Risk Assessment for the Spread of Flavescence Dorée-Related Phytoplasmas from Alder to Grapevine by Alternative Insect Vectors in Germany
Abstract
:1. Introduction
2. Materials and Methods
2.1. Insect and Plant Material
2.2. Transmission Trials
2.3. Survival Experiments
2.4. DNA Extraction
2.5. Phytoplasma Detection
2.6. Amplification and Sequencing of Genetic Markers
2.7. Phytoplasma Quantification
2.8. Statistical Analysis
3. Results
3.1. Dual-Choice Test
3.2. Transmission Test with Plant-Transferred Insects
3.3. Single Insect—Single Plant Transmission Tests
3.4. Survival Analysis
3.5. Phytoplasma Quantification
3.6. Genotyping of Phytoplasmas in Insects and Plants
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lee, I.-M.; Davis, R.E.; Gundersen-Rindal, D.E. Phytoplasma: Phytopathogenic mollicutes. Annu. Rev. Microbiol. 2000, 54, 221–255. [Google Scholar] [CrossRef] [PubMed]
- Lee, I.-M.; Gundersen-Rindal, D.E.; Bertaccini, A. Phytoplasma: Ecology and genomic diversity. Phytopathology 1998, 88, 1359–1366. [Google Scholar] [CrossRef] [PubMed]
- Martini, M.; Murari, E.; Mori, N.; Bertaccini, A. Identification and epidemic distribution of two Flavescence dorée—related phytoplasmas in Veneto (Italy). Plant Dis. 1999, 83, 925–930. [Google Scholar] [CrossRef]
- Arnaud, G.; Malembic-Maher, S.; Salar, P.; Bonnet, P.; Maixner, M.; Marcone, C.; Boudon-Padieu, E.; Foissac, X. Multilocus sequence typing confirms the close genetic interrelatedness of three distinct Flavescence dorée phytoplasma strain clusters and group 16SrV phytoplasmas infecting grapevine and alder in Europe. Appl. Environ. Microbiol. 2007, 73, 4001–4010. [Google Scholar] [CrossRef] [PubMed]
- Malembic-Maher, S.; Salar, P.; Filippin, L.; Carle, P.; Angelini, E.; Foissac, X. Genetic diversity of European phytoplasmas of the 16SrV taxonomic group and proposal of ‘Candidatus Phytoplasma rubi’. Int. J. Syst. Evol. Microbiol. 2011, 61, 2129–2134. [Google Scholar] [CrossRef]
- Schvester, D.; Carle, P.; Moutous, G. Sur la transmission de la Flavescence dorée des vignes par une cicadelle. C. R. Séances Acad. Agric. Fr. 1961, 47, 1021–1024. [Google Scholar]
- Mehle, N.; Seljak, G.; Rupar, M.; Ravnikar, M.; Dermastia, M. The first detection of a phytoplasma from the 16SrV (Elm yellows) group in the mosaic leafhopper Orientus ishidae. New Dis. Rep. 2010, 22, 11. [Google Scholar] [CrossRef]
- Filippin, L.; Jović, J.; Cvrković, T.; Forte, V.; Clair, D.; Toševski, I.; Boudon-Padieu, E.; Borgo, M.; Angelini, E. Molecular characteristics of phytoplasmas associated with Flavescence dorée in clematis and grapevine and preliminary results on the role of Dictyophara europaea as a vector. Plant Pathol. 2009, 58, 826–837. [Google Scholar] [CrossRef]
- Lessio, F.; Picciau, L.; Gonella, E.; Mandrioli, M.; Tota, F.; Alma, A. The mosaic leafhopper Orientus ishidae: Host plants, spatial distribution, infectivity, and transmission of 16SrV phytoplasmas to vines. Bull. Insectol. 2016, 69, 277–289. [Google Scholar]
- Casati, P.; Jermini, M.; Quaglino, F.; Corbani, G.; Schaerer, S.; Passera, A.; Bianco, P.A.; Rigamonti, I.E. New insights on Flavescence dorée phytoplasma ecology in the vineyard agro-ecosystem in southern Switzerland. Ann. Appl. Biol. 2017, 171, 37–51. [Google Scholar] [CrossRef]
- Malembic-Maher, S.; Desqué, D.; Khalil, D.; Salar, P.; Bergey, B.; Danet, J.-L.; Duret, S.; Dubrana-Ourabah, M.-P.; Beven, L.; Ember, I.; et al. When a Palearctic bacterium meets a Nearctic insect vector: Genetic and ecological insights into the emergence of the grapevine Flavescence dorée epidemics in Europe. PLoS Pathog. 2020, 16, e1007967. [Google Scholar] [CrossRef] [PubMed]
- Rigamonti, I.E.; Salvetti, M.; Girgenti, P.; Bianco, P.A.; Quaglino, F. Investigation on Flavescence dorée in Nort-Western Italy Identifies Map-M54 (16SrV-D/Map-FD2) as the only Phytoplasma Genotype in Vitis vinifera L. and Reveals the Presence of New Putative Reservoir Plants. Biology 2023, 12, 1216. [Google Scholar] [CrossRef] [PubMed]
- Maixner, M.; Reinert, W.; Darimont, H. Transmission of grapevine yellows by Oncopsis alni (Schrank) (Auchenorrhyncha: Macropsinae). Vitis 2000, 39, 83–84. [Google Scholar]
- Maixner, M.; Ahrens, U.; Seemüller, E. Detection of the German Grapevine Yellows (Vergilbungskrankheit) Mlo in Grapevine, Alternative Hosts and a Vector by a Specific Pcr Procedure. Eur. J. Plant Pathol. 1995, 101, 241–250. [Google Scholar] [CrossRef]
- Angelini, E.; Clair, D.; Borgo, M.; Bertaccini, A.; Boudon-Padieu, E. Flavescence dorée in France and Italy—Occurrence of closely related phytoplasma isolates and their near relationships to Palatinate Grapevine Yellows and an Alder Yellows Phytoplasma. Vitis 2001, 40, 79–86. [Google Scholar]
- Angelini, E.; Negrisolo, E.; Clair, D.; Borgo, M.; Boudon-Padieu, E. Phylogenetic relationships among Flavescence dorée strains and related phytoplasmas determined by heteroduplex mobility assay and sequence of ribosomal and Nonribosomal DNA. Plant Pathol. 2003, 52, 663–672. [Google Scholar] [CrossRef]
- Maixner, M.; Reinert, W. Oncopsis alni (Schrank) (Auchenorrhyncha: Cicadellidae) as a vector of the alder yellows phytoplasma of Alnus glutinosa (L.) Gaertn. Eur. J. Plant Pathol. 1999, 105, 87–94. [Google Scholar] [CrossRef]
- Jarausch, W.; Bischoff, F.; Runne, M.; Trapp, M. GIS-basierte Risikoanalyse zur Ausbreitung von Flavescence dorée- Phytoplasmen von Wildhabitaten in angrenzende Weinberge. Julius-Kuehn-Archiv 2018, 461, 343–344. [Google Scholar]
- Jarausch, B.; Biancu, S.; Kugler, S.; Wetzel, T.; Baumann, M.; Winterhagen, P.; Jarausch, W.; Kortekamp, A.; Maixner, M. First Report of Flavescence dorée -Related Phytoplasma in a Productive Vineyard in Germany. Plant Dis. 2021, 105, 3285. [Google Scholar] [CrossRef]
- Jarausch, B.; Biancu, S.; Lang, F.; Desque, D.; Salar, P.; Jarausch, W.; Xavier, F.; Sylvie, M.-M.; Maixner, M. Study of the epidemiology of “Flavescence dorée(FD)-related phytoplasmas and potential vectors in a FD-free area. Phytopathog. Mollicutes 2019, 9, 59–60. [Google Scholar] [CrossRef]
- Kunz, G.; Nickel, H.; Niedringhaus, R. Fotoatlas der Zikaden Deutschlands; Wissenschaftlich-Akademischer-Buchvertrieb-Fründt: Osnabrück, Germany, 2011; p. 293. [Google Scholar]
- Biedermann, R.; Niedringhaus, R. Die Zikaden Deutschlands—Bestimmungstafeln für alle Arten; Wissenschaftlich-Akademischer-Buchvertrieb-Fündt: Osnabrück, Germany, 2004; p. 410. [Google Scholar]
- EPPO. PM 7/079: Grapevine Flavescence dorée phytoplasma. Bull. OEPP/EPPO 2016, 46, 78–83. [Google Scholar] [CrossRef]
- Doyle, J.J.; Doyle, J.L. Isolation of plant DNA from fresh tissue. Focus 1990, 12, 13–15. [Google Scholar]
- Boudon-Padieu, E.; Bejat, A.; Clair, D.; Larrue, J.; Borgo, M.; Bertotto, L.; Angelini, E. Grapevine yellows: Comparison of different procedures for DNA extraction and amplification with PCR for routine diagnosis of phytoplasmas in grapevine. Vitis 2003, 42, 141–149. [Google Scholar]
- Lorenz, K.-H.; Schneider, B.; Ahrens, U.; Seemuller, E. Detection of the apple proliferation and pear decline phytoplasmas by PCR amplification of ribosomal and nonribosomal DNA. Phytopathology 1995, 85, 771–776. [Google Scholar] [CrossRef]
- Schneider, B.; Seemüller, E.; Smart, C.D.; Kirkpatrick, B.C. Phylogenetic classification of plant pathogenic mycoplasma-like organisms or phytoplasmas. In Molecular and Diagnostic Procedures in Mycoplasmology; Razin, S., Tully, J.G., Eds.; Academic Press: San Diego, CA, USA, 1995; Volume I, pp. 369–380. [Google Scholar]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Klein, J.P.; Moeschberger, M.L. Survival Analysis—Techniques for Censored and Truncated Data; Springer Science & Business Media: New York, NY, USA, 2003. [Google Scholar]
- Therneau, T.M.; Grambsch, P.M. Modeling Survival Data: Extending the Cox Model; Springer: New York, NY, USA, 2000. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing. Available online: https://www.R-project.org/ (accessed on 19 September 2023).
- Therneau, T. A Package for Survival Analysis in R. R Package Version 3.4-0. Available online: https://CRAN.R-project.org/package=survival (accessed on 20 September 2023).
- Kassambra, A. Survminer: Drawing Survival Curves Using ‘ggplot2’. R Package Version 0.4.9. Available online: https://CRAN.R-project.org/package=survminer (accessed on 20 September 2023).
- Lenth, R. Emmeans: Estimated Marginal Means, Aka Least-Squares Means. R Package Version 1.8.6. Available online: https://CRAN.R-project.org/package=emmeans (accessed on 20 September 2023).
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Patil, I. Visualizations with statistical details: The ‘ggstatsplot’ approach. J. Open Source Softw. 2021, 6, 3167. [Google Scholar] [CrossRef]
- Mehle, N.; Rupar, M.; Seljak, G.; Ravnikar, M.; Dermastia, M. Molucular diversity of ‘Flavescence dorée’ phytoplasma strains in Slovenia. Bull. Insectology 2011, 64, 29–30. [Google Scholar]
- Rizzoli, A.; Belgeri, E.; Jermini, M.; Conedera, M.; Filippin, L.; Angelini, E. Alnus glutinosa and Orientus ishidae (Matsumura, 1902) share phytoplasma genotypes linked to the ‘Flavescence dorée’ epidemics. J. Appl. Entomol. 2021, 145, 1015–1028. [Google Scholar] [CrossRef]
- Auriol, A.; Salar, P.; Pedemay, S.; Lusseau, T.; Desque, D.; Lacaze, D.; Bocquart, M.; Levillain, M.; Bey, J.-S.; Pienne, P.; et al. Origin of isolated cases of “flavescence doree” in North-East of France: Search for reservoir plants and insect vectors in semi-natural habitats near vineyards. Phytopathog. Mollicutes 2023, 13, 45–46. [Google Scholar] [CrossRef]
- Guglielmino, A. Observations on the genus Orientus (Rhynchota Cicadomorpha Cicadellidae) and description of a new species: O. amurensis n. sp. from Russia (Amur Region and Maritime Territory) and China (Liaoning Province). Marbg. Entomol. Publ. 2005, 3, 99–110. [Google Scholar]
- Klejdysz, T.; Zwolińska, A.; Walczak, M.; Kobiałka, M. The first record of a potential pest Orientus ishidae (Matsumura, 1902) (Hemiptera: Cicadellidae) in Poland. J. Plant Prot. Res. 2017, 57, 107–112. [Google Scholar] [CrossRef]
- Nickel, H. First addendum to the Leafhoppers and Planthoppers of Germany (Hemiptera: Auchenorrhyncha). Cicadina 2010, 11, 107–122. [Google Scholar]
- Parise, G. Notes on the biology of Orientus ishidae (Matsumura, 1902) in Piedmont (Italy): (Hemiptera: Cicadellidae: Deltocephalinae). Cicadina 2017, 17, 19–28. [Google Scholar]
- Nickel, H.; Remane, R. Artenliste der Zikaden Deutschlands, mit Angaben zu Nährpflanzen, Nahrungsbreite, Lebenszyklen, Areal und Gefährdung (Hemiptera, Fulgoromorpha et Cicadomorpha). Beitr. Zikadenkunde 2002, 5, 27–64. [Google Scholar]
- Chuche, J.; Backus, E.A.; Thiery, D.; Sauvion, N. First finding of a dual-meaning X wave for phloem and xylem fluid ingestion: Characterization of Scaphoideus titanus (Hemiptera: Cicadellidae) EPG waveforms. J. Insect Physiol. 2017, 102, 50–61. [Google Scholar] [CrossRef]
- Bressan, A.; Girolami, V.; Boudon-Padieu, É. Reduced fitness of the leafhopper vector Scaphoideus titanus exposed to Flavescence dorée phytoplasma. Entomol. Exp. Appl. 2005, 115, 283–290. [Google Scholar] [CrossRef]
- D’Amelio, R.; Palermo, S.; Marzachi, C.; Bosco, D. Influence of Chrysanthemum yellows phytoplasma on the fitness of two of its leafhopper vectors, Macrosteles quadripunctulatus and Euscelidius variegatus. Bull. Insectology 2008, 61, 349–354. [Google Scholar]
- Bosco, D.; D’Amelio, R. Transmission Specificity and Competition of Multiple Phytoplasmas in the Insect Vector; CAB International: Wallingford, UK, 2010. [Google Scholar]
- Weintraub, P.G.; Beanland, L. Insect vectors of phytoplasmas. Annu. Rev. Entomol. 2006, 51, 91–111. [Google Scholar] [CrossRef]
- Marzachi, C.; Bosco, D. Relative quantification of chrysanthemum yellows (16Sr I) phytoplasma in its plant and insect host using real-time polymerase chain reaction. Mol. Biotechnol. 2005, 30, 117–128. [Google Scholar] [CrossRef]
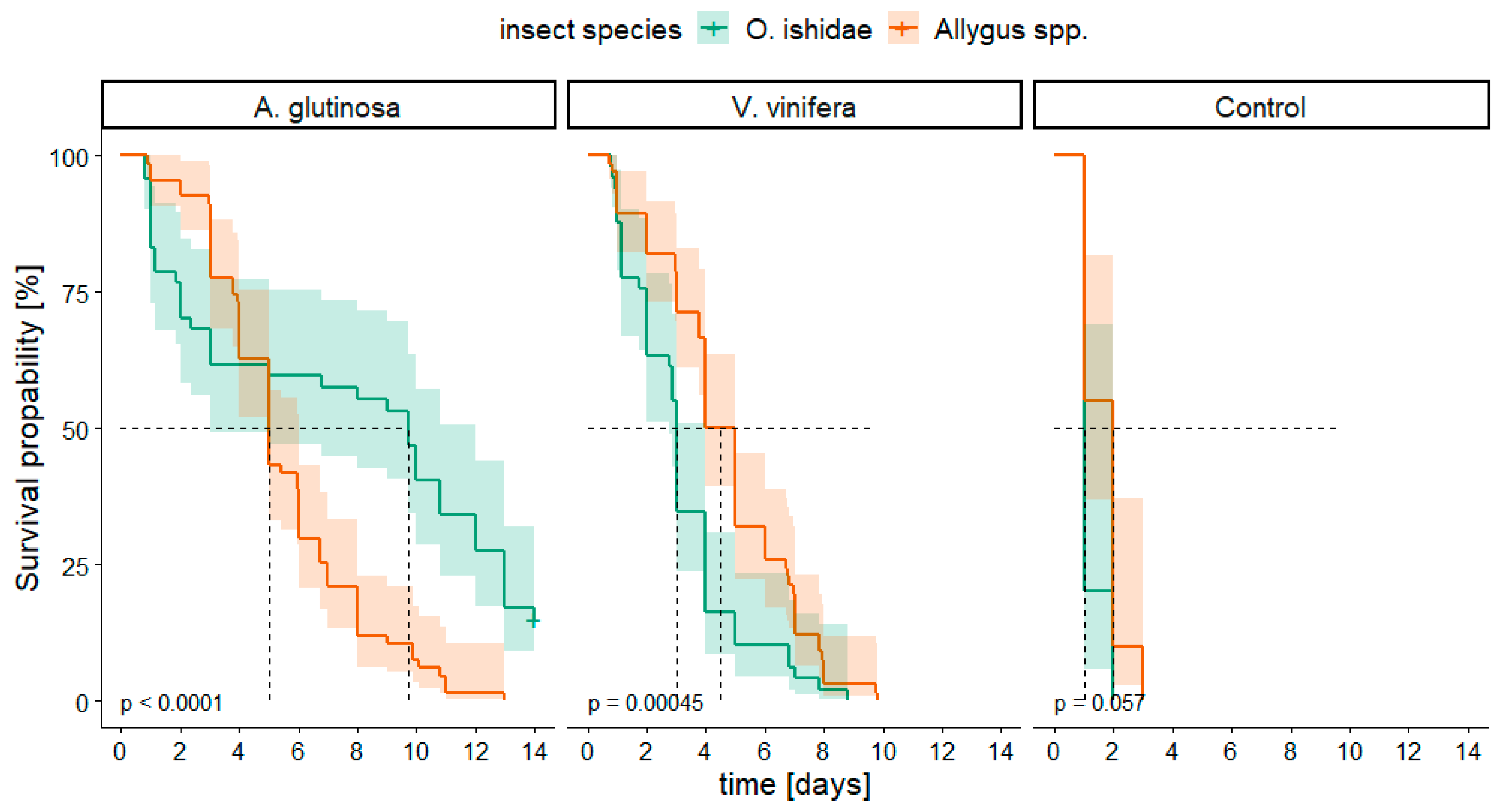
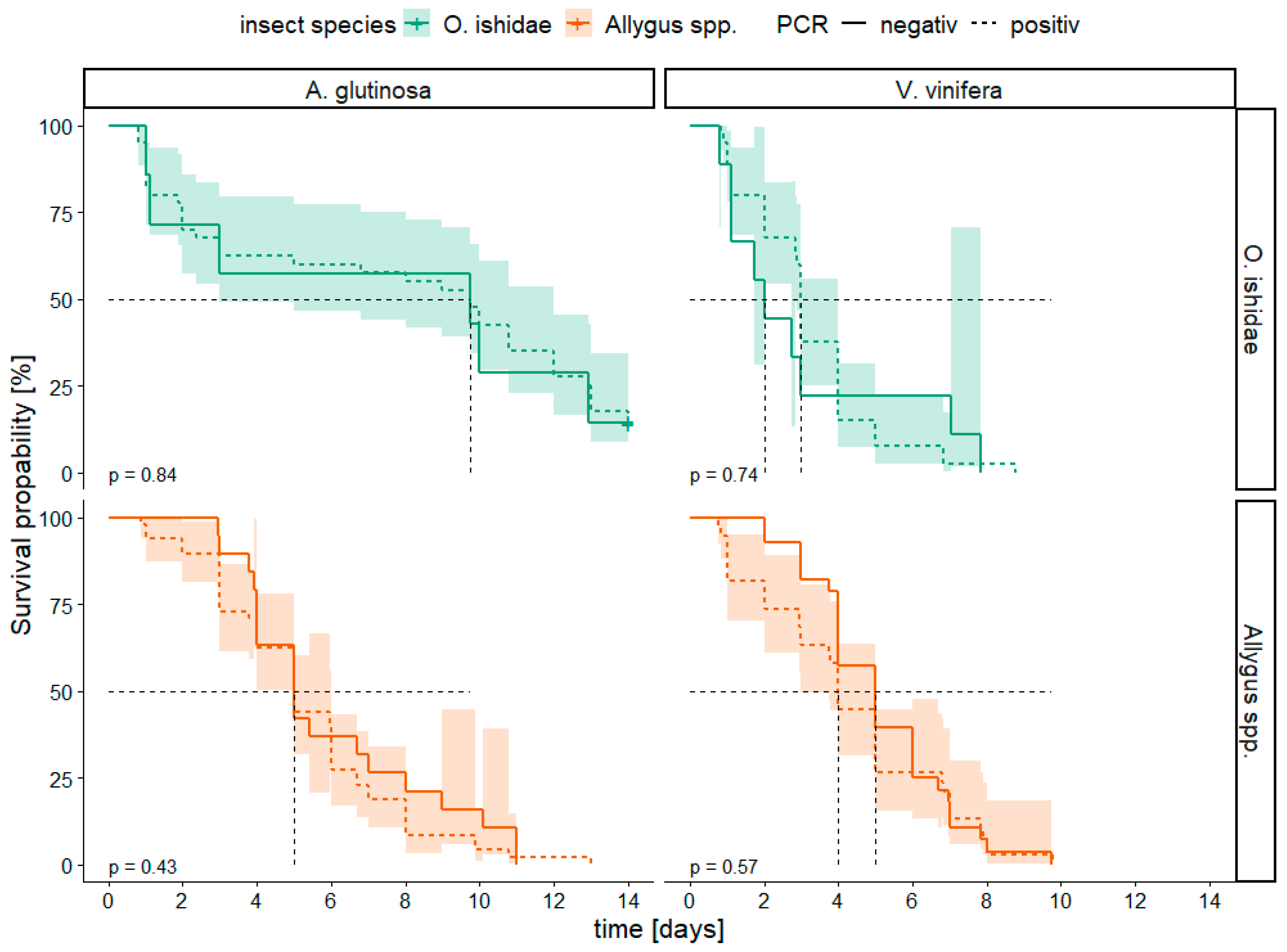
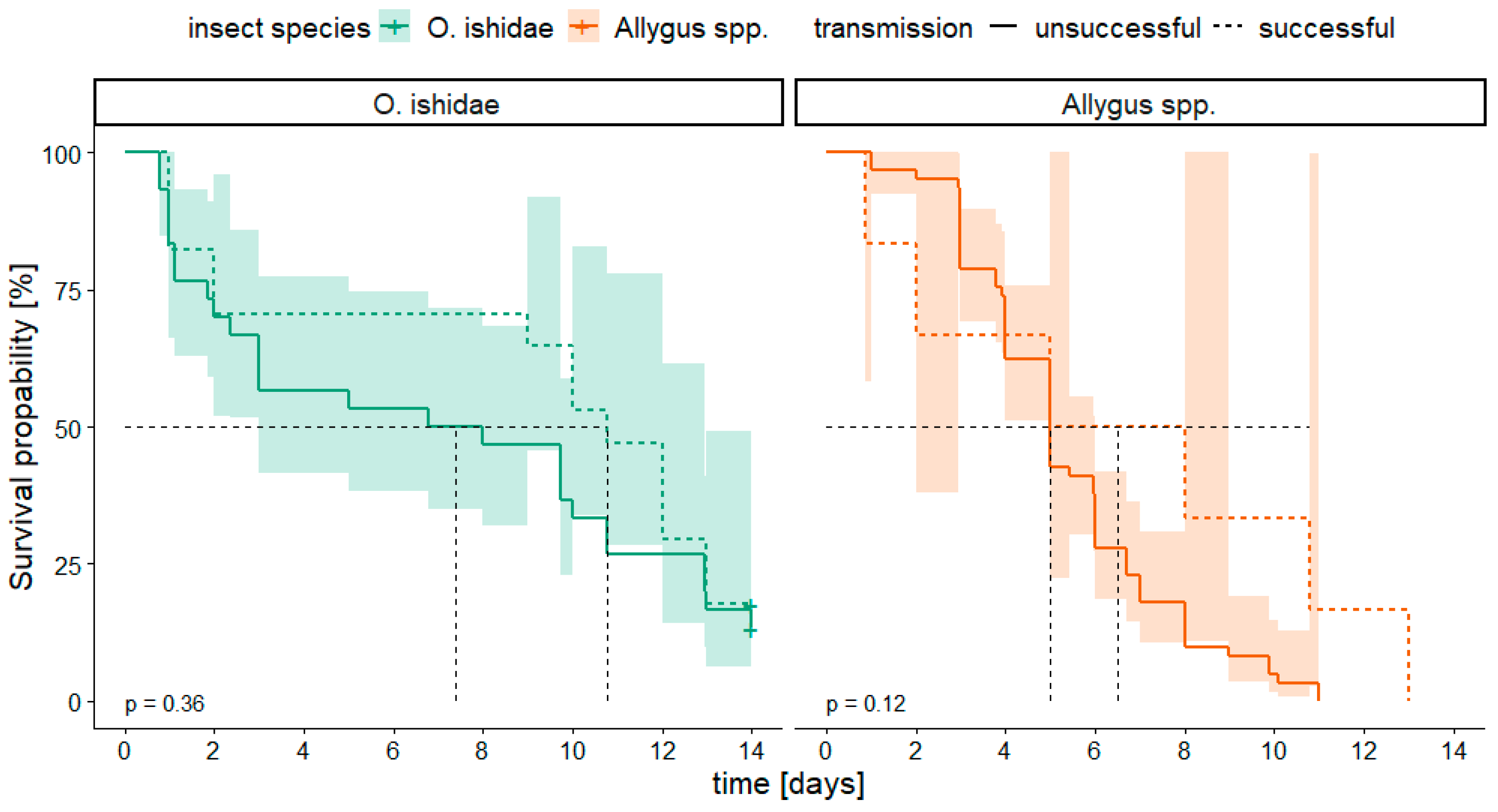

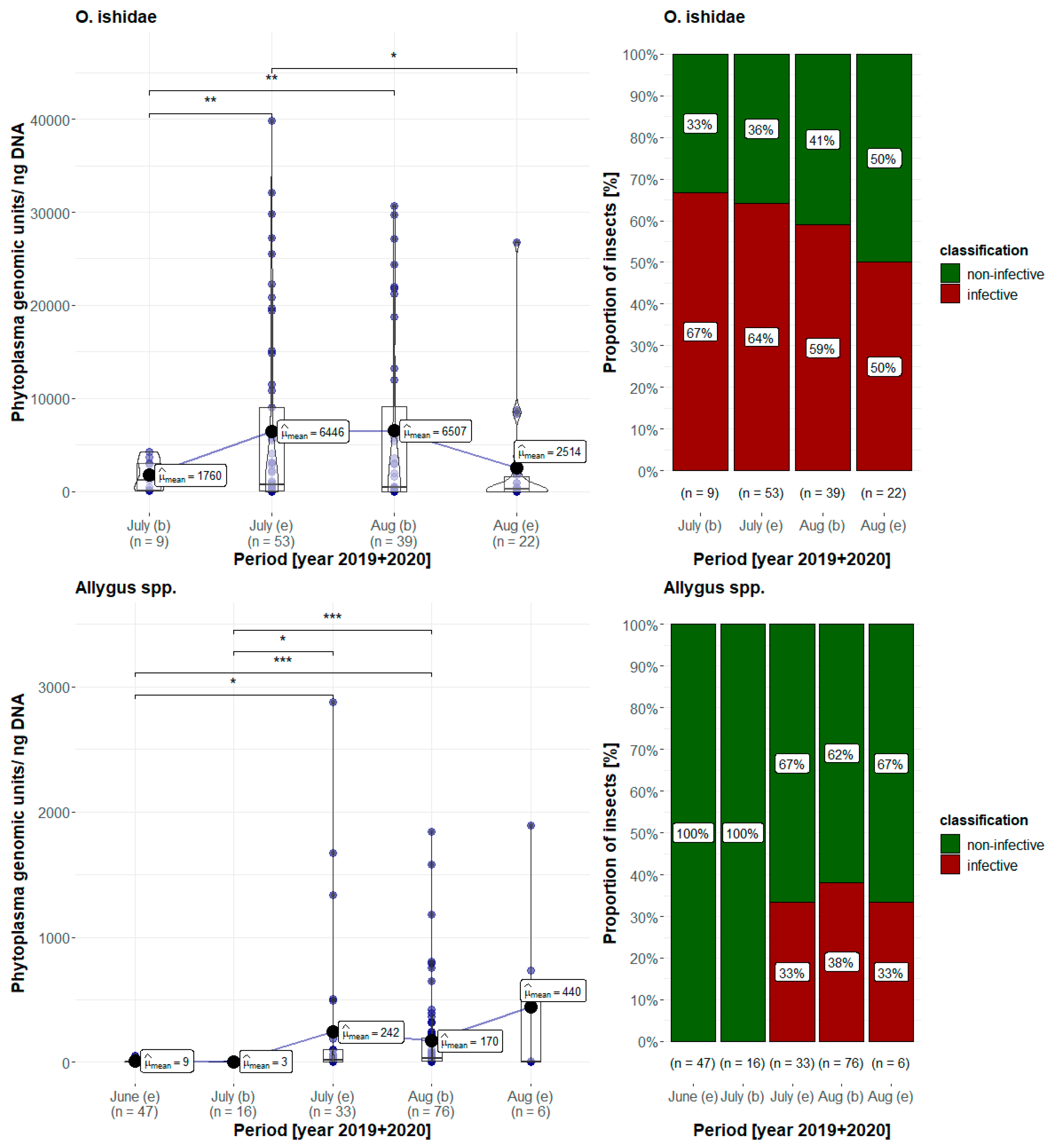
| 2017 | number of plants PCR+/ number of plants tested | number of insects caged | number of insects PCR+/ number of insects tested |
| Allygus spp. | - | 36 | 14/28 (50%) |
| A. glutinosa | 2/4 (50%) | - | - |
| V. vinifera | 0/4 (0%) | - | - |
| O. ishidae | - | 41 | 13/26 (50%) |
| A. glutinosa | 5/6 (83%) | - | - |
| V. vinifera | 1/6 (17%) | - | - |
| 2018 | number of plants PCR+/ number of plants tested | number of insects caged | number of insects PCR+/number of insects tested |
| Allygus spp. | - | 23 | 6/19 (32%) |
| A. glutinosa | 1/2 (50%) | - | - |
| V. vinifera | 0/2 (0%) | - | - |
| O. ishidae | - | 52 | 14/44 (32%) |
| A. glutinosa | 1/4 (25%) | - | - |
| V. vinifera | 0/3 (0%) | - | - |
| 2017 | number of plants PCR+/ number of plants tested | number of insects caged | number of insects PCR+/ number of insects tested |
| 1st test plant A. glutinosa | |||
| Allygus spp. | 4/5 (80%) | 42 | 7/15 (47%) |
| O. ishidae | 5/5 (100%) | 85 | 15/18 (83%) |
| Transfer to 2nd test plant V. vinifera | |||
| Allygus spp. | 0/5 (0%) | 27 | 14/27 (52%) |
| O. ishidae | 0/5 (0%) | 67 | 48/60 (80%) |
| 2018 | number of plants PCR+/ number of plants tested | number of insects caged | number of insects PCR+/ number of insects tested |
| 1st test plant A. glutinosa | |||
| Allygus spp. | 3/7 (43%) | 30 | 3/8 (38%) |
| O. ishidae | 8/12 (67%) | 94 | 14/25 (56%) |
| Transfer to 2nd test plant V. vinifera | |||
| Allygus spp. | 0/7 (0%) | 19 | 8/15 (53%) |
| O. ishidae | 0/12 (0%) | 64 | 27/55 (49%) |
| 2019 | number of plants PCR+/ number of plants tested | number of insects caged | number of insects PCR+/ number of insects tested |
| 1st test plant A. glutinosa | |||
| Allygus spp. | 2/7 (29%) | 55 | 13/18 (72%) |
| O. ishidae | 11/14 (79%) | 152 | 15/38 (39%) |
| Transfer to 2nd test plant V. vinifera | |||
| Allygus spp. | 0/7 (0%) | 36 | 4/29 (14%) |
| O. ishidae | 0/14 (0%) | 114 | 51/113 (45%) |
| 2019 | number of plants PCR+/ number of plants tested | number of insects PCR+/ number of insects tested |
| A. glutinosa | ||
| Allygus spp. | 2/17 (12%) | 11/17 (65%) |
| O. ishidae | 1/18 (16%) | 14/18 (78%) |
| V. vinifera | ||
| Allygus spp. | 0/17 (0%) | 12/17 (71%) |
| O. ishidae | 0/19 (0%) | 12/19 (63%) |
| 2020 | number of plants PCR+/ number of plants tested | number of insects PCR+/ number of insects tested |
| A. glutinosa | ||
| Allygus spp. | 7/50 (14%) | 36/50 (72%) |
| O.ishidae | 13/30 (43%) | 28/30 (93%) |
| V. vinifera | ||
| Allygus spp. | 0/49 (0%) | 24/49 (49%) |
| O. ishidae | 4/30 (13%) | 26/30 (87%) |
| IAP in Days | Number of Trials with Infected Insects | Number of Infective Insects | Number of Transmissions | Transmission Rate of Infective Insects |
|---|---|---|---|---|
| (a) Allygus spp. | ||||
| 1 | 2 | 0 | 0 | - |
| 2 | 2 | 1 | 1 | 100% |
| 3 | 6 | 0 | 0 | - |
| 4 | 4 | 1 | 0 | 0% |
| 5 | 9 | 2 | 2 | 100% |
| 6–8 | 13 | 4 | 3 | 75% |
| 8–10 | 0 | 0 | 0 | - |
| >10 | 1 | 1 | 1 | 100% |
| (b) O. ishidae | ||||
| 1 | 5 | 4 | 2 | 50% |
| 2 | 3 | 2 | 2 | 100% |
| 3 | 0 | 0 | 0 | - |
| 4 | 0 | 0 | 0 | - |
| 5 | 1 | 1 | 0 | 0% |
| 6–8 | 1 | 1 | 0 | 0% |
| 8–10 | 3 | 3 | 3 | 100% |
| >10 | 13 | 9 | 8 | 89% |
| Map Genotypes | ||||||||
|---|---|---|---|---|---|---|---|---|
| FD2 | PGY | |||||||
| Species | Number Analyzed | M38 | M14 | M39 | M47 | M48 | M53 | M110 |
| Allygus spp. | 65 | 56 | 2 | 1 | 2 | 1 | 2 | 1 |
| O. ishidae | 189 | 188 | 1 | |||||
| A. glutinosa | 40 | 39 | 1 | |||||
| V. vinifera | 5 | 5 | ||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jarausch, B.; Markheiser, A.; Jarausch, W.; Biancu, S.; Kugler, S.; Runne, M.; Maixner, M. Risk Assessment for the Spread of Flavescence Dorée-Related Phytoplasmas from Alder to Grapevine by Alternative Insect Vectors in Germany. Microorganisms 2023, 11, 2766. https://doi.org/10.3390/microorganisms11112766
Jarausch B, Markheiser A, Jarausch W, Biancu S, Kugler S, Runne M, Maixner M. Risk Assessment for the Spread of Flavescence Dorée-Related Phytoplasmas from Alder to Grapevine by Alternative Insect Vectors in Germany. Microorganisms. 2023; 11(11):2766. https://doi.org/10.3390/microorganisms11112766
Chicago/Turabian StyleJarausch, Barbara, Anna Markheiser, Wolfgang Jarausch, Sandra Biancu, Sanela Kugler, Miriam Runne, and Michael Maixner. 2023. "Risk Assessment for the Spread of Flavescence Dorée-Related Phytoplasmas from Alder to Grapevine by Alternative Insect Vectors in Germany" Microorganisms 11, no. 11: 2766. https://doi.org/10.3390/microorganisms11112766
APA StyleJarausch, B., Markheiser, A., Jarausch, W., Biancu, S., Kugler, S., Runne, M., & Maixner, M. (2023). Risk Assessment for the Spread of Flavescence Dorée-Related Phytoplasmas from Alder to Grapevine by Alternative Insect Vectors in Germany. Microorganisms, 11(11), 2766. https://doi.org/10.3390/microorganisms11112766






