickinson GmbH, Heidelberg, Germany) under aerobic conditions at 36 °C for 48 h. Colonies of the reference strain were suspended in 5 mL of 0.9% isotonic saline solution until a turbidity of 1.0 McFarland units (MFU) was measured by a densitometer (Densimat, bioMérieux, Marcy-I’Etoile, France). This cell suspension was ten-fold serially diluted with isotonic saline solution. Viable cell counts were determined using the plate count technique as described previously [32]. Culture conditions of incubated Columbia sheep blood plates were adjusted for S. suis to 36 °C for 48 h under aerobic conditions. DNA was extracted from each cell suspension using two distinct procedures.
For thermal extraction, 50 µL of each dilution was transferred to 450 µL of variplex LPTV buffer (AmplexDiagnostics GmbH, Gars Bahnhof, Germany) in a 2 mL microcentrifuge tube. Subsequently, each tube was mixed by vortexing and boiled for 10 min at 100 °C on the Eppendorf ThermoMixer® C (Eppendorf AG, Hamburg, Germany). After cooling down to room temperature for 10 min, the inoculated LPTV buffer was directly used as a template for LAMP. This procedure is further referred to as the LPTV boiling method. Enzymatic extraction and DNA purification were performed using the DNeasy® Blood & Tissue Kit (Qiagen GmbH, Hilden, Germany). One milliliter of each dilution was transferred to a 1.5 mL microcentrifuge tube and centrifuged at 10,000× g for 5 min. After discarding the supernatant, the bacterial pellet was resuspended in 180 μL enzymatic lysis buffer, mixed by vortexing, and incubated on a thermoshaker (Thermomixer Comfort, Eppendorf AG, Hamburg, Germany) at 37 °C for 1 h (1000 rpm). Subsequently, 25 μL of Proteinase K and 200 μL of Buffer AL were added to each tube and mixed by vortexing. The mixture was incubated at 56 °C for 1 h (1000 rpm). All further steps were carried out according to the manufacturer’s instructions (Qiagen GmbH, Hilden, Germany). The elution volume was set to 200 μL. Each eluate served as a template for the LAMP reaction.
2.8. Real-Time PCR Assay
Detection of S. suis by real-time PCR was performed using the BactoReal® Kit Streptococcus suis (ingenetix GmbH, Vienna, Austria) based on the fibrinogen binding protein gene (fbpS). The real-time PCR was performed according to the kit manufacturer’s instructions using the Applied Biosystems 7500 Real-Time PCR System (Life Technologies GmbH, Darmstadt, Germany). Ct-values were interpreted according to the kit manufacturer’s instructions.
2.9. Detection Limits of LAMP in Artificially Contaminated Brain and Joint Samples
The preparation of a tenfold dilution series up to dilution level 10−8 of the S. suis DSM 9682 strain and the determination of viable cell counts were conducted as described above. Brain and joint tissue of pigs without clinical signs of a S. suis infection were sampled during routine necropsy using sterile, PCR inhibitor-free viscose swabs (nerbe plus GmbH & Co. KG, Winsen/Luhe, Germany). In dead pigs, the head was separated from the body, the scalp removed, and the cranial cavity opened by removing the skullcap using a disinfected saw. Subsequently, a swab sample was flat-inserted through the leptomeninx in the superficial brain tissue of both cerebral hemispheres. The joints were opened using a clean knife. Through an incision parallel to the joint space, the superficial skin and surrounding supporting tissue were separated, so that the joint cavity was exposed without touching the articulating surfaces. After careful insertion of a swab in the joint cavity, synovia was collected and the synovialis was brushed. Due to easy accessibility, the elbow, or alternatively, the tarsal joint, was sampled. As a sample matrix, two brain or joint swabs per prepared cell dilution level were taken. In addition, one swab per sampled brain or joint was provided to ensure the absence of S. suis via the cultural investigation as described below. Samples that did not produce any S. suis suspecting colonies on agar plates were selected for artificial contamination experiments.
Using the LPTV boiling method for DNA extraction, eight swab samples per sample type were rotated and squeezed out in one tube containing 450 µL of LPTV buffer. Subsequently, 50 µL of S. suis cell suspension was taken from each dilution level and added to the prepared LPTV buffer. Further steps were carried out as described previously. For DNA extraction with the DNeasy® Blood & Tissue kit, the remaining eight swab samples per sample type were suspended in 500 μL of enzymatic lysis buffer in a microcentrifuge tube, followed by a vortexing step. Subsequently, 1 mL of each dilution level was transferred to a 1.5 mL microcentrifuge tube and centrifuged at 10,000× g for 5 min. The supernatant was carefully discarded. The cell pellet was resuspended with 180 μL of the enzymatic lysis buffer, including tissue components, and again mixed by vortexing. Further extraction was performed as described previously. Templates were tested using LAMP and real-time PCR as described previously. Experiments were performed in triplicate.
2.10. Detection of S. suis in Field Samples
Pigs with clinical or pathological evidence of a systemic S. suis infection were selected for the collection of clinical field samples. The brain and joints of these animals were sampled during diagnostic necropsies, as described above. For each brain and joint sampled, one swab was used for cultural investigation. For this purpose, the swab sample was fractionally smeared on Columbia agar containing 5% sheep blood (Becton Dickinson GmbH, Heidelberg, Germany), Chocolate Agar (Blood Agar No. 2 Base, Becton Dickinson GmbH, Heidelberg, Germany), Columbia CNA agar with 5% sheep blood (Becton Dickinson GmbH, Heidelberg, Germany), and Gassner agar (Oxoid Deuschland GmbH, Wesel, Germany), followed by incubation under aerobic or microaerobic (Chocolate agar) conditions at 36 °C for 24 h until first inspection and a further aerobic incubation until 48 h. Potentially pathogenic bacteria were selected based on their morphology, color, odor, media-specific growth, and haemolysis behavior and identified by using a specific biochemical substrate test series. In the case of the detection of S. suis, isolates were further cryobanked and serotyped using in-house PCR with modifications by Silva et al. and Kerdsin et al. (refer to Protocol S1) [33,34]. This conventional multiplex PCR was based on the gdh-gene and was able to differentiate the five capsular serotypes (cps-types) 1, 2, 4, 7, and 9 [33,34]. In addition, virulence-associated factors were determined as genes for the muramidase-released protein (mrp), suilysin (sly), extracellular factor (epf), and arginine deiminase (arcA) [33,34]. The PCR was suitable to identify the most common invasive serotypes in Germany [9]. All isolates carrying gdh or at least one virulence-associated factor gene without any of the five cps-types are further referred to as “non-typable S. suis”.
Two additional swabs per sampled brain or joint were stored at minus 20 °C until DNA extraction. For extraction using the LPTV boiling method, one tissue swab was suspended in 500 μL LPTV buffer and further processed as described above. For enzymatic extraction using the DNeasy® Blood & Tissue Kit, a tissue swab was suspended in 500 μL of isotonic saline solution, vortexed, and centrifuged at 10,000× g for 5 min. Subsequently, the supernatant was discarded, and the pellet was resuspended in 180 µL of enzymatic lysis buffer. Further steps were performed as described previously. Both DNA templates were examined by LAMP and real-time PCR. Cultural investigation with subsequent PCR-based serotyping as well as real-time PCR after DNeasy® Blood & Tissue Kit extraction served as reference methods. In addition, cryobanked S. suis isolates from clinical samples were cultured as described above. Using a sterile inoculation loop, 5 to 10 colonies from pure subcultures were suspended in 500 μL of LPTV buffer. DNA was extracted using the LPTV boiling method and all templates were examined by thrA-LAMP.
2.11. Data Management and Statistical Analyses
Original data from LAMP and real-time PCR were viewed and processed using the Genie® Explorer software (Version 2.0.7.11, OptiGene Ltd., Horsham, UK) and the 7500 Software for 7500 and 7500 Fast Real-time PCR Systems (Version 2.3, Life Technologies GmbH, Darmstadt, Germany), respectively. Statistical analyses were accomplished using Microsoft Office Excel 2016 (Microsoft Corporation, Redmond, WA, USA). Sensitivity and specificity, as well as positive and negative predictive values, each with 95% confidence intervals, were calculated using SAS 9.4m7 with the SAS Enterprise Guide, version 7.1 (SAS Institute Inc., Cary, NC, USA). The calculation of diagnostic quality criteria was performed as suggested by the U.S. Food and Drug Administration (FDA) [35].
3. Results
3.1. Optimized LAMP Reaction Conditions and Analytical Sensitivity
As the first step for assay optimization, the optimal reaction temperature of the standard and concentrated primer mixes was determined to be 65 °C for both primer mixes (fastest mean detection times, minimal standard deviation). At this temperature, a positive fluorescence signal was measured on average after 10.38 min when using the standard primer mix and after 7.83 min when using the concentrated primer mix. DNA-based analytical sensitivity was determined as 0.5 pg DNA per reaction mixture for both primer mixes. In contrast to the standard primer mix, even 50 fg of reference strain DNA could be successfully detected in 2 of 3 replicates when using the concentrated primer mix. In addition, the concentrated primer mix achieved overall faster mean detection times across all dilution levels. Considering these results, all further LAMP reactions were performed using the concentrated primer mix at a reaction temperature of 65 °C. Samples were considered positive for S. suis if both a fluorescence signal and a specific melting temperature of 84.63 ± 1 °C were detectable.
3.2. Analytical Specificity of thrA-Based LAMP
Among all 104 S. suis strains tested in this study, over 99% of isolates showed a positive fluorescence signal when performing the thrA-based LAMP. The diversity of serotypes and virulence-associated factor genes of these isolates is shown in Table 2. The isolates originated from clinically relevant organs (brain and cerebrospinal fluid, joint, heart, serosa, kidney, lung), as well as from naturally colonized tissues (bronchus, tonsil). The only S. suis isolate that showed no amplification by thrA-LAMP was identified as serotype 7, unusually without any of the detectable virulence-associated factor genes. Detailed biochemical identification using the API® 20 STREP system (bioMérieux, Marcy-I’Etoile, France) determined this isolate as S. suis biotype II with “good identification” (95.2 percentage of identification and T = 0.64). Further MALDI-TOF MS analysis of this strain at SAN Group Biotech Germany GmbH (Höltinghausen, Germany) identified this isolate as Streptococcus sp. Positive fluorescence signals for the other strains were measured within 7.17 to 11.60 min after starting the amplification process. Melting temperatures of corresponding LAMP products ranged from 84.19 °C to 84.89 °C, indicating species-specific fluorescence signals. All of the 116 non-S. suis strains (Table 3) were tested negative using thrA-LAMP.

Table 2.
Inclusivity testing of different serotypes and corresponding virulence-associated factor genes of S. suis isolates by thrA-LAMP.

Table 3.
Exclusivity testing of non-target species by thrA-LAMP.
3.3. Bacterial Cell-Based Detection Limit of thrA-LAMP
Bacterial cell-based detection limits were determined by the lowest mean viable cell count in a ten-fold serial dilution of S. suis strain DSM 9682 that could be successfully detected in triplicate. Using the LPTV boiling method for DNA extraction, a mean detection limit of 1.08 CFU/reaction (2.2 × 103 CFU/mL) was achieved. Kit extraction resulted in a higher mean detection limit of 53.8 CFU/reaction (2.2 × 103 CFU/mL).
3.4. Evaluation of Brain and Joint Matrix Effects on the Detection Limit of thrA-LAMP
Detection limits of the thrA-based LAMP after DNA extraction using the LPTV boiling method and DNeasy® Blood & Tissue Kit were determined by spiking experiments on artificially contaminated brain and joint swabs (Table 4). The detection limits were determined by the lowest mean viable cell count that could be successfully detected in triplicate. A detection limit of 104–105 CFU per swab was achieved by LAMP when DNA was extracted using the LPTV boiling method on brain swabs. When the same templates were examined by real-time PCR, almost no amplification of the target or the internal positive control was observed (Table S1). DNA extraction using the DNeasy® Blood & Tissue Kit improved the detection limit of thrA-LAMP and real-time PCR to 103–104 CFU/swab. Performing the LPTV boiling method on spiked joint swabs, the detection limit of LAMP was 103–104 CFU/swab. By applying kit extraction to joint swabs, LAMP became even less sensitive, with a detection limit of 104–105 CFU per swab. Compared to real-time PCR, LAMP had an at least 10-fold higher detection limit when examining joint samples.

Table 4.
Total number of positive signals achieved by thrA-LAMP and real-time PCR using LPTV-boiling method and the DNeasy® Blood & Tissue Kit to extract DNA from spiked brain and joint samples.
3.5. Diagnostic Quality Criteria for Detection of S. suis in Brain and Joint Swabs
Diagnostic quality criteria of thrA-LAMP compared to cultural investigation with subsequent PCR-based serotyping as well as real-time PCR after DNeasy® Blood & Tissue Kit extraction are summarized below (Table 5 and Table 6). The brains of 49 pigs were examined. S. suis was successfully isolated from 25 brains. 30 brain samples were positive in a real-time PCR examination. Using the LPTV boiling method, the thrA-based LAMP assay was able to correctly identify 22 of the culture-positive and 23 of the culture-negative brain samples. When real-time PCR served as a reference method, thrA-LAMP correctly identified 23 of the PCR-positive and all PCR-negative brain samples. When brain swabs were extracted and purified using the DNeasy® Blood & Tissue Kit, all culture-positive samples were correctly identified by LAMP, but the number of correctly detected culture-negative samples decreased to 21. Twenty-eight PCR-positive brain samples were identified as such by thrA-LAMP using kit extraction. The detection time median of LAMP was 18.45 min when using the LPTV boiling method. Applying kit extraction reduced the detection time median to 10.68 min (e.g., Figure 1a,b).

Table 5.
Diagnostic quality criteria of thrA-LAMP using LPTV boiling method and DNeasy® Blood & Tissue Kit for DNA extraction of brain swabs with cultural investigation as reference method.

Table 6.
Diagnostic quality criteria of thrA-LAMP using LPTV boiling method and DNeasy® Blood & Tissue Kit for DNA extraction of brain swabs with real-time PCR as reference method.
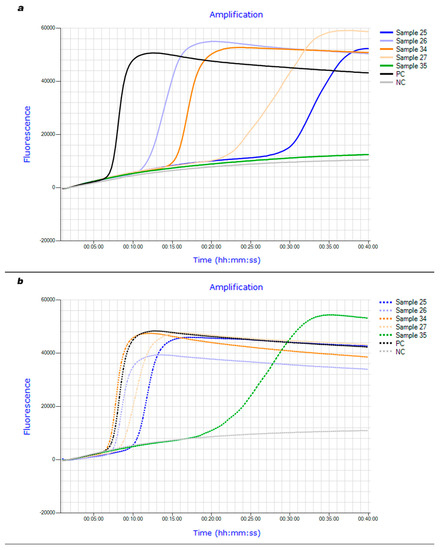
Figure 1.
(a): Fluorescence curves of thrA-LAMP examining brain field samples after DNA extraction using LPTV boiling method. (b): Fluorescence curves of thrA-LAMP examining brain field samples after DNA extraction using DNeasy® Blood & Tissue Kit.
Joint swabs were obtained from 34 different pigs. Of these, ten swabs were positive in the cultural investigation, and 23 joint swabs were tested positive by real-time PCR. In combination with the LPTV boiling method, thrA-LAMP was only able to correctly identify one joint swab as S. suis positive. As already observed for brain swabs, DNA extraction and purification using the DNeasy® Blood & Tissue Kit increased LAMP sensitivity to 100% (cultural investigation) and 82.61% (real-time PCR), respectively. Performing kit extraction, the specificity of thrA-LAMP was lower when examining culture-negative (58.33%) than PCR-negative joint swabs (90.91%). The one joint sample detected by LAMP using the LPTV boiling method showed a positive fluorescence signal after 18.63 min. The detection time median was 11.63 min when kit extraction was performed.
All 35 S. suis isolates retrieved from clinical samples gave a positive result when performing thrA-LAMP when DNA was directly extracted from bacterial colonies using the LPTV boiling method. The detection time median for a positive fluorescence signal was 6.28 min. The melting temperatures of all LAMP products were specific and ranged between 84.34 and 85.07 °C.
4. Discussion
In this study, the first S. suis-specific LAMP assay was developed and validated for rapid detection of this pathogen in brain and joint samples of clinically infected pigs. The assay was based on the thrA gene encoding the enzyme aspartokinase/homoserine dehydrogenase [36]. Since King et al. referred to thrA as a housekeeping gene, this region can be considered to be a highly conserved and specific genomic target for the S. suis species [36]. In the present study, the thrA-LAMP was able to successfully detect 99% of the S. suis strains, representing the globally predominant serotypes found in pigs [10]. Furthermore, no cross-reactions with non-target species were observed. By standardizing the template DNA concentration to 0.1 ng/μL, varying detection times of S. suis strains were caused mainly by polymorphisms of the thrA gene sequence. Considering the given range of detection times, it can be assumed that the thrA-sequence shares high similarity within the S. suis species. The only S. suis strain that was not detected by thrA-LAMP was biochemically confirmed using the API® 20 STREP system. Nevertheless, further MALDI-TOF MS analysis was inconclusive, as it identified this strain as Streptococcus sp. Multitest systems, such as API®, have been reported to misidentify S. suis strains, resulting in false-negative results [37,38]. MALDI-TOF MS analysis, on the other hand, tended to give false-positive results, especially in the classification of tonsil isolates [39]. Since the results of this study did not reflect any of the described cases, it seemed that the investigated strain was not a typical representative of the S. suis species [3]. Average nucleotide identity (ANI) analysis could be used for confident identification of this isolate [39]. Moreover, no reliable prediction can be made about its virulence and clinical relevance as this strain was isolated from the tonsil of a healthy carrier [40]. Consequently, the suitability of thrA-LAMP for the detection of clinically relevant S. suis strains remained unrestricted. The previous S. suis-LAMP assays developed by Zhang et al. and Meng et al. only targeted serotype-specific genes without covering the worldwide serotype distribution found in diseased pigs and humans [10,21,29,41]. While the recN-based LAMP developed by Arai et al. was able to detect all serotypes according to the currently valid taxonomy, a smaller number of 54 S. suis strains, as well as 19 non-target strains, were used for analytical specificity testing. Furthermore, for most serotypes, only one representative strain was tested without considering virulence-associated factor genes, so the data on inclusivity may not be entirely valid [23]. By including twice as many S. suis isolates and covering multiple representatives per serotype with distinct virulence-associated factor gene patterns, the thrA-LAMP was successfully tested on a population with higher genetic variability.
For the application of LAMP under field conditions, a fast and easy-to-use DNA extraction method was required [42]. The developed LPTV boiling method can be performed with a heating block in less than 20 min, thus requiring only minimal laboratory equipment. The detection limit of the thrA-LAMP assay using the LPTV boiling method for DNA extraction was compared to using purified templates obtained from the DNeasy® Blood & Tissue Kit. Performing the LPTV boiling method, thrA-LAMP showed a lower detection limit (1.08 CFU/reaction) compared to DNA extraction by the DNeasy® Blood & Tissue Kit (53.8 CFU/reaction). Although the absence of a purification step can negatively affect the detection limit, since possible inhibitors are not eliminated [43,44], the thrA-LAMP assay proved to be robust against potentially interfering components. Previous studies have already shown that thermal cell disruption using a boiling method could achieve higher DNA yields than commercial extraction kits based on enzymatic cell treatment [45]. The lower DNA yields of silica membrane-based DNA extraction methods could also be the consequence of insufficient DNA retention [46]. In consideration of these results, the LPTV boiling method was preferred for DNA extraction to gain the highest possible sensitivity from thrA-LAMP. In general, comparison of cell-based detection limits between previously published S. suis-LAMP assays was limited by the availability of data, differences in DNA extraction methods regarding washing steps and boiling time, as well as primer set design. Using the LPTV boiling method, the thrA-LAMP was less sensitive than comparable S. suis-LAMP assays (Meng et al.: 18.4 CFU/mL, Arai et al.: 58.3 CFU/mL, Huy et al.: 100 CFU/mL) [22,23,29]. However, because the detection limit for none of these LAMP assays was determined in a spiking experiment, the application-relevant sensitivity in the presence of the sample matrix remains unknown [22,23,29].
Spiking experiments were carried out to evaluate the detection limit of the thrA-LAMP in the presence of relevant tissue matrices. The effect of impure and purified templates on the detection rate of LAMP was assessed by comparing the LPTV boiling method and the DNeasy® Blood & Tissue Kit for DNA extraction. Examining unpurified brain templates generated by the LPTV boiling method, thrA-LAMP showed a superior detection limit compared to real-time PCR. The missing signal of the internal positive control of the PCR-negative samples indicated the presence of inhibitors as described in the manufacturer’s manual. So far, an interfering effect on DNA amplification has only been described for native cerebrospinal fluid but not for the brain tissue itself [47]. Furthermore, boiling cerebrospinal fluid has been found to be insufficient for removing potential PCR inhibitors, especially when contaminated with blood [47,48]. The inhibitory effect of the brain tissue could have been caused by lipids or small amounts of residual blood [27,43]. The superior resistance of the thrA-LAMP assay against containing molecular inhibitors, highlights the robustness of the LAMP method over real-time PCR [26,27,28]. Using the DNeasy® Blood & Tissue Kit for brain swabs, thrA-LAMP showed a 10-fold improved detection limit. This is most likely due to the elimination of potentially inhibiting brain tissue components during kit purification [49]. The ability of comparable silica membrane-based DNA extraction kits to generate high-purity eluates from brain tissue was previously demonstrated [50]. In the case of spiked joint swaps, the LPTV boiling method led to a lower detection limit of thrA-LAMP compared to brain swabs. Inhibitory matrix effects of sterile synovial fluid components, such as heme or glycoproteins with acidic polysaccharides, have already been described for PCR and real-time PCR [51,52]. Contrary to the spiked brain swabs, the detection limit of thrA-LAMP did not improve after the elimination of potential inhibitors using the DNA extraction kit. Surprisingly, thrA-LAMP showed a 10-fold higher detection limit compared to real-time PCR when examining impure joint templates, despite PCR being referred to as the more susceptible method [26,27,28]. This result suggests that this assumption does not apply universally and that the performance of both methods depends on the matrix under investigation. In summary, the spiking experiment showed the most promising results for the detection of S. suis in brain and joint swabs by thrA-LAMP using the LPTV boiling method for DNA extraction. By using the DNeasy® blood and tissue kit, the detection limit of thrA-LAMP could be improved for brain swabs but not for joint swabs.
Diagnostic quality criteria of thrA-LAMP were determined using cultural investigation and real-time PCR combined with the DNeasy® blood and tissue kits as reference methods. The thrA-based LAMP assay achieved high sensitivity (88%) and specificity (95.83%) applying the LPTV boiling method on brain swabs using cultural investigation as reference method. When real-time PCR served as a reference method, the sensitivity of thrA-LAMP decreased to 76.67%, but specificity increased to 100%. These differences between reference methods can be explained by the ability of real-time PCR to detect DNA of non-viable bacteria in culture-negative samples, resulting in higher numbers of true-positive samples [53]. Similar observations were made in previous studies in which PCR detected a higher number of S. suis-positive cerebrospinal fluid samples compared to culturing [54,55]. Since all clinical specimens were collected from animals with clinical or pathological evidence of S. suis-infection, it is possible that animals were treated with antibiotics prior to sampling, resulting in a reduction of vital bacteria and an unsuccessful cultural investigation [54]. Furthermore, it should be noted that about 45% of culturally negative brain and about 88% of culturally negative joint samples originated from organs with inflammatory findings (Table S2). The formation of biofilms in infected tissues as a strategy to evade host immunity has been demonstrated for S. suis [56,57] and was described as a reason for false negative results in cultural testing in both veterinary and human medicine [58,59]. Application of thrA-LAMP in combination with the LPTV boiling method on joint swabs showed unexpectedly poor sensitivity, regardless of the viewed reference method. Surprisingly, and contrary to the results of the spiking experiment, LAMP showed a strong improvement in diagnostic sensitivity when clinical joint swabs were processed by kit extraction. Similarly, Kuipers et al. previously showed that silica membrane-based DNA extraction of synovial fluid samples from patients with rheumatoid arthritis obtained higher PCR sensitivity compared to simpler heat-based methods [51]. While only healthy joints were sampled for spiking experiments, field samples were mostly obtained from joints showing evidence of inflammation (85%, Table S2). Thus, it is possible that the products of inflammation could have an additional inhibitory effect, which could not be accounted for in the spiking experiment. Larger quantities of cell debris resulting from bacterial and host cells in the context of inflammation could have increased the previously described inhibitory effect [43,48]. Furthermore, the resulting higher amounts of released leukocyte or host DNA could be involved in the inhibitory effect proposed for the inflammation [60,61,62]. Both brain and joint field samples were largely obtained from inflamed organs. The difference in sensitivity observed for clinical brain and joint samples extracted by the LPTV boiling method could be due to the higher proportion of inflamed joints (85%, Table S2) compared to brains (55%, Table S2) that were sampled. Also, tissue-dependent differences in immune responses, such as the frequency of certain immune cells like plasma cells, could be responsible for the varying sensitivities observed for the targeted organs [63,64]. An inflammation-induced increase in synovial IgG levels by plasma cells could have been responsible for tissue-specific inhibition, which was further enhanced by boiling together with target DNA during the LPTV boiling method [65,66,67,68]. Eliminating these potential inhibitors by applying the DNeasy® Blood and Tissue kit could be the reason for the improvement of thrA-LAMP sensitivity observed for clinical joint swabs [51,52]. Many S. suis-LAMP assays have not been evaluated against a defined reference method, making it difficult to evaluate the diagnostic quality criteria of thrA-LAMP [22,23,29]. Only the cps2J-LAMP of Zhang et al. was compared to a real-time PCR targeting the same gene. In that study, examination of 66 clinical samples of pigs and humans, including nasal swabs, blood serum, and cerebrospinal fluid, revealed 96.3% sensitivity and 100% specificity of cps2J-LAMP, showing similar results to thrA-LAMP using kit extraction and real-time PCR as reference methods [21]. Finally, all isolates recovered from field samples were correctly identified by the thrA-LAMP assay in combination with the LPTV boiling method. Thus, this method could be used for rapid confirmation of S. suis-suspected colonies as part of a cultural investigation.
5. Conclusions
The thrA-based LAMP assay established in this study allows specific identification of S. suis, including the variety of clinically relevant serotypes. The thrA-LAMP was found to be suitable for rapid detection of S. suis from brain swabs using the LPTV boiling method for extracting DNA. This easy-to-perform assay requires only a little equipment, making it well suited for field diagnostics. Combining the thrA-LAMP with the DNeasy® Blood and Tissue Kit increases sensitivity and allows application to brain and joint swabs. In future field studies, thrA-LAMP and LPTV boiling method should be optimized for samples taken from living pigs, such as heparinized blood, synovia, cerebrospinal fluid, and tonsillar swabs, in order to avoid the necessity of a necropsy. Future validation of the thrA-LAMP assay regarding human cerebrospinal fluid and synovial punctates would supplement a procedure applicable for intra vitam diagnostics of exposed humans.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/microorganisms11102447/s1, Protocol S1: Detailed procedure for serotyping of S. suis isolates by in-house multiplex PCR; Table S1: Detection limits of thrA-LAMP and real-time PCR using LPTV boiling method and DNeasy® Blood & Tissue Kit on artificially contaminated brain and joint samples; Table S2: Examination of field samples by cultural investigation, thrA-LAMP and real-time PCR using LPTV boiling method and DNeasy® Blood & Tissue Kit.
Author Contributions
Conceptualization, I.H.-P. and A.A.; Formal analysis, J.H. and K.R.; Investigation, J.H.; Methodology, J.H. and A.K.; Project administration, I.H.-P. and A.A.; Resources, I.H.-P. and A.A.; Supervision, A.K.; Validation, J.H. and A.K.; Visualization, J.H.; Writing—original draft, J.H.; Writing—review & editing, A.K., K.R., I.H.-P. and A.A. All authors have read and agreed to the published version of the manuscript.
Funding
This Open Access publication was funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation)—491094227 “Open Access Publication Funding” and the University of Veterinary Medicine Hannover, Foundation.
Institutional Review Board Statement
This study was approved by the ethics committee of the Institute for Animal Hygiene, Animal Welfare and Farm Animal Behaviour (ITTN) (University of Veterinary Medicine Hannover, Foundation). Project identification code: TVA-2021-V-93. Date of approval: 16 December 2021. Contact: Institute for Animal Hygiene, Animal Welfare and Farm Animal Behaviour (ITTN), University of Veterinary Medicine Hannover, 30173 Hannover, Germany.
Data Availability Statement
The data presented in this study are available in the presented article or Supplementary Material.
Acknowledgments
We wish to thank Mechthild Sieve, Simone Schwermann-Jäger and Brigitte Meisenheimer for her excellent technical assistance.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Gottschalk, M.; Segura, M. Streptococcosis. In Diseases of Swine, 11th ed.; Zimmermann, J.J., Karriker, L.A., Ramirez, A., Schwartz, K.J., Stevenson, G.W., Zhang, J., Eds.; WILEY Blackwell: Hoboken, NJ, USA, 2019; pp. 934–950. [Google Scholar]
- Staats, J.J.; Feder, I.; Okwumabua, O.; Chengappa, M.M. Streptococcus suis: Past and present. Vet. Res. Commun. 1997, 21, 381–407. [Google Scholar] [CrossRef] [PubMed]
- Okura, M.; Osaki, M.; Nomoto, R.; Arai, S.; Osawa, R.; Sekizaki, T.; Takamatsu, D. Current Taxonomical Situation of Streptococcus suis. Pathogens 2016, 5, 45. [Google Scholar] [CrossRef] [PubMed]
- Tien, L.H.T.; Nishibori, T.; Nishitani, Y.; Nomoto, R.; Osawa, R. Reappraisal of the taxonomy of Streptococcus suis serotypes 20, 22, 26, and 33 based on DNA-DNA homology and sodA and recN phylogenies. Vet. Microbiol. 2013, 162, 842–849. [Google Scholar] [CrossRef] [PubMed]
- Nomoto, R.; Maruyama, F.; Ishida, S.; Tohya, M.; Sekizaki, T.; Osawa, R. Reappraisal of the taxonomy of Streptococcus suis serotypes 20, 22 and 26: Streptococcus parasuis sp. nov. Int. J. Syst. Evol. Microbiol. 2015, 65, 438–443. [Google Scholar] [CrossRef] [PubMed]
- Guo, G.; Wang, Z.; Li, Q.; Yu, Y.; Li, Y.; Tan, Z.; Zhang, W. Genomic characterization of Streptococcus parasuis, a close relative of Streptococcus suis and also a potential opportunistic zoonotic pathogen. BMC Genom. 2022, 23, 469. [Google Scholar] [CrossRef] [PubMed]
- Tohya, M.; Arai, S.; Tomida, J.; Watanabe, T.; Kawamura, Y.; Katsumi, M.; Ushimizu, M.; Ishida-Kuroki, K.; Yoshizumi, M.; Uzawa, Y.; et al. Defining the taxonomic status of Streptococcus suis serotype 33: The proposal for Streptococcus ruminantium sp. nov. Int. J. Syst. Evol. Microbiol. 2017, 67, 3660–3665. [Google Scholar] [CrossRef]
- Hill, J.E.; Gottschalk, M.; Brousseau, R.; Harel, J.; Hemmingsen, S.M.; Goh, S.H. Biochemical analysis, cpn60 and 16S rDNA sequence data indicate that Streptococcus suis serotypes 32 and 34, isolated from pigs, are Streptococcus orisratti. Vet. Microbiol. 2005, 107, 63–69. [Google Scholar] [CrossRef]
- Prüfer, T.L.; Rohde, J.; Verspohl, J.; Rohde, M.; de Greeff, A.; Willenborg, J.; Valentin-Weigand, P. Molecular typing of Streptococcus suis strains isolated from diseased and healthy pigs between 1996–2016. PLoS ONE 2019, 14, e0210801. [Google Scholar] [CrossRef]
- Goyette-Desjardins, G.; Auger, J.P.; Xu, J.; Segura, M.; Gottschalk, M. Streptococcus suis, an important pig pathogen and emerging zoonotic agent-an update on the worldwide distribution based on serotyping and sequence typing. Emerg. Microbes Infect. 2014, 3, e45. [Google Scholar] [CrossRef]
- Haas, B.; Grenier, D. Understanding the virulence of Streptococcus suis: A veterinary, medical, and economic challenge. Med. Mal. Infect. 2018, 48, 159–166. [Google Scholar] [CrossRef]
- Dutkiewicz, J.; Sroka, J.; Zając, V.; Wasiński, B.; Cisak, E.; Sawczyn, A.; Kloc, A.; Wójcik-Fatla, A. Streptococcus suis: A re-emerging pathogen associated with occupational exposure to pigs or pork products. Part I—Epidemiology. Ann. Agric. Environ. Med. 2017, 24, 683–695. [Google Scholar] [CrossRef]
- Hopkins, D.; Poljak, Z.; Farzan, A.; Friendship, R. Factors contributing to mortality during a Streptoccocus suis outbreak in nursery pigs. Can. Vet. J. 2018, 59, 623–630. [Google Scholar]
- Neila-Ibáñez, C.; Casal, J.; Hennig-Pauka, I.; Stockhofe-Zurwieden, N.; Gottschalk, M.; Migura-García, L.; Pailler-García, L.; Napp, S. Stochastic Assessment of the Economic Impact of Streptococcus suis-Associated Disease in German, Dutch and Spanish Swine Farms. Front. Vet. Sci. 2021, 8, 676002. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Zhang, H.; Wu, Z.; Wang, S.; Cao, M.; Hu, D.; Wang, C. Streptococcus suis infection: An emerging/reemerging challenge of bacterial infectious diseases? Virulence 2014, 5, 477–497. [Google Scholar] [CrossRef] [PubMed]
- Wangsomboonsiri, W.; Luksananun, T.; Saksornchai, S.; Ketwong, K.; Sungkanuparph, S. Streptococcus suis infection and risk factors for mortality. J. Infect. 2008, 57, 392–396. [Google Scholar] [CrossRef] [PubMed]
- Lun, Z.R.; Wang, Q.P.; Chen, X.G.; Li, A.X.; Zhu, X.Q. Streptococcus suis: An emerging zoonotic pathogen. Lancet Infect. Dis. 2007, 7, 201–209. [Google Scholar] [CrossRef]
- Okwumabua, O.; O’Connor, M.; Shull, E. A polymerase chain reaction (PCR) assay specific for Streptococcus suis based on the gene encoding the glutamate dehydrogenase. FEMS Microbiol. Lett. 2003, 218, 79–84. [Google Scholar] [CrossRef]
- Xia, X.; Wang, X.; Wei, X.; Jiang, J.; Hu, J. Methods for the detection and characterization of Streptococcus suis: From conventional bacterial culture methods to immunosensors. Antonie Van Leeuwenhoek 2018, 111, 2233–2247. [Google Scholar] [CrossRef] [PubMed]
- Ishida, S.; le Tien, H.T.; Osawa, R.; Tohya, M.; Nomoto, R.; Kawamura, Y.; Takahashi, T.; Kikuchi, N.; Kikuchi, K.; Sekizaki, T. Development of an appropriate PCR system for the reclassification of Streptococcus suis. J. Microbiol. Methods 2014, 107, 66–70. [Google Scholar] [CrossRef]
- Zhang, J.; Zhu, J.; Ren, H.; Zhu, S.; Zhao, P.; Zhang, F.; Lv, H.; Hu, D.; Hao, L.; Geng, M.; et al. Rapid Visual Detection of Highly Pathogenic Streptococcus suis Serotype 2 Isolates by Use of Loop-Mediated Isothermal Amplification. J. Clin. Microbiol. 2013, 51, 3250–3256. [Google Scholar] [CrossRef]
- Huy, N.T.; Hang, T.T.; Boamah, D.; Lan, N.T.; Van Thanh, P.; Watanabe, K.; Huong, V.T.; Kikuchi, M.; Ariyoshi, K.; Morita, K.; et al. Development of a single-tube loop-mediated isothermal amplification assay for detection of four pathogens of bacterial meningitis. FEMS Microbiol. Lett. 2012, 337, 25–30. [Google Scholar] [CrossRef] [PubMed]
- Arai, S.; Tohya, M.; Yamada, R.; Osawa, R.; Nomoto, R.; Kawamura, Y.; Sekizaki, T. Development of loop-mediated isothermal amplification to detect Streptococcus suis and its application to retail pork meat in Japan. Int. J. Food Microbiol. 2015, 208, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Notomi, T.; Okayama, H.; Masubuchi, H.; Yonekawa, T.; Watanabe, K.; Amino, N.; Hase, T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000, 28, E63. [Google Scholar] [CrossRef] [PubMed]
- Wong, Y.P.; Othman, S.; Lau, Y.L.; Radu, S.; Chee, H.Y. Loop-mediated isothermal amplification (LAMP): A versatile technique for detection of micro-organisms. J. Appl. Microbiol. 2018, 124, 626–643. [Google Scholar] [CrossRef] [PubMed]
- Kaneko, H.; Kawana, T.; Fukushima, E.; Suzutani, T. Tolerance of loop-mediated isothermal amplification to a culture medium and biological substances. J. Biochem. Biophys. Methods 2007, 70, 499–501. [Google Scholar] [CrossRef] [PubMed]
- Francois, P.; Tangomo, M.; Hibbs, J.; Bonetti, E.J.; Boehme, C.C.; Notomi, T.; Perkins, M.D.; Schrenzel, J. Robustness of a loop-mediated isothermal amplification reaction for diagnostic applications. FEMS Immunol. Med. Microbiol. 2011, 62, 41–48. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Wang, F.; Prinyawiwatkul, W.; Ge, B. Robustness of Salmonella loop-mediated isothermal amplification assays for food applications. J. Appl. Microbiol. 2014, 116, 81–88. [Google Scholar] [CrossRef]
- Meng, J.; Li, C.; Wang, Y.; Bian, Z.; Chu, P.; Zhai, S.; Yang, D.; Song, S.; Li, Y.; Jiang, Z.; et al. Accelerated loop-mediated isothermal amplification method for the rapid detection of Streptococcus suis serotypes 2 and 14 based on single nucleotide polymorphisms. Front. Cell Infect. Microbiol. 2022, 12, 1034762. [Google Scholar] [CrossRef]
- Boonyong, N.; Kaewmongkol, S.; Khunbutsri, D.; Satchasataporn, K.; Meekhanon, N. Contamination of Streptococcus suis in pork and edible pig organs in central Thailand. Vet. World 2019, 12, 165–169. [Google Scholar] [CrossRef]
- Nagamine, K.; Hase, T.; Notomi, T. Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol. Cell Probes 2002, 16, 223–229. [Google Scholar] [CrossRef]
- Kreitlow, A.; Becker, A.; Ahmed, M.F.E.; Kittler, S.; Schotte, U.; Plötz, M.; Abdulmawjood, A. Combined Loop-Mediated Isothermal Amplification Assays for Rapid Detection and One-Step Differentiation of Campylobacter jejuni and Campylobacter coli in Meat Products. Front. Microbiol. 2021, 12, 668824. [Google Scholar] [CrossRef] [PubMed]
- Silva, L.M.; Baums, C.G.; Rehm, T.; Wisselink, H.J.; Goethe, R.; Valentin-Weigand, P. Virulence-associated gene profiling of Streptococcus suis isolates by PCR. Vet. Microbiol. 2006, 115, 117–127. [Google Scholar] [CrossRef] [PubMed]
- Kerdsin, A.; Akeda, Y.; Hatrongjit, R.; Detchawna, U.; Sekizaki, T.; Hamada, S.; Gottschalk, M.; Oishi, K. Streptococcus suis serotyping by a new multiplex PCR. J. Med. Microbiol. 2014, 63, 824–830. [Google Scholar] [CrossRef] [PubMed]
- Meier, K. Statistical Guidance on Reporting Results from Studies Evaluating Diagnostic Tests—Guidance for Industry and FDA Staff; FDA: White Oak, MD, USA, 2007; p. 39. [Google Scholar]
- King, S.J.; Leigh, J.A.; Heath, P.J.; Luque, I.; Tarradas, C.; Dowson, C.G.; Whatmore, A.M. Development of a multilocus sequence typing scheme for the pig pathogen Streptococcus suis: Identification of virulent clones and potential capsular serotype exchange. J. Clin. Microbiol. 2002, 40, 3671–3680. [Google Scholar] [CrossRef] [PubMed]
- Gottschalk, M.; Xu, J.; Calzas, C.; Segura, M. Streptococcus suis: A new emerging or an old neglected zoonotic pathogen? Future Microbiol. 2010, 5, 371–391. [Google Scholar] [CrossRef] [PubMed]
- Gottschalk, M.; Higgins, R.; Jacques, M.; Beaudoin, M.; Henrichsen, J. Isolation and characterization of Streptococcus suis capsular types 9–22. J. Vet. Diagn. Investig. 1991, 3, 60–65. [Google Scholar] [CrossRef]
- Werinder, A.; Aspán, A.; Söderlund, R.; Backhans, A.; Sjölund, M.; Guss, B.; Jacobson, M. Whole-Genome Sequencing Evaluation of MALDI-TOF MS as a Species Identification Tool for Streptococcus suis. J. Clin. Microbiol. 2021, 59, e0129721. [Google Scholar] [CrossRef]
- Fittipaldi, N.; Segura, M.; Grenier, D.; Gottschalk, M. Virulence factors involved in the pathogenesis of the infection caused by the swine pathogen and zoonotic agent Streptococcus suis. Future Microbiol. 2012, 7, 259–279. [Google Scholar] [CrossRef]
- Kerdsin, A. Human Streptococcus suis Infections in Thailand: Epidemiology, Clinical Features, Genotypes, and Susceptibility. Trop. Med. Infect. Dis. 2022, 7, 359. [Google Scholar] [CrossRef]
- Mori, Y.; Kanda, H.; Notomi, T. Loop-mediated isothermal amplification (LAMP): Recent progress in research and development. J. Infect. Chemother. 2013, 19, 404–411. [Google Scholar] [CrossRef]
- Wilson, I.G. Inhibition and facilitation of nucleic acid amplification. Appl. Environ. Microbiol. 1997, 63, 3741–3751. [Google Scholar] [CrossRef] [PubMed]
- Papatheodorou, S.A.; Halvatsiotis, P.; Houhoula, D. A comparison of different DNA extraction methods and molecular techniques for the detection and identification of foodborne pathogens. AIMS Microbiol. 2021, 7, 304–319. [Google Scholar] [CrossRef] [PubMed]
- Amagliani, G.; Giammarini, C.; Omiccioli, E.; Brandi, G.; Magnani, M. Detection of Listeria monocytogenes using a commercial PCR kit and different DNA extraction methods. Food Control 2007, 18, 1137–1142. [Google Scholar] [CrossRef]
- Kemp, B.M.; Winters, M.; Monroe, C.; Barta, J.L. How much DNA is lost? Measuring DNA loss of short-tandem-repeat length fragments targeted by the PowerPlex 16® system using the Qiagen MinElute purification kit. Hum. Biol. 2014, 86, 313–329. [Google Scholar] [CrossRef] [PubMed]
- Dennett, C.; Klapper, P.E.; Cleator, G.M.; Lewis, A.G. CSF pretreatment and the diagnosis of herpes encephalitis using the polymerase chain reaction. J. Virol. Methods 1991, 34, 101–104. [Google Scholar] [CrossRef]
- Alfonso, Y.; Fraga, J.; Cox, R.; Bandera, F.; Pomier, O.; Fonseca, C.; Ginorio, D.; Torres, G.; Capo, V. Comparison of four DNA extraction methods from cerebrospinal fluid for the detection of Toxoplasma gondii by polymerase chain reaction in AIDS patients. Med. Sci. Monit. 2008, 14, Mt1–Mt6. [Google Scholar] [PubMed]
- QIAGEN. DNeasy® Blood & Tissue Handbook; Qiagen GmbH: Hilden, Germany, 2020; p. 8. Available online: https://www.qiagen.com/gb/resources/resourcedetail?id=68f29296-5a9f-40fa-8b3d-1c148d0b3030&lang=en (accessed on 13 May 2021).
- Funabashi, K.S.; Barcelos, D.; Visoná, I.; Silva, M.S.; Sousa, M.L.; de Franco, M.F.; Iwamura, E.S. DNA extraction and molecular analysis of non-tumoral liver, spleen, and brain from autopsy samples: The effect of formalin fixation and paraffin embedding. Pathol. Res. Pract. 2012, 208, 584–591. [Google Scholar] [CrossRef]
- Kuipers, J.G.; Nietfeld, L.; Dreses-Werringloer, U.; Koehler, L.; Wollenhaupt, J.; Zeidler, H.; Hammer, M. Optimised sample preparation of synovial fluid for detection of Chlamydia trachomatis DNA by polymerase chain reaction. Ann. Rheum. Dis. 1999, 58, 103–108. [Google Scholar] [CrossRef]
- Schneeweiss, W.; Stanek, C.; Wagner, M.; Hein, I. Inhibitor-free DNA for real-time PCR analysis of synovial fluid from horses, cattle and pigs. Vet. Microbiol. 2007, 121, 189–193. [Google Scholar] [CrossRef]
- Giuliano, C.; Patel, C.R.; Kale-Pradhan, P.B. A Guide to Bacterial Culture Identification And Results Interpretation. Pharm. Ther. 2019, 44, 192–200. [Google Scholar]
- Nga, T.V.; Nghia, H.D.; Tu, T.P.; Diep, T.S.; Mai, N.T.; Chau, T.T.; Sinh, D.X.; Phu, N.H.; Nga, T.T.; Chau, N.V.; et al. Real-time PCR for detection of Streptococcus suis serotype 2 in cerebrospinal fluid of human patients with meningitis. Diagn. Microbiol. Infect. Dis. 2011, 70, 461–467. [Google Scholar] [CrossRef] [PubMed]
- Mai, N.T.; Hoa, N.T.; Nga, T.V.; Linh, D.; Chau, T.T.; Sinh, D.X.; Phu, N.H.; Chuong, L.V.; Diep, T.S.; Campbell, J.; et al. Streptococcus suis meningitis in adults in Vietnam. Clin. Infect. Dis. 2008, 46, 659–667. [Google Scholar] [CrossRef] [PubMed]
- Ma, F.; Yi, L.; Yu, N.; Wang, G.; Ma, Z.; Lin, H.; Fan, H. Streptococcus suis Serotype 2 Biofilms Inhibit the Formation of Neutrophil Extracellular Traps. Front. Cell Infect. Microbiol. 2017, 7, 86. [Google Scholar] [CrossRef] [PubMed]
- Dutkiewicz, J.; Zając, V.; Sroka, J.; Wasiński, B.; Cisak, E.; Sawczyn, A.; Kloc, A.; Wójcik-Fatla, A. Streptococcus suis: A re-emerging pathogen associated with occupational exposure to pigs or pork products. Part II—Pathogenesis. Ann. Agric. Environ. Med. 2018, 25, 186–203. [Google Scholar] [CrossRef]
- Abdullahi, U.F.; Igwenagu, E.; Mu’azu, A.; Aliyu, S.; Umar, M.I. Intrigues of biofilm: A perspective in veterinary medicine. Vet. World 2016, 9, 12–18. [Google Scholar] [CrossRef]
- Fux, C.A.; Stoodley, P.; Hall-Stoodley, L.; Costerton, J.W. Bacterial biofilms: A diagnostic and therapeutic challenge. Expert Rev. Anti-Infect. Ther. 2003, 1, 667–683. [Google Scholar] [CrossRef]
- Morata, P.; Queipo-Ortuño, M.I.; de Dios Colmenero, J. Strategy for optimizing DNA amplification in a peripheral blood PCR assay used for diagnosis of human brucellosis. J. Clin. Microbiol. 1998, 36, 2443–2446. [Google Scholar] [CrossRef]
- Cogswell, F.B.; Bantar, C.E.; Hughes, T.G.; Gu, Y.; Philipp, M.T. Host DNA can interfere with detection of Borrelia burgdorferi in skin biopsy specimens by PCR. J. Clin. Microbiol. 1996, 34, 980–982. [Google Scholar] [CrossRef]
- Radomski, N.; Kreitmann, L.; McIntosh, F.; Behr, M.A. The critical role of DNA extraction for detection of mycobacteria in tissues. PLoS ONE 2013, 8, e78749. [Google Scholar] [CrossRef]
- Hammerschmitt, M.E.; Schwertz, C.I.; Lopes, B.C.; Pereira, P.R.; Frandoloso, R.; Driemeier, D. Clinical and pathological aspects of an outbreak of Streptococcus suis serotype 9 infection in pigs. Pesqui. Veterinária Bras. 2022, 42. [Google Scholar] [CrossRef]
- Vasconcelos, D.; Middleton, D.M.; Chirino-Trejo, J.M. Lesions caused by natural infection with Streptococcus suis type 9 in weaned pigs. J. Vet. Diagn. Investig. 1994, 6, 335–341. [Google Scholar] [CrossRef] [PubMed]
- Munthe, E.; Natvig, J.B. Immunglobulin classes, subclasses and complexes of IgG rheumatoid factor in rheumatoid plasma cells. Clin. Exp. Immunol. 1972, 12, 55–70. [Google Scholar]
- Timoney, J.F.; Yarkoni, U. Immunoglobulins IgG and IgM in synovial fluids of swine with Erysipelothrix polyarthritis. Vet. Microbiol. 1976, 1, 467–474. [Google Scholar] [CrossRef]
- Al-Soud, W.A.; Jönsson, L.J.; Râdström, P. Identification and characterization of immunoglobulin G in blood as a major inhibitor of diagnostic PCR. J. Clin. Microbiol. 2000, 38, 345–350. [Google Scholar] [CrossRef] [PubMed]
- Queipo-Ortuño, M.I.; De Dios Colmenero, J.; Macias, M.; Bravo, M.J.; Morata, P. Preparation of bacterial DNA template by boiling and effect of immunoglobulin G as an inhibitor in real-time PCR for serum samples from patients with brucellosis. Clin. Vaccine Immunol. 2008, 15, 293–296. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).