Occurrence of Neonatal Necrotizing Enterocolitis in Premature Neonates and Gut Microbiota: A Case–Control Prospective Multicenter Study
Abstract
:1. Introduction
2. Materials and Methods
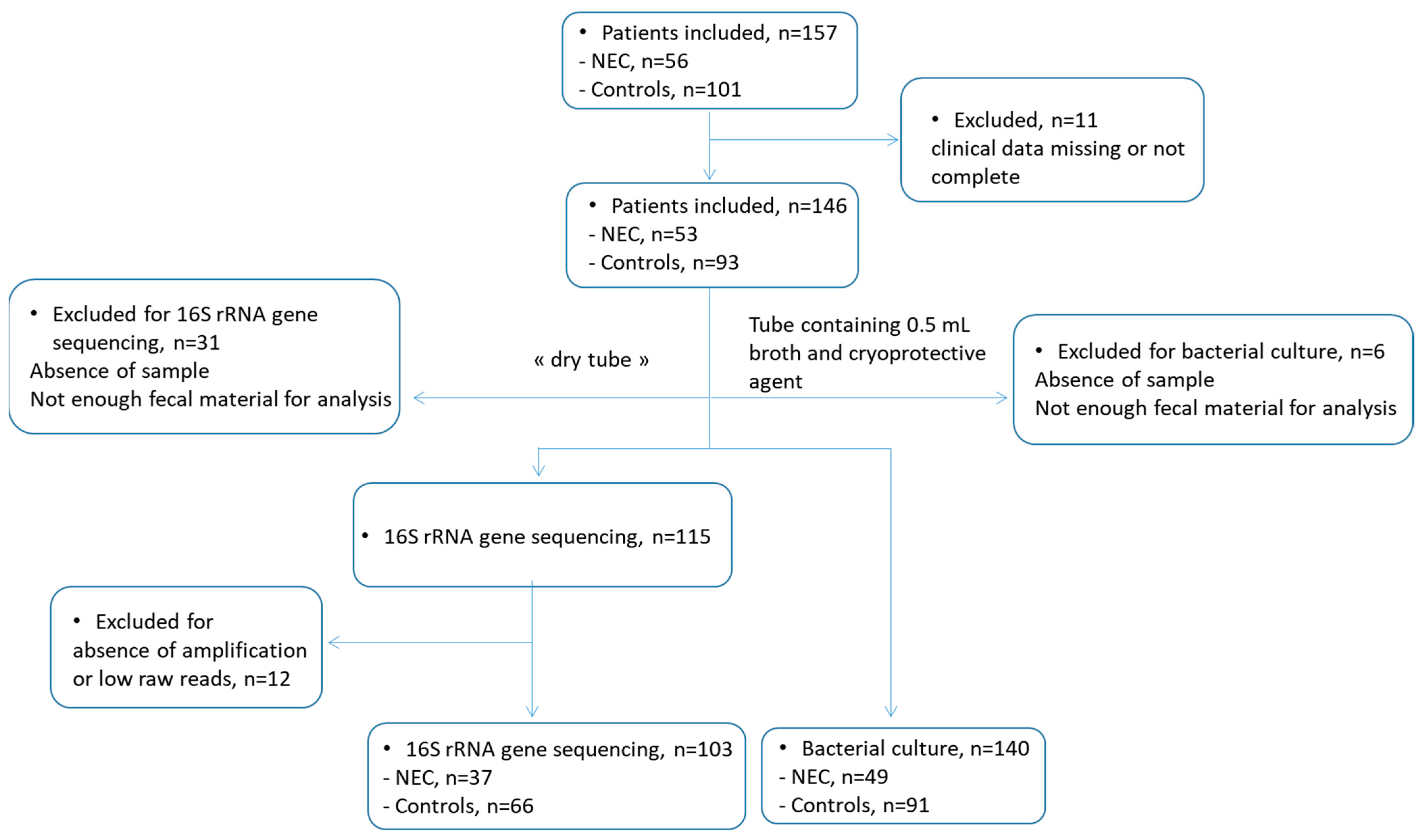
2.1. Study Design and Patients
2.2. Fecal Samples
2.3. DNA Extraction, Sequencing, and Microbiota Analysis
2.4. E. coli, C. butyricum, and C. neonatale Strain Isolation and Identification
2.5. Phylogenetic and Virulence Characterization of E. coli Strains
2.6. Antimicrobial Susceptibility
2.7. Statistical Analysis
3. Results
3.1. Population Characteristics
3.2. Gut Microbiota Analysis
3.3. Gut Microbiota in Preterm Neonates with or without NEC
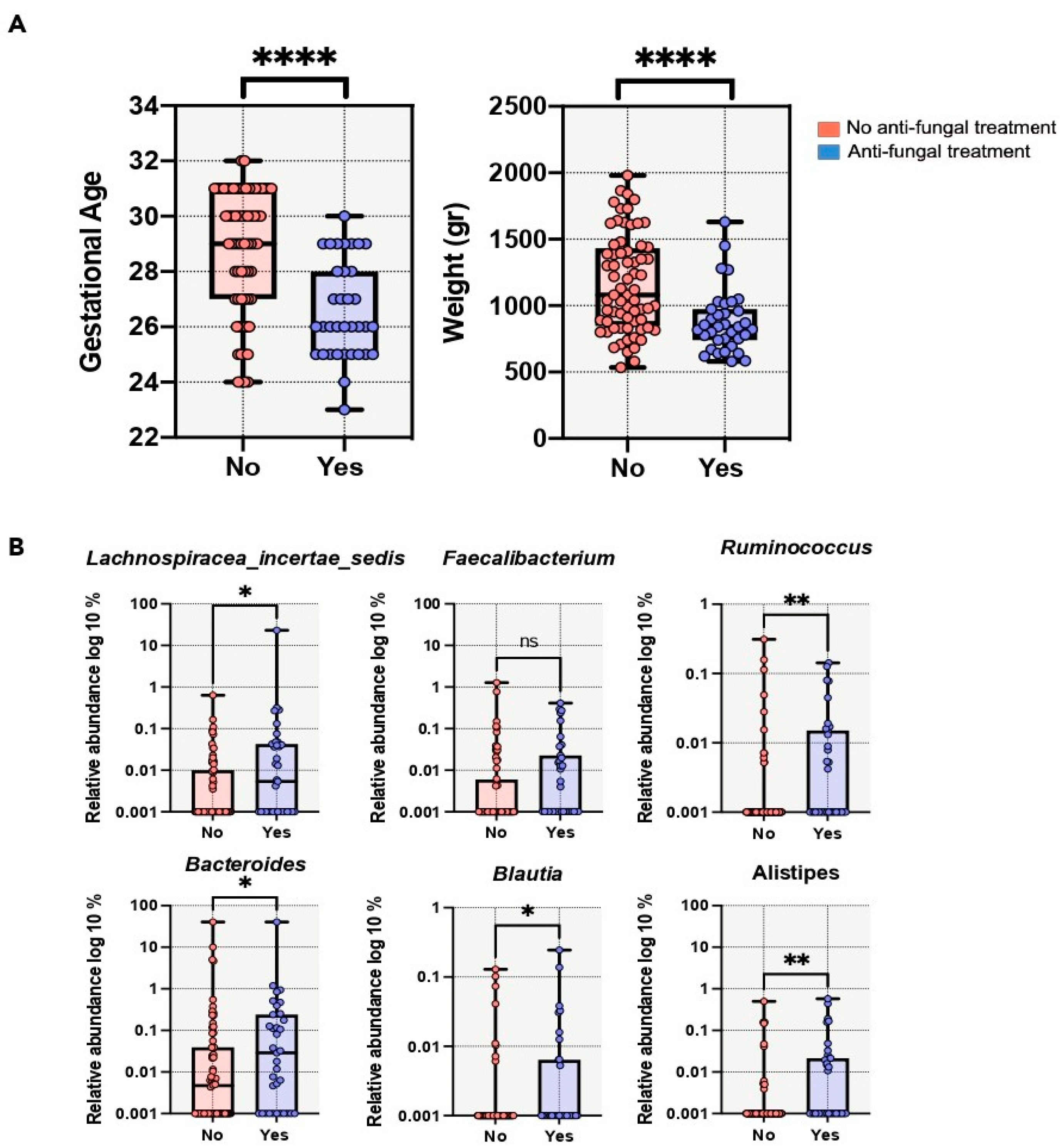
3.4. Perinatal Factors and Gut Microbiota
3.5. E. coli Isolation, Characterization, and Antimicrobial Susceptibility Levels
3.6. C. butyricum and C. neonatale Isolation and Antimicrobial Susceptibility Levels
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Heron, M. Deaths: Leading Causes for 2016. Natl. Vital Stat. Rep. 2018, 67, 1–76. [Google Scholar] [PubMed]
- Battersby, C.; Santhalingam, T.; Costeloe, K.; Modi, N. Incidence of Neonatal Necrotising Enterocolitis in High-Income Countries: A Systematic Review. Arch. Dis. Child Fetal Neonatal Ed. 2018, 103, F182–F189. [Google Scholar] [CrossRef] [PubMed]
- Bazacliu, C.; Neu, J. Pathophysiology of Necrotizing Enterocolitis: An Update. Curr. Pediatr. Rev. 2019, 15, 68–87. [Google Scholar] [CrossRef] [PubMed]
- Neu, J. Necrotizing Enterocolitis: The Future. Neonatology 2020, 117, 240–244. [Google Scholar] [CrossRef] [PubMed]
- Neu, J.; Walker, W.A. Necrotizing Enterocolitis. N. Engl. J. Med. 2011, 364, 255–264. [Google Scholar] [CrossRef]
- Morowitz, M.J.; Poroyko, V.; Caplan, M.; Alverdy, J.; Liu, D.C. Redefining the Role of Intestinal Microbes in the Pathogenesis of Necrotizing Enterocolitis. Pediatrics 2010, 125, 777–785. [Google Scholar] [CrossRef]
- Wang, Y.; Hoenig, J.D.; Malin, K.J.; Qamar, S.; Petrof, E.O.; Sun, J.; Antonopoulos, D.A.; Chang, E.B.; Claud, E.C. 16S RRNA Gene-Based Analysis of Fecal Microbiota from Preterm Infants with and without Necrotizing Enterocolitis. ISME J. 2009, 3, 944–954. [Google Scholar] [CrossRef]
- Mai, V.; Young, C.M.; Ukhanova, M.; Wang, X.; Sun, Y.; Casella, G.; Theriaque, D.; Li, N.; Sharma, R.; Hudak, M.; et al. Fecal Microbiota in Premature Infants Prior to Necrotizing Enterocolitis. PLoS ONE 2011, 6, e20647. [Google Scholar] [CrossRef]
- Claud, E.C.; Keegan, K.P.; Brulc, J.M.; Lu, L.; Bartels, D.; Glass, E.; Chang, E.B.; Meyer, F.; Antonopoulos, D.A. Bacterial Community Structure and Functional Contributions to Emergence of Health or Necrotizing Enterocolitis in Preterm Infants. Microbiome 2013, 1, 20. [Google Scholar] [CrossRef]
- Morrow, A.L.; Lagomarcino, A.J.; Schibler, K.R.; Taft, D.H.; Yu, Z.; Wang, B.; Altaye, M.; Wagner, M.; Gevers, D.; Ward, D.V.; et al. Early Microbial and Metabolomic Signatures Predict Later Onset of Necrotizing Enterocolitis in Preterm Infants. Microbiome 2013, 1, 13. [Google Scholar] [CrossRef]
- Ward, D.V.; Scholz, M.; Zolfo, M.; Taft, D.H.; Schibler, K.R.; Tett, A.; Segata, N.; Morrow, A.L. Metagenomic Sequencing with Strain-Level Resolution Implicates Uropathogenic E. coli in Necrotizing Enterocolitis and Mortality in Preterm Infants. Cell Rep. 2016, 14, 2912–2924. [Google Scholar] [CrossRef] [PubMed]
- Normann, E.; Fahlén, A.; Engstrand, L.; Lilja, H.E. Intestinal Microbial Profiles in Extremely Preterm Infants with and without Necrotizing Enterocolitis. Acta Paediatr. 2013, 102, 129–136. [Google Scholar] [CrossRef] [PubMed]
- Raveh-Sadka, T.; Thomas, B.C.; Singh, A.; Firek, B.; Brooks, B.; Castelle, C.J.; Sharon, I.; Baker, R.; Good, M.; Morowitz, M.J.; et al. Gut Bacteria Are Rarely Shared by Co-Hospitalized Premature Infants, Regardless of Necrotizing Enterocolitis Development. Elife 2015, 4, e05477. [Google Scholar] [CrossRef] [PubMed]
- Cassir, N.; Benamar, S.; Khalil, J.B.; Croce, O.; Saint-Faust, M.; Jacquot, A.; Million, M.; Azza, S.; Armstrong, N.; Henry, M.; et al. Clostridium butyricum Strains and Dysbiosis Linked to Necrotizing Enterocolitis in Preterm Neonates. Clin. Infect. Dis. 2015, 61, 1107–1115. [Google Scholar] [CrossRef]
- Roze, J.C.; Ancel, P.Y.; Lepage, P.; Martin-Marchand, L.; Al, N.Z.; Delannoy, J.; Picaud, J.C.; Lapillonne, A.; Aires, J.; Durox, M.; et al. Nutritional Strategies and Gut Microbiota Composition as Risk Factors for Necrotizing Enterocolitis in Very-Preterm Infants. Am. J. Clin. Nutr. 2017, 106, 821–830. [Google Scholar] [CrossRef]
- Sim, K.; Shaw, A.G.; Randell, P.; Cox, M.J.; McClure, Z.E.; Li, M.S.; Haddad, M.; Langford, P.R.; Cookson, W.O.; Moffatt, M.F.; et al. Dysbiosis Anticipating Necrotizing Enterocolitis in Very Premature Infants. Clin. Infect. Dis. 2015, 60, 389–397. [Google Scholar] [CrossRef]
- Zhou, Y.; Shan, G.; Sodergren, E.; Weinstock, G.; Walker, W.A.; Gregory, K.E. Longitudinal Analysis of the Premature Infant Intestinal Microbiome Prior to Necrotizing Enterocolitis: A Case-Control Study. PLoS ONE 2015, 10, e0118632. [Google Scholar] [CrossRef]
- Coggins, S.A.; Wynn, J.L.; Weitkamp, J.H. Infectious Causes of Necrotizing Enterocolitis. Clin. Perinatol. 2015, 42, 133–154, ix. [Google Scholar] [CrossRef]
- Pammi, M.; Cope, J.; Tarr, P.I.; Warner, B.B.; Morrow, A.L.; Mai, V.; Gregory, K.E.; Kroll, J.S.; McMurtry, V.; Ferris, M.J.; et al. Intestinal Dysbiosis in Preterm Infants Preceding Necrotizing Enterocolitis: A Systematic Review and Meta-Analysis. Microbiome 2017, 5, 31. [Google Scholar] [CrossRef]
- Tarracchini, C.; Milani, C.; Longhi, G.; Fontana, F.; Mancabelli, L.; Pintus, R.; Lugli, G.A.; Alessandri, G.; Anzalone, R.; Viappiani, A.; et al. Unraveling the Microbiome of Necrotizing Enterocolitis: Insights in Novel Microbial and Metabolomic Biomarkers. Microbiol. Spectr. 2021, 9, e0117621. [Google Scholar] [CrossRef]
- Bernard, K.; Burdz, T.; Wiebe, D.; Alfa, M.; Bernier, A.-M. Clostridium neonatale Sp. Nov. Linked to Necrotizing Enterocolitis in Neonates and a Clarification of Species Assignable to the Genus Clostridium (Prazmowski 1880) Emend. Lawson and Rainey 2016. Int. J. Syst. Evol. Microbiol. 2018, 68, 2416–2423. [Google Scholar] [CrossRef] [PubMed]
- Cassir, N.; Grandvuillemin, I.; Boxberger, M.; Jardot, P.; Boubred, F.; La Scola, B. Case Report: Clostridium neonatale Bacteremia in a Preterm Neonate With Necrotizing Enterocolitis. Front. Pediatr. 2021, 9, 771467. [Google Scholar] [CrossRef] [PubMed]
- Hosny, M.; Bou Khalil, J.Y.; Caputo, A.; Abdallah, R.A.; Levasseur, A.; Colson, P.; Cassir, N.; La Scola, B. Multidisciplinary Evaluation of Clostridium butyricum Clonality Isolated from Preterm Neonates with Necrotizing Enterocolitis in South France between 2009 and 2017. Sci. Rep. 2019, 9, 2077. [Google Scholar] [CrossRef] [PubMed]
- Olm, M.R.; Bhattacharya, N.; Crits-Christoph, A.; Firek, B.A.; Baker, R.; Song, Y.S.; Morowitz, M.J.; Banfield, J.F. Necrotizing Enterocolitis Is Preceded by Increased Gut Bacterial Replication, Klebsiella, and Fimbriae-Encoding Bacteria. Sci. Adv. 2019, 5, eaax5727. [Google Scholar] [CrossRef]
- Lepage, P.; Seksik, P.; Sutren, M.; de la Cochetiere, M.F.; Jian, R.; Marteau, P.; Dore, J. Biodiversity of the Mucosa-Associated Microbiota Is Stable along the Distal Digestive Tract in Healthy Individuals and Patients with IBD. Inflamm. Bowel. Dis. 2005, 11, 473–480. [Google Scholar] [CrossRef]
- Furet, J.P.; Firmesse, O.; Gourmelon, M.; Bridonneau, C.; Tap, J.; Mondot, S.; Dore, J.; Corthier, G. Comparative Assessment of Human and Farm Animal Faecal Microbiota Using Real-Time Quantitative PCR. FEMS Microbiol. Ecol. 2009, 68, 351–362. [Google Scholar] [CrossRef]
- Wilson, K.H.; Blitchington, R.B.; Greene, R.C. Amplification of Bacterial 16S Ribosomal DNA with Polymerase Chain Reaction. J. Clin. Microbiol. 1990, 28, 1942–1946. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Pena, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME Allows Analysis of High-Throughput Community Sequencing Data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Cole, J.R.; Wang, Q.; Cardenas, E.; Fish, J.; Chai, B.; Farris, R.J.; Kulam-Syed-Mohideen, A.S.; McGarrell, D.M.; Marsh, T.; Garrity, G.M.; et al. The Ribosomal Database Project: Improved Alignments and New Tools for RRNA Analysis. Nucleic Acids Res. 2009, 37, D141–D145. [Google Scholar] [CrossRef]
- Ferraris, L.; Butel, M.J.; Campeotto, F.; Vodovar, M.; Roze, J.C.; Aires, J. Clostridia in Premature Neonates’ Gut: Incidence, Antibiotic Susceptibility, and Perinatal Determinants Influencing Colonization. PLoS ONE 2012, 7, e30594. [Google Scholar] [CrossRef]
- Bouvet, P.; Ferraris, L.; Dauphin, B.; Popoff, M.R.; Butel, M.J.; Aires, J. 16S RRNA Gene Sequencing, Multilocus Sequence Analysis, and Mass Spectrometry Identification of the Proposed New Species “Clostridium neonatale”. J. Clin. Microbiol. 2014, 52, 4129–4136. [Google Scholar] [CrossRef] [PubMed]
- Clermont, O.; Christenson, J.K.; Denamur, E.; Gordon, D.M. The Clermont Escherichia coli Phylo-Typing Method Revisited: Improvement of Specificity and Detection of New Phylo-Groups. Environ. Microbiol. Rep. 2013, 5, 58–65. [Google Scholar] [CrossRef] [PubMed]
- Chapman, T.A.; Wu, X.Y.; Barchia, I.; Bettelheim, K.A.; Driesen, S.; Trott, D.; Wilson, M.; Chin, J.J. Comparison of Virulence Gene Profiles of Escherichia coli Strains Isolated from Healthy and Diarrheic Swine. Appl. Environ. Microbiol. 2006, 72, 4782–4795. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.R.; O’Bryan, T.T. Detection of the Escherichia coli Group 2 Polysaccharide Capsule Synthesis Gene kpsM by a Rapid and Specific PCR-Based Assay. J. Clin. Microbiol. 2004, 42, 1773–1776. [Google Scholar] [CrossRef] [PubMed]
- The European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters, Version 13.0. 2023. Available online: http://www.eucast.org (accessed on 20 April 2023).
- Benjamini, Y.; Drai, D.; Elmer, G.; Kafkafi, N.; Golani, I. Controlling the False Discovery Rate in Behavior Genetics Research. Behav. Brain Res. 2001, 125, 279–284. [Google Scholar] [CrossRef]
- Arumugam, M.; Raes, J.; Pelletier, E.; Le, P.D.; Yamada, T.; Mende, D.R.; Fernandes, G.R.; Tap, J.; Bruls, T.; Batto, J.M.; et al. Enterotypes of the Human Gut Microbiome. Nature 2011, 473, 174–180. [Google Scholar] [CrossRef]
- Cuna, A.; Morowitz, M.J.; Ahmed, I.; Umar, S.; Sampath, V. Dynamics of the Preterm Gut Microbiome in Health and Disease. Am. J. Physiol. Gastrointest. Liver Physiol. 2021, 320, G411–G419. [Google Scholar] [CrossRef]
- Lee, J.K.-F.; Hern Tan, L.T.; Ramadas, A.; Ab Mutalib, N.-S.; Lee, L.-H. Exploring the Role of Gut Bacteria in Health and Disease in Preterm Neonates. Int. J. Environ. Res. Public Health 2020, 17, 6963. [Google Scholar] [CrossRef]
- Fouhy, F.; Guinane, C.M.; Hussey, S.; Wall, R.; Ryan, C.A.; Dempsey, E.M.; Murphy, B.; Ross, R.P.; Fitzgerald, G.F.; Stanton, C.; et al. High-Throughput Sequencing Reveals the Incomplete, Short-Term Recovery of Infant Gut Microbiota Following Parenteral Antibiotic Treatment with Ampicillin and Gentamicin. Antimicrob. Agents Chemother. 2012, 56, 5811–5820. [Google Scholar] [CrossRef]
- Korpela, K.; Salonen, A.; Virta, L.J.; Kekkonen, R.A.; De Vos, W.M. Association of Early-Life Antibiotic Use and Protective Effects of Breastfeeding: Role of the Intestinal Microbiota. JAMA Pediatr. 2016, 170, 750–757. [Google Scholar] [CrossRef]
- Esaiassen, E.; Hjerde, E.; Cavanagh, J.P.; Pedersen, T.; Andresen, J.H.; Rettedal, S.I.; Støen, R.; Nakstad, B.; Willassen, N.P.; Klingenberg, C. Effects of Probiotic Supplementation on the Gut Microbiota and Antibiotic Resistome Development in Preterm Infants. Front. Pediatr. 2018, 6, 347. [Google Scholar] [CrossRef] [PubMed]
- Mshvildadze, M.; Neu, J.; Shuster, J.; Theriaque, D.; Li, N.; Mai, V. Intestinal Microbial Ecology in Premature Infants Assessed with Non-Culture-Based Techniques. J. Pediatr. 2010, 156, 20–25. [Google Scholar] [CrossRef] [PubMed]
- Boccia, D.; Stolfi, I.; Lana, S.; Moro, M.L. Nosocomial Necrotising Enterocolitis Outbreaks: Epidemiology and Control Measures. Eur. J. Pediatr. 2001, 160, 385–391. [Google Scholar] [CrossRef]
- Hoy, C.M.; Wood, C.M.; Hawkey, P.M.; Puntis, J.W.L. Duodenal Microflora in Very-Low-Birth-Weight Neonates and Relation to Necrotizing Enterocolitis. J. Clin. Microbiol. 2000, 68, 4539–4547. [Google Scholar] [CrossRef] [PubMed]
- Dierikx, T.H.; Deianova, N.; Groen, J.; Vijlbrief, D.C.; Hulzebos, C.; de Boode, W.P.; d’Haens, E.J.; Cossey, V.; Kramer, B.W.; van Weissenbruch, M.M.; et al. Association between Duration of Early Empiric Antibiotics and Necrotizing Enterocolitis and Late-Onset Sepsis in Preterm Infants: A Multicenter Cohort Study. Eur. J. Pediatr. 2022, 181, 3715–3724. [Google Scholar] [CrossRef]
- Gibson, M.K.; Wang, B.; Ahmadi, S.; Burnham, C.-A.D.; Tarr, P.I.; Warner, B.B.; Dantas, G. Developmental Dynamics of the Preterm Infant Gut Microbiota and Antibiotic Resistome. Nat. Microbiol. 2016, 1, 16024. [Google Scholar] [CrossRef]
- Greenwood, C.; Morrow, A.L.; Lagomarcino, A.J.; Altaye, M.; Taft, D.H.; Yu, Z.; Newburg, D.S.; Ward, D.V.; Schibler, K.R. Early Empiric Antibiotic Use in Preterm Infants Is Associated with Lower Bacterial Diversity and Higher Relative Abundance of Enterobacter. J. Pediatr. 2014, 165, 23–29. [Google Scholar] [CrossRef]
- Li, Y.; Shen, R.L.; Ayede, A.I.; Berrington, J.; Bloomfield, F.H.; Busari, O.O.; Cormack, B.E.; Embleton, N.D.; van Goudoever, J.B.; Greisen, G.; et al. Early Use of Antibiotics Is Associated with a Lower Incidence of Necrotizing Enterocolitis in Preterm, Very Low Birth Weight Infants: The NEOMUNE-NeoNutriNet Cohort Study. J. Pediatr. 2020, 227, 128–134.e2. [Google Scholar] [CrossRef]
- Silverman, M.A.; Konnikova, L.; Gerber, J.S. Impact of Antibiotics on Necrotizing Enterocolitis and Antibiotic-Associated Diarrhea. Gastroenterol. Clin. N. Am. 2017, 46, 61–76. [Google Scholar] [CrossRef]
- Gill, E.M.; Jung, K.; Qvist, N.; Ellebaek, M.B. Antibiotics in the Medical and Surgical Treatment of Necrotizing Enterocolitis. A Systematic Review. BMC Pediatr. 2022, 22, 66. [Google Scholar] [CrossRef]
- Eberhart, M.; Grisold, A.; Lavorato, M.; Resch, E.; Trobisch, A.; Resch, B. Extended-Spectrum Beta-Lactamase (ESBL) Producing Enterobacterales in Stool Surveillance Cultures of Preterm Infants Are No Risk Factor for Necrotizing Enterocolitis: A Retrospective Case-Control Study over 12 Years. Infection 2020, 48, 853–860. [Google Scholar] [CrossRef] [PubMed]
- Mesa, V.; Monot, M.; Ferraris, L.; Popoff, M.; Mazuet, C.; Barbut, F.; Delannoy, J.; Dupuy, B.; Butel, M.J.; Aires, J. Core-, Pan- and Accessory Genome Analyses of Clostridium neonatale: Insights into Genetic Diversity. Microb. Genom. 2022, 8, 000813. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, M.L.; Limon, J.J.; Bar, A.S.; Leal, C.A.; Gargus, M.; Tang, J.; Brown, J.; Funari, V.A.; Wang, H.L.; Crother, T.R.; et al. Immunological Consequences of Intestinal Fungal Dysbiosis. Cell Host Microbe 2016, 19, 865–873. [Google Scholar] [CrossRef] [PubMed]


| Characteristics | NEC Cases (n = 53) | Controls (n = 93) | p Value |
|---|---|---|---|
| Gestational age (weeks); median (quartiles) | 28 ± 2; 29.0 (26–30) | 28 ± 2; 29.0 (26–30) | 0.85 |
| Birth weight (g); median (quartiles) | 1077 ± 359; 1000 (820–1280) | 1062 ± 303; 980 (840–1335) | 0.97 |
| Human milk—no. (%) | 39 (73) | 81 (87) | 0.54 |
| Vaginal delivery—no. (%) | 21 (40) | 38 (41) | 1 |
| Maternal antibiotic therapy—no. (%) | 24 (45) | 39 (41) | 0.60 |
| 19 (36) | 25 (27) | 0.26 |
| 19 (36) | 34 (36) | 1 |
| Neonatal antibiotic therapy—no. (%) | 48 (90) | 68 (73) | 0.01 |
| 29 (55) | 54 (58) | 0.70 |
| duration (days) [min–max] | 3 ± 1.5 [1–7] | 4 ± 2.2 [1–10] | 0.9 |
| 45 (83) | 48 (52) | <0.0001 |
| duration (days) [min–max] | 12 ± 6.7 [3–34] | 10 ± 8.0 [2–39] | 0.07 |
| Antifungal treatment—no. (%) | 15 (28) | 26 (28) | 1 |
| duration (days) [min–max] | 9 ± 9.6 [1–29] | 12 ± 10.0 [1–35] | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aires, J.; Ilhan, Z.E.; Nicolas, L.; Ferraris, L.; Delannoy, J.; Bredel, M.; Chauvire-Drouard, A.; Barbut, F.; Rozé, J.-C.; Lepage, P.; et al. Occurrence of Neonatal Necrotizing Enterocolitis in Premature Neonates and Gut Microbiota: A Case–Control Prospective Multicenter Study. Microorganisms 2023, 11, 2457. https://doi.org/10.3390/microorganisms11102457
Aires J, Ilhan ZE, Nicolas L, Ferraris L, Delannoy J, Bredel M, Chauvire-Drouard A, Barbut F, Rozé J-C, Lepage P, et al. Occurrence of Neonatal Necrotizing Enterocolitis in Premature Neonates and Gut Microbiota: A Case–Control Prospective Multicenter Study. Microorganisms. 2023; 11(10):2457. https://doi.org/10.3390/microorganisms11102457
Chicago/Turabian StyleAires, Julio, Zehra Esra Ilhan, Lancelot Nicolas, Laurent Ferraris, Johanne Delannoy, Maxime Bredel, Anne Chauvire-Drouard, Frédéric Barbut, Jean-Christophe Rozé, Patricia Lepage, and et al. 2023. "Occurrence of Neonatal Necrotizing Enterocolitis in Premature Neonates and Gut Microbiota: A Case–Control Prospective Multicenter Study" Microorganisms 11, no. 10: 2457. https://doi.org/10.3390/microorganisms11102457
APA StyleAires, J., Ilhan, Z. E., Nicolas, L., Ferraris, L., Delannoy, J., Bredel, M., Chauvire-Drouard, A., Barbut, F., Rozé, J.-C., Lepage, P., Butel, M.-J., & ClosNEC Study Group. (2023). Occurrence of Neonatal Necrotizing Enterocolitis in Premature Neonates and Gut Microbiota: A Case–Control Prospective Multicenter Study. Microorganisms, 11(10), 2457. https://doi.org/10.3390/microorganisms11102457





