Abstract
Emerging research underscores the substantial link between gut flora and various inflammatory skin diseases. We hypothesize that there exists a complex gut–skin axis, possibly affecting the progression of conditions such as eczema, acne, psoriasis, and rosacea. However, the precise nature of the causal connection between gut flora and skin diseases remains unestablished. In this study, we started by compiling summary data from genome-wide association studies (GWAS) featuring 211 unique gut microbiota and four types of skin conditions. We scrutinized these data across different taxonomic strata. Subsequently, we leveraged Mendelian randomization (MR) to ascertain if there is a causal link between gut microbiota and these skin conditions. We also performed a bidirectional MR analysis to identify the causality’s direction. By utilizing Mendelian randomization, we identified 26 causal connections between the gut microbiome and four recognized inflammatory skin conditions, including 9 positive and 17 negative causal directions. Additional sensitivity analyses of these results revealed no evidence of pleiotropy or heterogeneity. Our MR analysis suggests a causal connection between gut microbiota and skin diseases, potentially providing groundbreaking perspectives for future mechanistic and clinical studies on microbiota-affected skin conditions.
1. Introduction
Skin and subcutaneous disorders ranked as the 18th leading cause of disability-adjusted life years (DALYs) globally in the 2013 global burden of disease report, imposing an economic burden amounting to hundreds of millions of USD annually [1]. The human gut harbors a diverse array of microbial communities, with gut flora playing a pivotal role in human health. This includes modulating the immune system, impacting nutrient absorption, and influencing disease progression [2]. The gut–skin axis (GSA) is an emerging concept that delineates the interplay between the gut microbiota and the skin. The connection between gut flora and skin health, as well as disease, has gained considerable attention in recent research. The relationship between particular inflammatory skin conditions and the microbiome is believed to be influenced by an impaired intestinal barrier, enhanced inflammatory mediators, and byproducts discharged by microorganisms [3]. Short-chain fatty acids (SCFAs) are the main products of intestinal microbiota and act as a crucial energy provider for enterocytes. They play crucial roles in mucosal protection, immune regulation, and metabolism across various tissues [4]. SCFAs, encompassing propionate, acetate salts, and butyrate, are noted to restrain the growth, mobility, and adherence of inflammatory cells, thus manifesting anti-inflammatory impacts [5]. While the majority of related studies are still in their exploratory stages, there have been reports suggesting an association between intestinal flora and certain inflammatory skin diseases, such as eczema [6], acne [7], psoriasis [8], and rosacea [9]. Current research on gut flora and skin diseases may be constrained by confounding factors, including the environment, diet, and antibiotics [3]. In conclusion, the presence and orientation of a causal connection between intestinal microbiota and inflammatory skin conditions continue to be ambiguous. Consequently, it is essential to delve into the potential causal link between gut flora and inflammatory skin diseases.
Mendelian randomization is a robust method that integrates data from GWAS in a pooled manner. MR is a regularly used mechanism for extrapolating potential causal links between exposure elements and complex results. It capitalizes on genetic variants significantly correlated with exposure as instrumental variables to deduce causality [10]. Owing to the random allocation of genes at the moment of conception, they remain unaffected by confounding factors. This unique attribute of MR enables the reduction of confounding influences. If a genetic variant is linked with an exposure, and this exposure is subsequently linked with an outcome, it logically infers that the genetic variant should also maintain a connection with the outcome. In this study, we employed a two-sample MR to explore the potential causal link between the structure of the gut microbiome and the susceptibility to skin diseases as previously described. We focused on four skin conditions, namely, eczema, acne, psoriasis, and rosacea, using data from public GWAS repositories. By employing a bidirectional MR approach, we aimed not only to ascertain if the gut microbiota influences the risk of these dermatological diseases by chance but also to determine whether a genetic predisposition to these skin conditions has a causal effect on the gut microbiota. Our goal is to highlight the role of gut microbiota in the development of dermatoses. Such comprehension could eventually set the stage for the development of novel therapeutic strategies, encompassing probiotic therapy, dietary alterations, and fecal microbiota transplantation (FMT).
2. Method
2.1. Exposure Data
We picked single nucleotide polymorphisms (SNPs) linked with human gut microbial composition as instrumental variables (IVs) from the Genome-Wide Association Studies (GWAS) dataset of the international consortium MiBioGen [11]. This consortium carried out a large-scale, multiethnic GWAS encompassing 18,340 participants from 24 cohorts across the United States, Canada, and other countries. The study scrutinized microbial composition using gene sequencing profiles against 16S rRNA genes. It incorporated a total of 211 taxonomic units, consisting of 131 genera, 35 families, 20 orders, 16 classes, and 9 phyla.
2.2. Outcome Data
We procured dermatological data on eczema, acne, and psoriasis from FinnGen version 8 (https://r8.risteys.finngen.fi/, accessed on 15 May 2023) and selected rosacea data from the IEU Open GWAS project (https://gwas.mrcieu.ac.uk/, accessed on 15 May 2023) (Supplementary Table S1).
2.3. Selection of Instrumental Variables
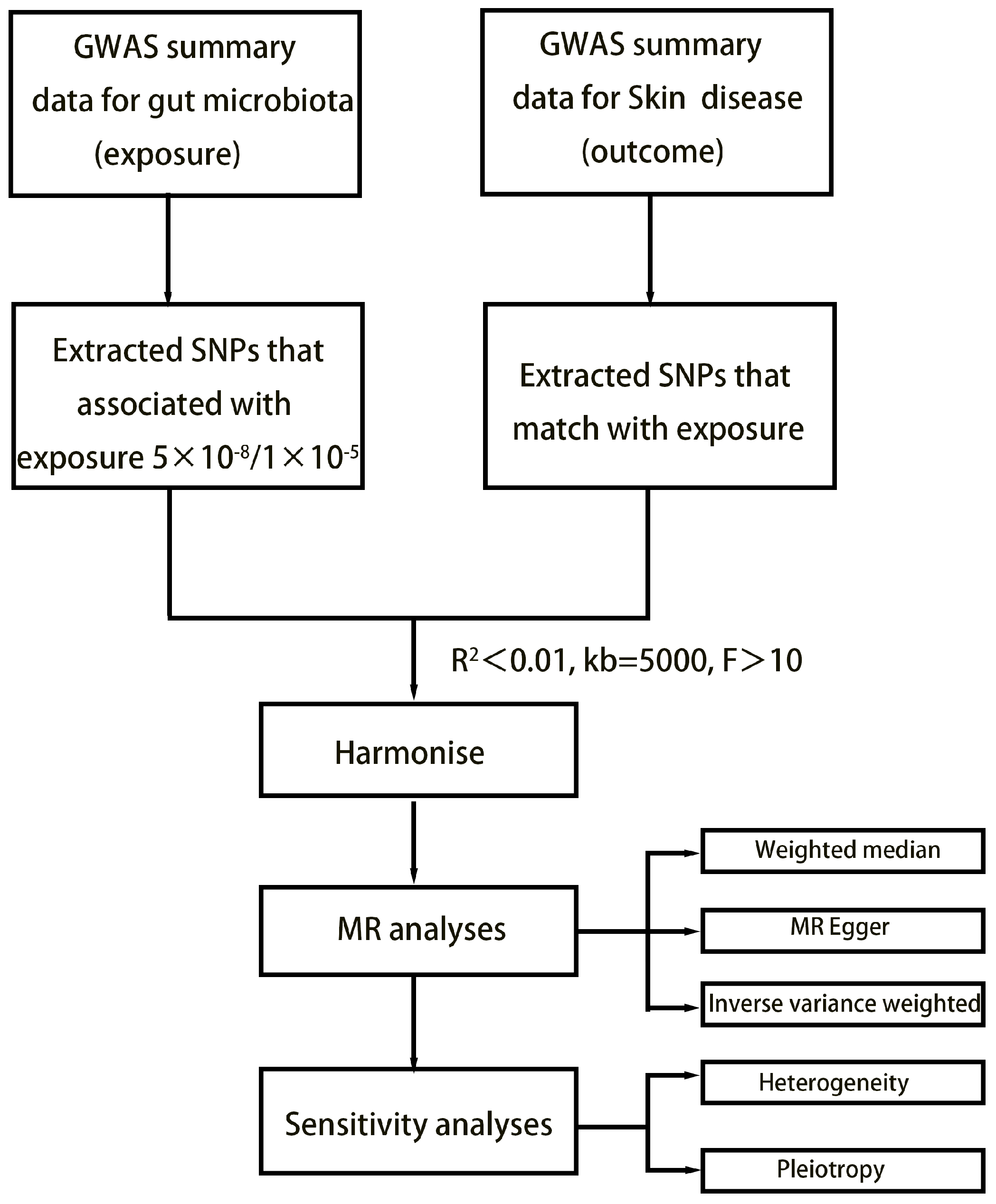
The study’s flowchart is illustrated in Figure 1. Concisely, the gut microbiota was employed as the exposure, while dermatological disease was used as the outcome. We selected one threshold for SNPs smaller than the genome-wide statistical significance threshold (5 × 10−8) as the IVs. Unfortunately, only a limited number of gut microbiota were selected as IVs following our SNP selection. To explore a wider range of relationships between dermatophytosis and gut microbiota for more comprehensive results, we established a second threshold to identify SNP significance levels smaller than the locus (1 × 10−5). We chose a secondary set of IVs to uncover additional potential causal relationships. We set the linkage disequilibrium correlation coefficients to R2 < 0.01 and a clumping distance > 5000 kb to ensure no linkage disequilibrium among the chosen IVs. To lessen the effect of weak instrument bias on causal inference, we applied the formula F = β2 exposure/SE2 exposure to evaluate the potency of the IVs and removed those with F < 10 [12]. Echo SNPs, which are those with identical base pairs on the forward and reverse strands (such as A/T and C/G), can be ambiguous when genotyping. Therefore, we eliminated these SNPs to reduce this potential source of error and thus enhance the accuracy of our analysis.
Figure 1.
Flowchart of MR analysis.
2.4. Statistical Analysis
Currently, several popular MR methods, including the inverse variance weighted (IVW) test [13], MR-PRESSO [14], MR-Egger regression [15], and weighted median estimator [16], are routinely utilized for MR analysis. According to related studies, the IVW method delivers more precise inferences of causality during the analysis. Hence, we chose the IVW method as the main approach for examining causality in the two-sample MR (TSMR) analysis [17]. For attributes that contained only one SNP, where the IVW test was not suitable, we employed the Wald ratio test to estimate causal impacts [18]. We utilized MR-PRESSO and MR-Egger regression tests to keep track of potential horizontal pleiotropy effects. A p (intercept) value < 0.05 would suggest the existence of horizontal pleiotropy. The heterogeneity among the chosen SNPs was measured using Cochran’s Q statistic, and a “leave-one-out” analysis was performed to exclude each instrumental SNP in turn to identify any potentially heterogeneous SNPs. We applied this method to ascertain the direction of causality in the MR analysis [19]. Subsequently, we executed a reverse MR analysis, using the same configurations and methods as those in the forward MR. To examine any potential reverse causation effects, we treated the four inflammatory skin diseases as exposure factors and gut flora as the outcome variable. All statistical analyses were conducted using the TwoSampleMR and MR-PRESSO packages in R software (version 4.2.2).
3. Results
3.1. SNP Selection
At the locus-wide significance level (p < 1 × 10−5), we identified a total of 2832 SNPs as IVs according to the selection criteria. Detailed information about the selected instrumental variables is presented in Supplementary Table S2. At the genome-wide statistical significance thresholds (p < 5 × 10−8), we identified a total of 30 SNPs as IVs (Supplementary Table S3).
3.2. Results of the TSMR Analysis (Genome-Wide Statistical Significance Threshold, p < 5 × 10−8)
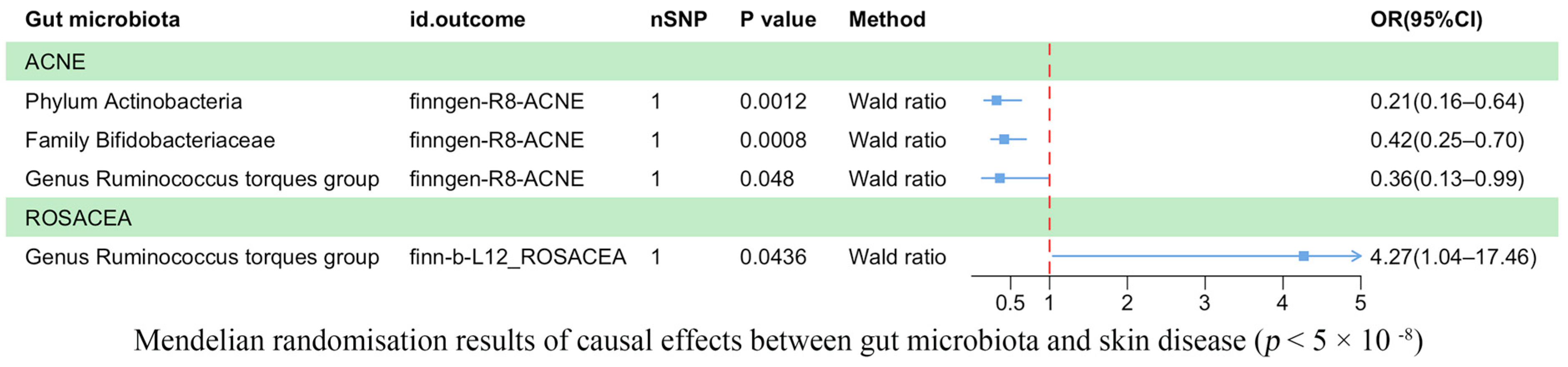
Our analysis revealed a causal correlation between acne and one phylum, one family, and one genus (Figure 2). Wald ratio estimates suggested that the phylum Actinobacteria (OR = 0.21, 95% CI: 0.16–0.64, p = 1.20 × 10−3), the family Bifidobacteriaceae (OR = 0.42, 95% CI: 0.25–0.70, p = 8.00 × 10−4), and the genus Ruminococcus torques group (OR = 0.36, 95% CI: 0.13–0.99, p = 4.80 × 10−2) may play a protective role against acne.
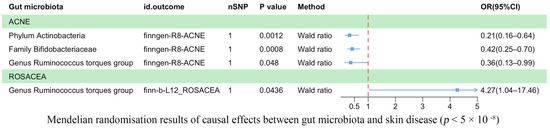
Figure 2.
Mendelian randomization results on the causal relationship between the gut microbiome and the risk of four inflammatory skin diseases (p < 5 × 10−8).
Rosacea was associated causally with one genus (Figure 2). Wald ratio estimates indicated that the genus Ruminococcus torques group was potentially linked with rosacea (OR = 4.27, 95% CI: 1.04–17.46, p = 4.36 × 10−2).
3.3. Results of the TSMR Analysis (Gene Locus Range Significance Levels, p < 1 × 10−5)
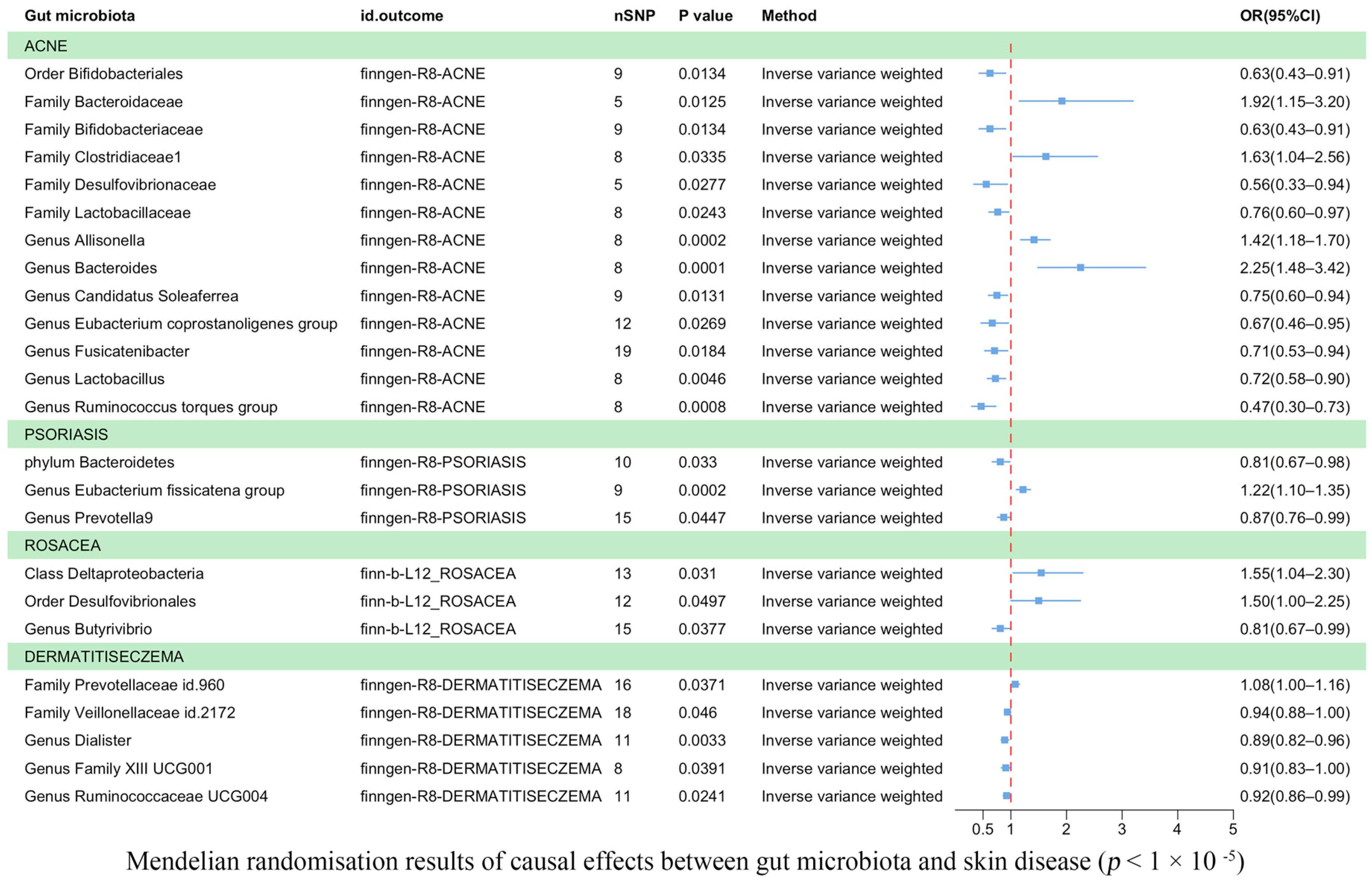
Eczema was found to be causally correlated with two bacterial families and three bacterial genera (Figure 3). According to IVW estimates, the family Veillonellaceae (OR = 0.94, 95% CI: 0.88–1.00, p = 4.60 × 10−2), the genus Dialister (OR = 0.89, 95% CI: 0.82–0.96, p = 3.30 × 10−3), the genus Family XIII UCG001 (OR = 0.91, 95% CI: 0.83–1.00, p = 3.91 × 10−2), and Ruminococcaceae UCG004 (OR = 0.92, 95% CI: 0.86–0.99, p = 2.41 × 10−2) were associated with a reduced risk of eczema. Conversely, the family Prevotellaceae (OR = 1.08, 95% CI: 1.00–1.16, p = 3.71 × 10−2) was potentially associated with an increased risk of eczema.
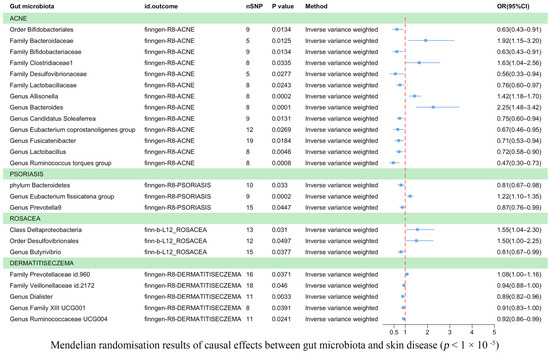
Figure 3.
Mendelian randomization results on the causal relationship between the gut microbiome and the risk of four inflammatory skin diseases (p < 1 × 10−5).
Acne was associated causally with one order, five families, and seven genera (Figure 3). IVW estimates showed that the order Bifidobacteriales (OR = 0.63, 95% CI: 0.43–0.91, p = 1.34 × 10−2), family Bifidobacteriaceae (OR = 0.63, 95% CI: 0.43–0.91, p = 1.34 × 10−2), family Desulfovibrionaceae (OR = 0.56, 95% CI: 0.33–0.94, p = 2.7 × 10−2), and family Lactobacillaceae (OR = 0.76, 95% CI: 0.60–0.97, p = 2.43 × 10−2) were potentially protective against acne. Similarly, the genera Candidatus Soleaferrea (OR = 0.75, 95% CI: 0.60–0.94, p = 1.31 × 10−2), Eubacterium coprostanoligenes (OR = 0.67, 95% CI: 0.46–0.95, p = 2.69 × 10−2), Fusicatenibacter (OR = 0.71, 95% CI: 0.53–0.94, p = 1.84 × 10−2), Lactobacillus (OR = 0.72, 95% CI: 0.58–0.90, p = 4.55 × 10−3), and Ruminococcus torques group (OR = 0.47, 95% CI: 0.30–0.73, p = 8.41 × 10−4) were also found to have a protective effect against acne. On the other hand, IVW estimates indicated that the family Bacteroidaceae (OR = 1.92, 95% CI: 1.15–3.20, p = 1.25 × 10−2), family Clostridiaceae1 (OR = 1.63, 95% CI: 1.04–2.56, p = 3.35 × 10−2), genus Allisonella (OR = 1.42, 95% CI: 1.18–1.70, p = 2.16 × 10−4), and genus Bacteroides (OR = 2.25, 95% CI: 1.48–3.42, p = 1.37 × 10−4) were potentially associated with an increased risk of acne.
Psoriasis was found to be causally correlated with one phylum and two genera (Figure 3). IVW estimates indicated that the phylum Bacteroidetes (OR = 0.81, 95% CI: 0.67–0.98, p = 3.30 × 10−2) and the genus Prevotella 9 (OR = 0.87, p = 4.47 × 10−2) were associated with a reduced risk of psoriasis. However, the genus Eubacterium fissicatena group (OR = 1.22, 95% CI: 1.10–1.35, p = 1.81 × 10−4) was potentially associated with an increased risk of psoriasis.
Rosacea was found to be correlated with one class, one order, and one genus (Figure 3). IVW estimates suggested that the class Deltaproteobacteria (OR = 1.55, 95% CI: 1.04–2.30, p = 3.10 × 10−2) and the order Desulfovibrionales (OR = 1.50, 95% CI: 1.00–2.25, p = 4.97 × 10−2) were potentially associated with an increased risk of rosacea. Conversely, the genus Butyrivibrio (OR = 0.81, 95% CI: 0.67–0.99, p = 3.77 × 10−2) was associated with a reduced risk of rosacea. In the Supplementary Material, we utilized scatter plots as a powerful data visualization tool to illustrate the relationship between gut flora and causal variables associated with skin diseases (Supplementary Figures S1–S4). Furthermore, all the significant results are outlined in Table 1, while a comprehensive list of results can be found in Supplementary Table S4.

Table 1.
Mendelian randomization (MR) results of causal effects between gut microbiome and four kinds of skin diseases.
3.4. Sensitivity Analyses
In our sensitivity assessments, no pleiotropy was detected in the causal estimates (Supplementary Table S5). Specifically, the MR-Egger intercept analysis did not uncover any signs of directional pleiotropy between inflammatory skin diseases and gut microbiota (Supplementary Table S6). Additionally, Cochran’s Q statistics revealed no significant heterogeneity (p > 0.05) (Supplementary Table S7). The leave-one-out assay did not show any outliers, indicating that the results were robust and not dependent on any individual gene variants (Supplementary Figures S5–S8). The MR Steiger directionality test also showed no abnormalities (Supplementary Table S8). In the Supplementary Materials, we include funnel plots to provide a visual representation of the instrument variable heterogeneity in our study. The distribution of points around the causal effect line provides insight into the validity of our instrumental variables. These plots aids in the identification and handling of potential heterogeneity, thereby enhancing the robustness of our Mendelian randomization analysis.
3.5. Reverse TSMR Analysis
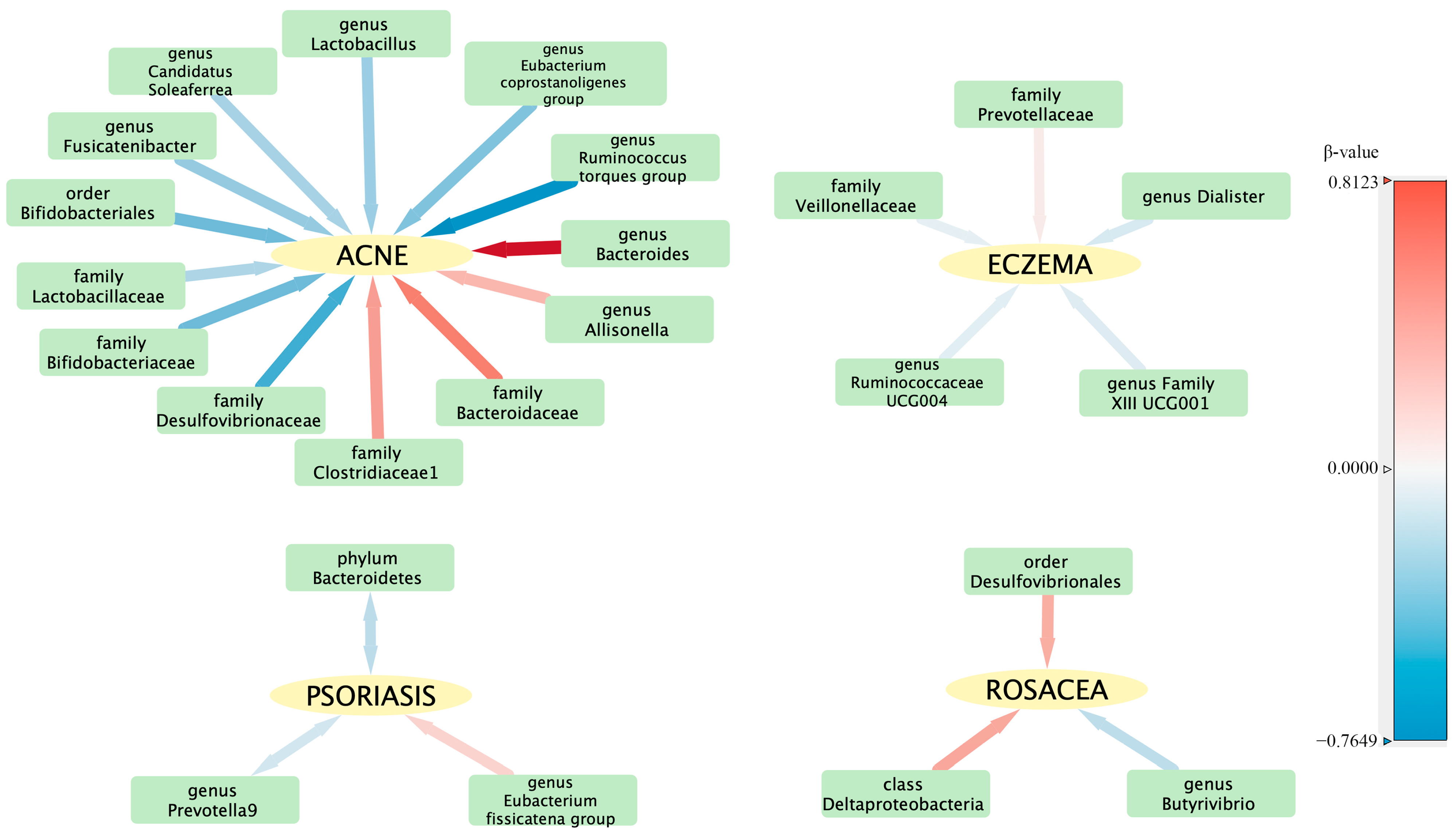
To evaluate any reverse causation effects, we used inflammatory skin diseases as the exposure and gut microbiota as the outcome. We discovered a bidirectional relationship between psoriasis and both the phylum Bacteroidetes and the genus Prevotella 9 (Table 2). The reverse causation sensitivity analysis did not reveal any abnormalities (Supplementary Table S9). In order to better understand the complex interactions between the intestinal flora and various skin diseases, a correlation network was constructed and is illustrated in Figure 4.

Table 2.
Bidirectional MR results of the causal effects between gut microbiome and skin disease.
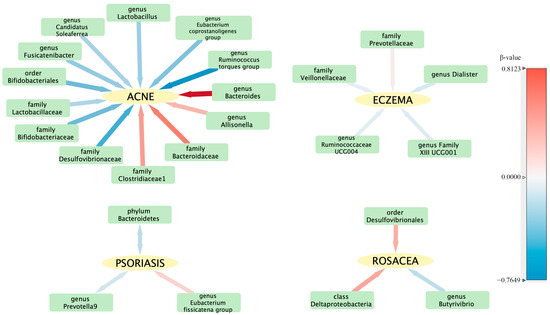
Figure 4.
Visualization of the correlation network between intestinal flora and skin diseases. The hue of each line signifies the magnitude of the β-value, while the thickness of each line represents its absolute value, illustrating the strength and direction of the relationship.
4. Discussion
This research signifies the first comprehensive effort to assess the causative association between gut microbiota and inflammatory skin conditions from a genetic perspective, using the largest genome-wide meta-analysis of gut microbes conducted by the MiBioGen consortium. The burgeoning body of research has unveiled evidence supporting a “skin–gut axis”, with our identification of 26 such causative relationships based on two-sample Mendelian randomization (MR) analysis.
Our two-sample MR (TSMR) research revealed that the genus Dialister serves as a protective factor for eczema, with the genus Dialister recognized as a propionic acid producer in the gut [20]. It has been found that individuals with eczema have significantly lower concentrations of SCFAs such as propionic acid than healthy individuals [21]. Propionic acid, an essential SCFA, inhibits inflammation induced by Th2 and Th17 bias in patients with eczema [22]. Thus, we propose that the protective influence of the Dialister genus group against eczema may be associated with SCFAs, particularly propionate. Ruminococcaceae are the primary SCFA producers in the gut microbiota [23]. Ruminococcaceae UCG004 demonstrated a protective effect against eczema. In one study, the creation of *AP-L01 microgels, which encapsulate Lactobacillus lactis LI01 in algal pectin (AP), alleviated acute liver injury in mice by boosting SCFA producers and reducing pathogenic microbes, thereby altering the gut microbiota. Ruminococcaceae UCG004 levels were elevated in the AP-LI01 administration group [24]. The family XIII UCG001 genus has been identified as a protective factor against eczema. Some research has found that Family XIII UCG001 plays a protective role in certain inflammatory conditions, such as scleritis and rheumatoid arthritis [25]. Further exploration is needed to understand the mechanism through which Family XIII UCG001 functions.
The order Bifidobacteriales serves as a protective factor against acne. Bifidobacterium is known to enhance the gut mucosal barrier and reduce the levels of lipopolysaccharide (LPS) in the intestine. There may be a certain link between LPS and acne. Studies have shown that LPS endotoxins are present in the blood of acne patients, and these patients exhibit high reactivity to LPS. A study involving 40 acne patients demonstrated the presence of LPS endotoxins in their blood and a high reactivity toward LPS [26]. The family Bifidobacteriaceae has been identified as a risk factor for acne. It has been associated with the serum concentrations of various inflammatory markers, including TNF-α and IL-6 [27]. The inflammatory factors IL-1β and TNF-α have a close relationship with the pathogenesis of acne [28]. The genus Bacteroides has been identified as a risk factor for acne and has been reported to be associated with various inflammatory conditions [29]. Toxins produced by Bacteroides have been shown to trigger the NF-κB and MAPK signaling pathways in activated B cells, leading to the secretion of interleukin-8 (IL-8) and TNF-α [30]. The genus Candidatus Soleaferrea is a protective factor against acne. Candidatus Soleaferrea is regarded as a beneficial microbiota, although research on it is currently limited. The genus Fusicatenibacter also serves as a protective factor against acne. Fusicatenibacter has been demonstrated to stimulate the production of the anti-inflammatory cytokine interleukin-10 (IL-10) in gut wall monocytes extracted from ulcerative colitis (UC) models in patients and mice. IL-10 can suppress inflammatory responses [31]. Upon stimulation by Propionibacterium acnes (P. acnes), the secretion of TNF-α and proinflammatory IL-8 is enhanced in peripheral blood mononuclear cells (PBMCs) of individuals with acne. In contrast, the secretion of anti-inflammatory IL-10 by the PBMCs of acne patients significantly decreases. Furthermore, the capability of CD14 cells in acne patients to engulf P. acnes bacteria is impaired, but the introduction of exogenous IL-10 to PBMC cultures can reinstate phagocytic activity [32]. Therefore, the genus Fusicatenibacter may play a protective role in acne through the secretion of IL-10. The genus Lactobacillus is a protective factor against acne. A randomized controlled study showed that supplementation with the probiotic Lactobacillus can decrease the gene expression of insulin-like growth factor 1 (IGF1) by 32% and increase the gene expression of forkhead box protein O1 (FOXO1) by 65%, improving the appearance of adult acne [33]. Other genera were not found to be mechanistically associated with the presence of acne, and later stages need to be further explored.
The phylum Bacteroidetes serves as a protective factor in psoriasis. A review of several studies revealed a decreased abundance of Bacteroidetes in psoriasis patients compared to healthy controls [34,35]. Interleukin 17 (IL-17), a proinflammatory cytokine, plays a pivotal role in the pathogenesis of psoriasis. A decrease in regulatory T cell (Treg) levels in psoriasis patients leads to an imbalance between effector T cells and suppressor T cells [36]. Studies on patients with obstructive sleep apnea (OSA) have shown reduced numbers of Bacteroidetes to be associated with Th17/Treg imbalance [37]. We propose that Bacteroidetes may have a similar role in psoriasis. The phylum Bacteroidetes is significant among the short-chain fatty acid-producing microflora in the gut, breaking down dietary fiber and other complex carbohydrates to produce short-chain fatty acids (SCFAs) such as butyric acid and propionic acid [38]. SCFAs exhibit anti-inflammatory characteristics in psoriasis and have the ability to activate regulatory T cells in the colon, aiding in maintaining their equilibrium [39]. The administration of SCFAs can ameliorate psoriasis symptoms by inhibiting histone deacetylase (HDAC) in Tregs and rejuvenating their activity. It has been found to decrease splenomegaly and IL-17 expression and to stimulate IL-10 and Foxp3 in the spleen [40]. The genus Prevotella 9, also protective against psoriasis, is part of the Bacteroidetes phylum. It exhibits high fermentability of cellulose and other polysaccharides and is adept at producing large amounts of SCFAs [41]. Therefore, we suggest that both Bacteroidetes and Prevotella 9 protect against psoriasis due to their SCFA production. The genus Eubacterium fissicatena group, a risk factor for psoriasis, is not yet clearly associated with a specific mechanism in the literature. We discovered a bidirectional relationship between psoriasis and Bacteroidetes, as well as Prevotella 9. However, this relationship may be influenced by other, uncontrolled confounding factors. This necessitates further study to confirm potential complex interactions.
The class Deltaproteobacteria and order Desulfovibrionales are both risk factors for rosacea. Both belong to the Proteobacteria phylum, with Desulfovibrionales being part of the Deltaproteobacteria class. This suggests that these bacteria may play an important role in rosacea. In a study on Crohn’s disease, an inflammatory bowel disease, researchers found Desulfovibrionales in the intestinal mucosa and submucosal tissues of patients with the disease, while the bacteria were not detected in healthy individuals. This suggests that Desulfovibrionales may contribute to the pathology of inflammatory bowel disease [42]. Given that rosacea often coexists with inflammatory bowel diseases such as ulcerative colitis and Crohn’s disease [43], the mechanisms related to these two microbiota in rosacea need to be further explored. The genus Ruminococcus torques group has been identified as a potential risk factor for rosacea. Research has indicated a positive correlation between this bacterial group and inflammatory markers such as IL-1β, IL-6, IFN-γ, and TNF-α [44]. These factors have been found to play a crucial role in the pathophysiology of rosacea [28,45].
In order to delve into the potential impact of specific substances within the gut microbiome on the development and treatment of skin diseases, utilizing artificial neural network technology to simulate and predict the generation of microbial secreted substances might be an effective approach [46]. After initial conclusions are drawn from the simulations, these predictions can be validated through in vitro or in vivo experiments. By thoroughly studying these molecular mechanisms, we can better understand how the gut microbiome influences the development and severity of inflammatory skin diseases. This understanding not only provides a theoretical foundation for aiding the treatment of inflammatory skin diseases by modulating the gut microbiome in the future but also helps us identify possible new drug targets, thereby developing new treatment methods. Ultimately, through this interdisciplinary research approach, we aim to provide new possibilities for controlling the symptoms of inflammatory skin diseases and improving the quality of life of patients.
This study has several strengths. First, it is the pioneer in employing bidirectional two-sample Mendelian randomization (TSMR) to elucidate a causal relationship between gut microbiota and inflammatory skin diseases. This relationship is not confounded by potential confounding factors or subjected to reverse causality. Second, we assessed instrumental variables in each group to ensure that no bias was introduced. Third, our findings were robust and validated through multiple analyses with consistent results. In addition, our results were verified for reliability via comprehensive sensitivity analyses. These findings add significantly to our understanding of the role of gut microbiota in skin health and disease, highlighting the importance of microbial balance in maintaining skin health. They also point toward the potential for future research to focus on developing microbiome-based therapies for the treatment of these skin conditions. Therefore, further studies are required to validate these relationships and to explore the mechanisms underpinning these links.
However, certain limitations should be noted. First, the bulk of the patient data in our GWAS summary were of European origin, with a limited amount of gut microbiota data coming from other ethnic groups. This could potentially introduce a bias in our estimates and limit the universal applicability of our results. Secondly, due to the limited number of instrumental variables included in the gut microbiota GWAS data and the lack of species-level data, it was not possible to ascertain if there was an overlap of participants in the GWAS data for the exposure and outcome variables. Additionally, given the extensive number and complex hierarchical structure of microbial taxa, adjustments for multiple comparisons, particularly global multiple corrections, might be overly conservative, potentially leading to an increased risk of Type II errors (false negatives). As such, negative results should be interpreted with caution, as they do not definitively rule out a causal relationship.
5. Conclusions
In conclusion, we conducted a comprehensive assessment of the causal relationships between gut microbiota and an array of inflammatory skin diseases. We found that acne was associated with two bacterial families and three genera, revealing one positive and four negative causal pathways. Furthermore, acne exhibited connections with one phylum, one order, five families, and seven genera, encompassing four positive and ten negative causal directions. Psoriasis demonstrated a correlation with one phylum and two genera, with one positive and two negative causal pathways identified. Rosacea was found to be associated with one bacterial class, one order, and two genera, indicating three positive and one negative causal connections. These findings underscore the complex interplay between gut microbiota and inflammatory skin conditions and highlight the potential of microbiota modulation as a novel therapeutic avenue. Further research is warranted to validate these associations and to elucidate the underlying molecular mechanisms. This will ultimately pave the way toward personalized microbiota-based interventions for the prevention and treatment of these common and debilitating skin disorders.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/microorganisms11102586/s1, Table S1: Overview of GWAS datasets for eczema, acne, psoriasis, and rosacea; Table S2: The genetic IVs used in TSMR analysis between gut microbiota and skin diseases (locus-wide significance, p < 5 × 10−8); Table S3: The genetic IVs used in TSMR analysis between gut microbiota and skin diseases (locus-wide significance, p < 1 × 10−5); Table S4: Causal effects of TSMR analysis between gut microbiota and skin diseases; Table S5: The results of MR-PRESSO test; Table S6: The results of MR-Egger intercept test; Table S7: Results of Cochrane’s Q test; Table S8: Results of MR Steiger directionality test; Table S9: Causal effects between skin diseases and identified bacterial taxa in the reverse TSMR analysis; Figure S1: Scatter plot analysis of the association between gut microbiota and eczema; Figure S2: Scatter plot analysis of the association between gut microbiota and acne; Figure S3: Scatter plot analysis of the association between gut microbiota and psoriasis; Figure S4: Scatter plot analysis of the association between gut microbiota and rosacea; Figure S5: Forest plots of SNPs associated with gut microbiota and eczema via the leave-one-out method; Figure S6: Forest plots of SNPs associated with gut microbiota and acne via the leave-one-out method; Figure S7: Forest plots of SNPs associated with gut microbiota and psoriasis via the leave-one-out method; Figure S8: Forest plots of SNPs associated with gut microbiota and rosacea via the leave-one-out method. A detailed funnel plot is provided in the Supplementary Material, where it is named using a combination of the specific skin disease under study and the corresponding “id.exposure” identifier.
Author Contributions
J.L. and J.G. were responsible for the design and initial drafting of the manuscript. Data collection was carried out collaboratively by J.L., J.G. and J.Y. P.C. and Y.D. undertook the task of creating the graphical representations. Y.L., M.W. and Y.W. provided mentorship and oversight, guiding the revision process of the article. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
The summary data for FINNGEN are readily accessible for download from the FinnGen version 8 website: https://r8.risteys.finngen.fi/ (accessed on 15 May 2023). Similarly, the summary data for IEU can be obtained from the official website: https://gwas.mrcieu.ac.uk/ (accessed on 15 May 2023). Other datasets generated and/or examined in the course of the present study are also publicly accessible. They are included within this published article and in its Supplementary Information file.
Acknowledgments
We express our gratitude to the MiBioGen consortium for their generous provision of GWAS data related to the intestinal flora. We also extend our heartfelt appreciation to both the participants and researchers involved in the FinnGen and IEU studies.
Conflicts of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as potential conflict of interest.
Abbreviations
| GWAS | Genome-wide association study |
| MR | Mendelian randomization |
| GSA | Gut–skin axis |
| SCFAs | Short-chain fatty acids |
| IVs | Instrumental variables |
| SNP | Single-nucleotide polymorphism |
| IVW | Inverse variance weighted |
References
- Karimkhani, C.; Dellavalle, R.P.; Coffeng, L.E.; Flohr, C.; Hay, R.J.; Langan, S.M.; Naghavi, M. Global Skin Disease Morbidity and Mortality: An Update from the Global Burden of Disease Study 2013. JAMA Dermatol. 2017, 153, 406–412. [Google Scholar] [CrossRef] [PubMed]
- Salem, I.; Ramser, A.; Isham, N.; Ghannoum, M.A. The Gut Microbiome as a Major Regulator of the Gut-Skin Axis. Front. Microbiol. 2018, 9, 1459. [Google Scholar] [CrossRef] [PubMed]
- Mahmud, M.R.; Akter, S.; Tamanna, S.K.; Mazumder, L.; Esti, I.Z.; Banerjee, S.; Pirttilä, A.M. Impact of gut microbiome on skin health: Gut-skin axis observed through the lenses of therapeutics and skin diseases. Gut Microbes 2022, 14, 2096995. [Google Scholar] [CrossRef]
- Willemsen, L.E.M.; Koetsier, M.A.; van Deventer, S.J.H.; Van Tol, E.A.F. Short chain fatty acids stimulate epithelial mucin 2 expression through differential effects on prostaglandin E1 and E2 production by intestinal myofibroblasts. Gut 2003, 52, 1442–1447. [Google Scholar] [CrossRef] [PubMed]
- Samuelson, D.R.; Welsh, D.A.; Shellito, J.E. Regulation of lung immunity and host defense by the intestinal microbiota. Front. Microbiol. 2015, 6, 1085. [Google Scholar] [CrossRef] [PubMed]
- Pothmann, A.; Illing, T.; Wiegand, C.; Hartmann, A.A.; Elsner, P. The Microbiome and Atopic Dermatitis: A Review. Am. J. Clin. Dermatol. 2019, 20, 749–761. [Google Scholar] [CrossRef]
- Lee, Y.B.; Byun, E.J.; Kim, H.S. Potential Role of the Microbiome in Acne: A Comprehensive Review. J. Clin. Med. 2019, 8, 987. [Google Scholar] [CrossRef]
- Yan, D.; Issa, N.; Afifi, L.; Jeon, C.; Chang, H.-W.; Liao, W. The Role of the Skin and Gut Microbiome in Psoriatic Disease. Curr. Dermatol. Rep. 2017, 6, 94–103. [Google Scholar] [CrossRef]
- Chen, Y.-J.; Lee, W.-H.; Ho, H.J.; Tseng, C.-H.; Wu, C.-Y. An altered fecal microbial profiling in rosacea patients compared to matched controls. J. Formos. Med. Assoc. 2020, 120 Pt 1, 256–264. [Google Scholar] [CrossRef]
- Dan, Y.-L.; Wang, P.; Cheng, Z.; Wu, Q.; Wang, X.-R.; Wang, D.-G.; Pan, H.-F. Circulating adiponectin levels and systemic lupus erythematosus: A two-sample Mendelian randomization study. Rheumatology 2020, 60, 940–946. [Google Scholar] [CrossRef]
- Kurilshikov, A.; Medina-Gomez, C.; Bacigalupe, R.; Radjabzadeh, D.; Wang, J.; Demirkan, A.; Le Roy, C.I.; Garay, J.A.R.; Finnicum, C.T.; Liu, X.; et al. Large-scale association analyses identify host factors influencing human gut microbiome composition. Nat. Genet. 2021, 53, 156–165. [Google Scholar] [CrossRef]
- Burgess, S.; Thompson, S.G.; CRP CHD Genetics Collaboration. Avoiding bias from weak instruments in Mendelian randomization studies. Int. J. Epidemiol. 2011, 40, 755–764. [Google Scholar] [CrossRef]
- Burgess, S.; Butterworth, A.; Thompson, S.G. Mendelian Randomization Analysis with Multiple Genetic Variants Using Summarized Data. Genet. Epidemiol. 2013, 37, 658–665. [Google Scholar] [CrossRef] [PubMed]
- Verbanck, M.; Chen, C.-Y.; Neale, B.; Do, R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat. Genet. 2018, 50, 693–698. [Google Scholar] [CrossRef] [PubMed]
- Bowden, J.; Davey Smith, G.; Burgess, S. Mendelian randomization with invalid instruments: Effect estimation and bias detection through Egger regression. Int. J. Epidemiol. 2015, 44, 512–525. [Google Scholar] [CrossRef]
- Bowden, J.; Smith, G.D.; Haycock, P.C.; Burgess, S. Consistent Estimation in Mendelian Randomization with Some Invalid Instruments Using a Weighted Median Estimator. Genet. Epidemiol. 2016, 40, 304–314. [Google Scholar] [CrossRef] [PubMed]
- Burgess, S.; Dudbridge, F.; Thompson, S.G. Combining information on multiple instrumental variables in Mendelian randomization: Comparison of allele score and summarized data methods. Stat. Med. 2016, 35, 1880–1906. [Google Scholar] [CrossRef] [PubMed]
- Pagoni, P.; Dimou, N.L.; Murphy, N.; Stergiakouli, E. Using Mendelian randomisation to assess causality in observational studies. BMJ Ment. Health 2019, 22, 67–71. [Google Scholar] [CrossRef]
- Bowden, J.; Hemani, G.; Smith, G.D. Invited Commentary: Detecting Individual and Global Horizontal Pleiotropy in Mendelian Randomization—A Job for the Humble Heterogeneity Statistic? Am. J. Epidemiol. 2018, 187, 2681–2685. [Google Scholar] [CrossRef]
- Kwon, M.-S.; Lim, S.K.; Jang, J.-Y.; Lee, J.; Park, H.K.; Kim, N.; Yun, M.; Shin, M.-Y.; Jo, H.E.; Oh, Y.J.; et al. Lactobacillus sakei WIKIM30 Ameliorates Atopic Dermatitis-Like Skin Lesions by Inducing Regulatory T Cells and Altering Gut Microbiota Structure in Mice. Front. Immunol. 2018, 9, 1905. [Google Scholar] [CrossRef]
- Morotomi, M.; Nagai, F.; Sakon, H.; Tanaka, R. Dialister succinatiphilus sp. nov. and Barnesiella intestinihominis sp. nov., isolated from human faeces. Int. J. Syst. Evol. Microbiol. 2008, 58 Pt 12, 2716–2720. [Google Scholar] [CrossRef]
- Arpaia, N.; Campbell, C.; Fan, X.; Dikiy, S.; Van Der Veeken, J.; DeRoos, P.; Liu, H.; Cross, J.R.; Pfeffer, K.; Coffer, P.J.; et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 2013, 504, 451–455. [Google Scholar] [CrossRef]
- Ohira, H.; Tsutsui, W.; Fujioka, Y. Are Short Chain Fatty Acids in Gut Microbiota Defensive Players for Inflammation and Atherosclerosis? J. Atheroscler. Thromb. 2017, 24, 660–672. [Google Scholar] [CrossRef]
- Zhuge, A.; Li, B.; Yuan, Y.; Lv, L.; Li, Y.; Wu, J.; Yang, L.; Bian, X.; Wang, K.; Wang, Q.; et al. Lactobacillus salivarius LI01 encapsulated in alginate-pectin microgels ameliorates d-galactosamine-induced acute liver injury in rats. Appl. Microbiol. Biotechnol. 2020, 104, 7437–7455. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Yang, L.; Zhao, L.; Bai, F.; Liu, X. Comparison of Intestinal Microbes in Noninfectious Anterior Scleritis Patients with and Without Rheumatoid Arthritis. Front. Microbiol. 2022, 13, 925929. [Google Scholar] [CrossRef]
- Bowe, W.P.; Logan, A.C. Acne vulgaris, probiotics and the gut-brain-skin axis-back to the future? Gut Pathog. 2011, 3, 1. [Google Scholar] [CrossRef] [PubMed]
- Kurokawa, I.; Layton, A.M.; Ogawa, R. Updated Treatment for Acne: Targeted Therapy Based on Pathogenesis. Dermatol. Ther. 2021, 11, 1129–1139. [Google Scholar] [CrossRef] [PubMed]
- Salamon, M.; Sysa-Jedrzejowska, A.; Lukamowicz, J.; Lukamowicz, M.; Swiatkowska, E.; Wozniacka, A. Concentration of selected cytokines in serum of patients with acne rosacea. Przegl. Lek. 2008, 65, 371–374. [Google Scholar]
- Zafar, H.; Saier, M.H., Jr. Gut Bacteroides species in health and disease. Gut Microbes 2021, 13, 1848158. [Google Scholar] [CrossRef]
- Kim, J.M.; Cho, S.J.; Oh, Y.; Jung, H.; Kim, Y.; Kim, N. Nuclear factor-kappa B activation pathway in intestinal epithelial cells is a major regulator of chemokine gene expression and neutrophil migration induced by Bacteroides fragilis enterotoxin. Clin. Exp. Immunol. 2002, 130, 59–66. [Google Scholar] [CrossRef]
- Takeshita, K.; Mizuno, S.; Mikami, Y.; Sujino, T.; Saigusa, K.; Matsuoka, K.; Naganuma, M.; Sato, T.; Takada, T.; Tsuji, H.; et al. A Single Species of Clostridium Subcluster XIVa Decreased in Ulcerative Colitis Patients. Inflamm. Bowel Dis. 2016, 22, 2802–2810. [Google Scholar] [CrossRef]
- Caillon, F.; O’connell, M.; Eady, E.; Jenkins, G.; Cove, J.; Layton, A.; Mountford, A. Interleukin-10 secretion from CD14+ peripheral blood mononuclear cells is downregulated in patients with acne vulgaris. Br. J. Dermatol. 2010, 162, 296–303. [Google Scholar] [CrossRef] [PubMed]
- Fabbrocini, G.; Bertona, M.; Picazo, Ó.; Pareja-Galeano, H.; Monfrecola, G.; Emanuele, E. Supplementation with Lactobacillus rhamnosus SP1 normalises skin expression of genes implicated in insulin signalling and improves adult acne. Benef. Microbes 2016, 7, 625–630. [Google Scholar] [CrossRef] [PubMed]
- Sikora, M.; Stec, A.; Chrabaszcz, M.; Knot, A.; Waskiel-Burnat, A.; Rakowska, A.; Olszewska, M.; Rudnicka, L. Gut Microbiome in Psoriasis: An Updated Review. Pathogens 2020, 9, 463. [Google Scholar] [CrossRef]
- Chen, Y.; Ho, H.J.; Tseng, C.; Lai, Z.; Shieh, J.; Wu, C. Intestinal microbiota profiling and predicted metabolic dysregulation in psoriasis patients. Exp. Dermatol. 2018, 27, 1336–1343. [Google Scholar] [CrossRef]
- Kim, J.; Moreno, A.; Krueger, J.G. The imbalance between Type 17 T-cells and regulatory immune cell subsets in psoriasis vulgaris. Front. Immunol. 2022, 13, 1005115. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Liu, Q.; Wu, H.; Tang, T.; Zhao, T.; Li, Z. The dysbiosis gut microbiota induces the alternation of metabolism and imbalance of Th17/Treg in OSA patients. Arch. Microbiol. 2022, 204, 217. [Google Scholar]
- Louis, P.; Scott, K.P.; Duncan, S.H.; Flint, H.J. Understanding the effects of diet on bacterial metabolism in the large intestine. J. Appl. Microbiol. 2007, 102, 1197–1208. [Google Scholar] [CrossRef]
- Chang, P.V.; Hao, L.; Offermanns, S.; Medzhitov, R. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc. Natl. Acad. Sci. USA 2014, 111, 2247–2252. [Google Scholar] [CrossRef]
- Schwarz, A.; Philippsen, R.; Schwarz, T. Induction of Regulatory T Cells and Correction of Cytokine Disbalance by Short-Chain Fatty Acids: Implications for Psoriasis Therapy. J. Investig. Dermatol. 2020, 141, 95–104.e2. [Google Scholar] [CrossRef]
- Salonen, A.; Lahti, L.; Salojärvi, J.; Holtrop, G.; Korpela, K.; Duncan, S.H.; Date, P.; Farquharson, F.; Johnstone, A.M.; Lobley, G.E.; et al. Impact of diet and individual variation on intestinal microbiota composition and fermentation products in obese men. ISME J. 2014, 8, 2218–2230. [Google Scholar] [CrossRef] [PubMed]
- Chiodini, R.J.; Dowd, S.E.; Chamberlin, W.M.; Galandiuk, S.; Davis, B.; Glassing, A. Microbial Population Differentials between Mucosal and Submucosal Intestinal Tissues in Advanced Crohn’s Disease of the Ileum. PLoS ONE 2015, 10, e0134382. [Google Scholar] [CrossRef] [PubMed]
- Holmes, A.D.; Spoendlin, J.; Chien, A.L.; Baldwin, H.; Chang, A.L.S. Evidence-based update on rosacea comorbidities and their common physiologic pathways. J. Am. Acad. Dermatol. 2018, 78, 156–166. [Google Scholar] [CrossRef] [PubMed]
- Zhen, Z.; Xia, L.; You, H.; Jingwei, Z.; Shasha, Y.; Xinyi, W.; Wenjing, L.; Xin, Z.; Chaomei, F. An Integrated Gut Microbiota and Network Pharmacology Study on Fuzi-Lizhong Pill for Treating Diarrhea-Predominant Irritable Bowel Syndrome. Front. Pharmacol. 2021, 12, 746923. [Google Scholar] [CrossRef] [PubMed]
- Holmes, A.D.; Steinhoff, M. Integrative concepts of rosacea pathophysiology, clinical presentation and new therapeutics. Exp. Dermatol. 2016, 26, 659–667. [Google Scholar] [CrossRef]
- Saber, W.I.A.; Ghoniem, A.A.; Al-Otibi, F.O.; El-Hersh, M.S.; Eldadamony, N.M.; Menaa, F.; Elattar, K.M. A comparative study using response surface methodology and artificial neural network towards optimized production of melanin by Aureobasidium pullulans AKW. Sci. Rep. 2023, 13, 13545. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).