Abstract
Calonectria pseudoreteaudii is an important causal agent of Eucalyptus leaf blight in southern China. This pathogen causes Eucalyptus tree disease across numerous regions in southern China. In addition to diseased leaves, C. pseudoreteaudii has occasionally been isolated from soil in Eucalyptus plantations. The aim of this study was to clarify whether C. pseudoreteaudii causing Eucalyptus leaf blight in China is mainly clonally reproduced and to determine the potential spreading mechanism of C. pseudoreteaudii between diseased leaves and soil. To this end, 10 polymorphic microsatellite markers were analyzed to detect the genetic diversity of 97 C. pseudoreteaudii isolates from diseased leaves and soil in a Eucalyptus plantation in Guangxi Zhuang Autonomous Region, southern China. The analysis showed that the genetic diversity of the isolates from both the diseased leaves and soil was high. However, the gene and genotype diversity of the C. pseudoreteaudii isolates from diseased leaves were higher than those of the isolates from the soil. Moreover, all genotypes detected in the isolates from the soil were also found in the isolates from the diseased leaves. Structural analyses did not show clear population structures related to the population substrates of the diseased leaves or soil, and molecular variance analyses indicated that no significant genetic differentiation existed between the diseased leaf and soil populations. These results suggest that C. pseudoreteaudii in soil spreads from diseased leaves, and that an asexual cycle is the primary reproductive mode in both diseased leaf and soil populations. This is the first study on the genetic diversity and population structure of C. pseudoreteaudii. The high genetic diversity and spread pathways of this pathogen may pose challenges in controlling the disease. C. pseudoreteaudii from both diseased leaves and soils in Eucalyptus plantations needs to be carefully monitored for disease control and management.
1. Introduction
Leaf blight caused by Calonectria spp. is one of the most prominent diseases of Eucalyptus trees in Southeast Asia and South America [1,2,3,4,5,6,7,8]. C. pseudoreteaudii is one of the dominant species isolated from blighted Eucalyptus leaves, especially among some genotypes of E. urophylla × E. grandis and E. urophylla × E. tereticornis in plantations and nurseries in southern China [4,5,8,9,10]. It is a heterothallic species that resides in the C. reteaudii species complex [6,10,11,12,13]. In China, C. pseudoreteaudii has been recovered from Eucalyptus in Fujian, Guangdong, Guangxi, and Hainan provinces [4,5,6,8,9,10,14]. C. pseudoreteaudii mainly infects Eucalyptus trees that are less than two years old in the high temperature and humidity season. In some regions of Guangdong, Guangxi, and Hainan provinces, C. pseudoreteaudii has caused severe disease and nearly all leaves of infected Eucalyptus trees have blighted and dropped [5,8,10]. In addition to Eucalyptus in China, C. pseudoreteaudii has been isolated from Eucalyptus hybrids in Vietnam, India, and Indonesia; Macadamia sp. in Vietnam, Lao, and China; and blueberry in China [13,15,16,17,18,19,20].
Disease symptoms caused by C. pseudoreteaudii in Eucalyptus plantations include leaf and shoot blight [4,6,8,9,10,12,13,14,21]. In the early stages, the pathogen mainly infects the lower and middle leaves of Eucalyptus trees, resulting in grayish, water-soaked spots. Subsequently, these spots spread rapidly, becoming extensive necrotic areas, resulting in stem and shoot rot and leaf blight [8,10]. In nurseries, C. pseudoreteaudii causes Eucalyptus seedling stem, cutting, and leaf rot [8,12,14,21]. Under high-temperature and -humidity conditions, white conidiophore masses with the typical morphological characteristics of Calonectria spp. are observed frequently on the leaves and shoots of Eucalyptus trees and seedlings [8,10].
In Eucalyptus plantations in China, C. pseudoreteaudii has mainly been isolated from diseased Eucalyptus tissues, and occasionally from the soil under diseased Eucalyptus trees [4,8]. Wang and Chen [8] performed the first systematic investigation of Eucalyptus leaf disease caused by Calonectria species on the Leizhou Peninsula of southern China. The results indicated that C. pseudoreteaudii (identified as C. pentaseptata before the taxonomic revisions by Liu et al. [6]) was widely isolated from the blighted leaves of a number of Eucalyptus genotypes planted in different regions, and 773 C. pseudoreteaudii isolates were obtained from 14 sampling sites [8]. However, Calonectria isolates were not isolated from the soil in Eucalyptus plantations in the investigation. Recently, Li et al. [4] conducted several disease surveys in regions in southern China planted with Eucalyptus threatened by Calonectria leaf blight. They found that in distant regions in southern China, C. pseudoreteaudii was commonly isolated from the blighted leaves of Eucalyptus, but seldom from the soil under the diseased Eucalyptus trees. The results showed that 243 C. pseudoreteaudii isolates were obtained from six of the nine sampled regions, whereas only four C. pseudoreteaudii isolates were isolated from two of the nine sampled regions [4].
In recent studies, we evaluated the genotype diversity of C. pseudoreteaudii based on sequences of the translation elongation factor 1-alpha (tef1), β-tubulin (tub2), calmodulin (cmdA), and histone H3 (his3) gene regions [8,10]. On the basis of a number of Eucalyptus genotypes in multiple geographic sites, our results indicated that the genotype diversity of C. pseudoreteaudii was very low [8,10]. Only two genotypes were detected among the 55 tested isolates recovered from diseased Eucalyptus leaves by Wang and Chen [8]. In another study by our group [10], two genotypes were identified in 66 C. pseudoreteaudii isolates from diseased leaves, and only one genotype was present in 53 C. pseudoreteaudii isolates obtained from soil. The genotypes identified in the diseased leaves included those from the soil [10].
On the basis of the results from previous studies [4,8,10], we were keen to clarify whether C. pseudoreteaudii that causes Eucalyptus leaf blight in China is mainly clonally reproduced with low genetic diversity. Furthermore, we expected to clarify the differences in the population diversity of C. pseudoreteaudii from diseased leaves and soil within the same Eucalyptus plantation, which will help us to understand the potential spread pathways of C. pseudoreteaudii between diseased leaves and soil.
Recently, C. pseudoreteaudii causing leaf blight on Eucalyptus trees in one plantation resulted in tree damage [10], and a number of C. pseudoreteaudii isolates from diseased leaves and soil under the diseased trees were obtained. The objectives of this study were to (i) investigate the genetic diversity and (ii) understand the differences in the population diversity of C. pseudoreteaudii isolates from diseased leaves and soil using 10 polymorphic microsatellite markers.
2. Materials and Methods
2.1. Calonectria pseudoreteaudii Isolates and DNA Extraction
A total of 97 C. pseudoreteaudii isolates obtained from the same one-year-old Eucalyptus plantation in Hepu County, Beihai region, Guangxi, southern China (21°33′19.8756″ N, 109°42′27.0792″ E), in October 2018 were used in this study (Table S1). These included 94 isolates from Wu and Chen [10] and three additional isolates (CSF15861, CSF15910, and CSF15984). All 97 isolates were deposited in the culture collection (CSF) of the Research Institute of Fast-Growing Trees (RIFT), Chinese Academy of Forestry (CAF), in Zhanjiang, Guangdong Province, China. All 97 isolates were identified via DNA sequence comparisons of the tef1, tub2, cmdA, and his3 gene regions ([10]; Table S1). Of the 97 isolates, 64 and 33 isolates were recovered from diseased Eucalyptus leaves and soil, respectively (Table S1). The 64 diseased leaf isolates were obtained from 64 Eucalyptus trees (one per tree). The 33 soil isolates were isolated from 18 soil samples under the diseased trees, and one or two isolates were obtained from each soil sample (Table S1). The trees in the one-year-old Eucalyptus plantation were five- to six-meters high, and the soil was relatively moist, as the sampling period was the rainy season in the plantation (Figure 1 in Wu and Chen [10]). The 64 isolates from the diseased leaves and 33 isolates from the soil were treated as two populations for further population studies.
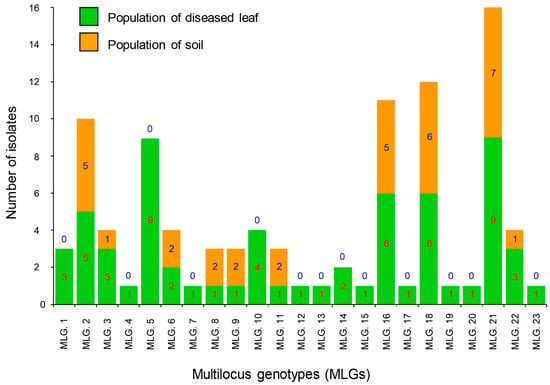
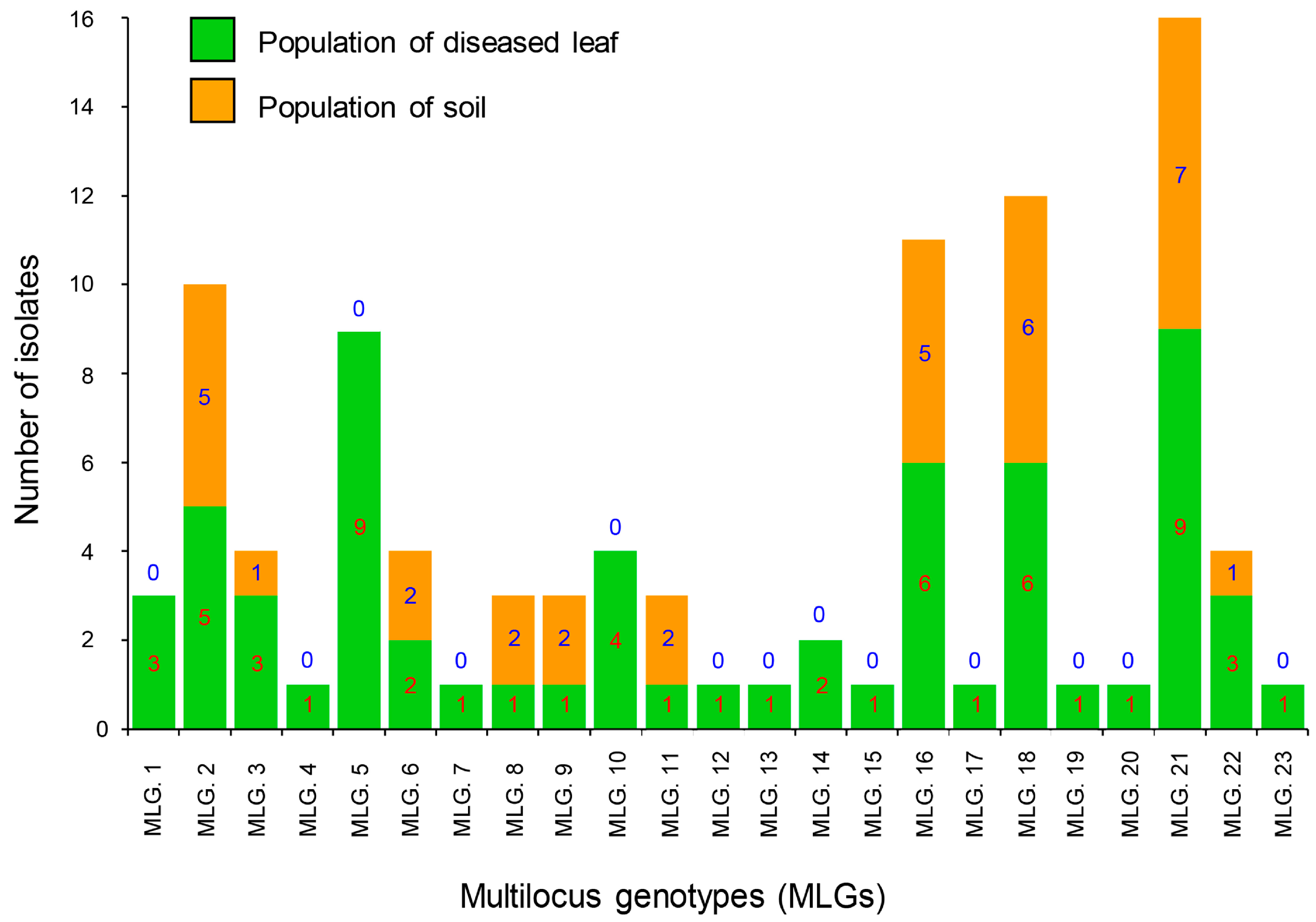
Figure 1.
Multilocus genotypes (MLGs) generated from two C. pseudoreteaudii populations from diseased leaves and soil. Twenty-three genotypes were detected in the full dataset. The number of individuals residing in each genotype is indicated in each bar graph.
To ensure that each culture represented a single individual, a single hyphal tip of each culture was moved to 2% malt extract agar (MEA) (20 g of agar powder, 20 g of malt extract powder, and 1000 µL of a 30 mg/mL streptomycin sulfate solution per liter of water) and agitated. Malt extract powder was purchased from the Beijing Shuangxuan microbial culture medium products factory, Beijing, China; agar powder was purchased from Beijing Solarbio Science & Technology Co., Ltd., Beijing, China; and the streptomycin sulfate solution was purchased from Shanghai Sango Biotech Co., Ltd., Shanghai, China, and incubated at room temperature. Mycelia were collected from 10-day-old cultures using a sterile scalpel and moved to 2 mL Eppendorf tubes for DNA extraction. Genomic DNA was extracted following the CTAB protocol [22]. In order to degrade the RNA, the extracted DNA was dissolved using 3 µL of RNase (10 mg/mL) and 30 µL of TE buffer (1 M tris-HCl and 0.5 M EDTA, pH 8.0), which were added to the extracted DNA at 37 °C for 1 h. The DNA’s concentration was measured using a NanoDrop 2000 spectrometer (Thermo Fisher Scientific, Waltham, MA, USA).
2.2. Microsatellite Locus Sequencing and Allele Scoring
Ten microsatellite markers with polymorphisms that were easily PCR-amplified and sequenced were used to genotype all of the C. pseudoreteaudii isolates [23]. The PCR reactions were performed as follows: 95 °C for 5 min, followed by 35 cycles of 95 °C for 30 s, 30 s at a 52 °C annealing temperature for each pair of primers, 72 °C for 1 min, and a final extension at 72 °C for 10 min. The PCR products were checked via agarose gel electrophoresis using a 2% agarose gel with 4S GelRed (Sangon Biotech Co., Ltd., Shanghai, China) and a 1× tris-acetate-EDTA (TAE) buffer at a constant voltage (80 V) for 40 min and viewed under UV light using a Molecular Imager Gel Doc XR system (Bio-Rad Laboratories, Inc., Hercules, CA, USA). To accurately score the alleles and confirm different alleles observed at each locus were because of the expansion or contraction of the microsatellite region instead of indels in the flanking regions, all of the amplified isolates were sequenced using Sanger sequencing. All of the PCR products were sequenced in the forward and reverse directions using corresponding primers from the Beijing Genomics Institute (Guangzhou, China). Different fragment sizes at each locus were considered to represent different alleles based on variations in the number of repeat motifs in the amplified microsatellite region. The combination of alleles from all ten genotyped markers for each isolate was regarded as a multilocus genotype (MLG). Sequences representing all alleles generated at a locus were deposited in GenBank.
2.3. Genotype Accumulation Curve
To test the sample number of each population and to confirm whether the number of loci used in the current study was sufficient for showing the genotypic diversity of C. pseudoreteaudii, the genotype accumulation curve of each population was plotted using the R package poppr [24]. The curve was assessed by randomly sampling one locus to n − 1 loci (n is the total number of loci), which counted the number of observed genotypes. The method was repeated 1000 times without a replacement to generate a distribution for each of the 10 sampled loci [24,25]. A curve reaching a plateau indicated that the loci were able to accurately represent the maximum number of MLGs present in the analyzed dataset.
2.4. Population Genetic Diversity Analyses
For each population from the diseased leaves and soil, GENALEX version 6.5 [26] was used to calculate the total number of alleles (Na), number of effective alleles (Nef) [27], number of MLGs, and Nei’s unbiased gene diversity (Hexp) [28]. The genotypic diversity, including the evenness (E), the number of expected MLGs for the smallest sample size based on rarefaction (eMLGs), the Shannon–Wiener index of the MLG diversity (H) [29], and Stoddart and Taylor’s index of MLG diversity (G) [30], was calculated using the R package poppr [24].
2.5. Population Structure, Minimum Spanning Network, and Molecular Variance Analyses
STRUCTURE 2.3.3 [31,32] was applied to assign individuals to clusters (K) based on their allele frequency per locus using the model-based Bayesian clustering approach. The full dataset of the 97 isolates from the diseased leaf and soil populations was submitted to STRUCTURE analyses with 20 independent iterations, with the K value ranging from 1 to 10, using the admixture model of 1,000,000 Markov Chain Monte Carlo (MCMC) iterations following a burn-in period of 250,000 iterations. The optimal number of clusters was evaluated by calculating the median values of lnPr(K) and ΔK using the online STRUCTURE HARVESTER platform [33]. The CLUMPAK (Clustering Markov Packager Across K) online platform was used to visualize the cluster pattern [34]. To determine the possible evolutionary relationships among the MLGs observed in the 97 individuals across the diseased leaf and soil populations, the poppr package was used to draw a minimum spanning network (MSN) based on Bruvo’s distance [24]. To evaluate the genetic differentiation between and within populations based on the sources (diseased leaves or soil) of the isolates, an analysis of the molecular variance [35] was conducted using GENALEX version 6.5.
2.6. Population Reproduction Mode
The standardized index of association (rBarD) was employed to determine whether there was random mating (rBarD close or equal to zero) or not (rBarD significantly greater than zero) in the populations. The significance of rBarD at p < 0.05 was evaluated by comparing the observed value of rBarD with the obtained from a total of 999 randomization samplings of the same dataset [36]. The clone-corrected datasets were used for the evaluation using the ia function in the R package poppr [24].
3. Results
3.1. Microsatellite Locus Sequencing and Allele Scoring
All 97 isolates were successfully amplified with 10 pairs of SSR markers. A total of 50 alleles were detected, with the 10 markers, ranging from 2 to 11 alleles per locus (mean 5.0). The gene diversity of the 10 loci ranged from 0.48 to 0.83, based on Nei’s indices, and the evenness of the alleles ranged from 0.66 to 0.96 (Table 1). Locus CPS156 had the highest number of observed alleles (n = 11), and its gene diversity was the highest (Hexp = 0.83). Locus CPS103 had the lowest number of observed alleles (n = 2) and the lowest gene diversity (Hexp = 0.48).

Table 1.
Details of the ten polymorphic microsatellite markers for genotyping the 97 C. pseudoreteaudii isolates in this study.
3.2. Genotype Accumulation Curve
The genotype accumulation curve shows that the set of nine microsatellite markers was adequate to detect 100% of the MLGs in the diseased leaf population (Supplementary Materials Figure S1). The genotype accumulation curve of the SSR data indicates that a plateau was reached with six markers in the soil population (Supplementary Materials Figure S2). Thus, 10 SSR markers were adequate to detect the genetic diversity of the isolates in each population.
3.3. Population Genetic Diversity Analyses
The gene diversity of the diseased leaf population (Hexp = 0.636) was higher than that of the soil population (Hexp = 0.581) (Table 2). Both the Shannon–Wiener index (H) and Stoddart and Taylor’s Index (G) show that the genotype diversity of the diseased leaf population (H = 2.795; G = 12.721) was higher than that of the soil population (H = 2.102; G = 7.118). The evenness values of the diseased leaf and soil populations were 0.80 and 0.84, respectively. The results indicate that the genotypes were relatively evenly distributed in the diseased leaf and soil populations.

Table 2.
Genetic diversity of C. pseudoreteaudii populations from diseased leaves and soil.
A total of 23 MLGs were determined across all 97 of the C. pseudoreteaudii isolates. Ten genotypes were detected in the diseased leaf and soil populations. The remaining 13 genotypes were detected only in the diseased leaf population (Figure 1 and Figure 2). MLG2, MLG16, MLG18, and MLG21 were dominant genotypes in both populations.
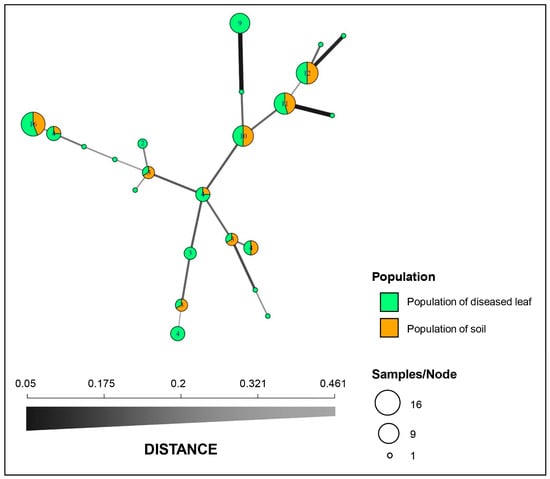
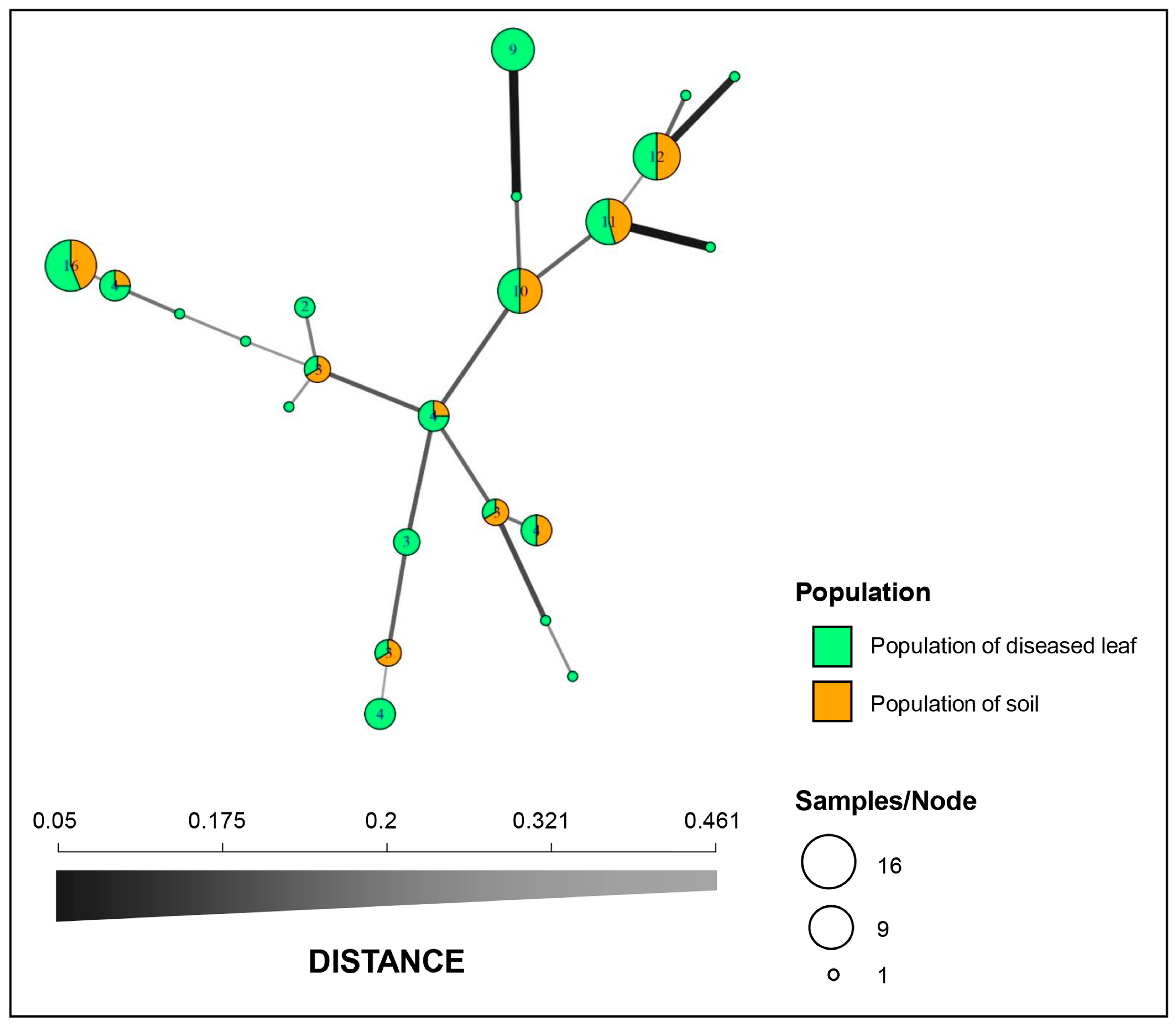
Figure 2.
Minimum spanning network (MSN) constructed with Bruvo’s genetic distance. Each node represents a single multilocus genotype (MLG), and the node size is directly proportional to the sample size. The thickness and shading of the lines represent the genetic distance between the two genotypes (a thicker line denotes a smaller genetic distance).
3.4. Population Structure, Minimum Spanning Network, and Molecular Variance Analyses
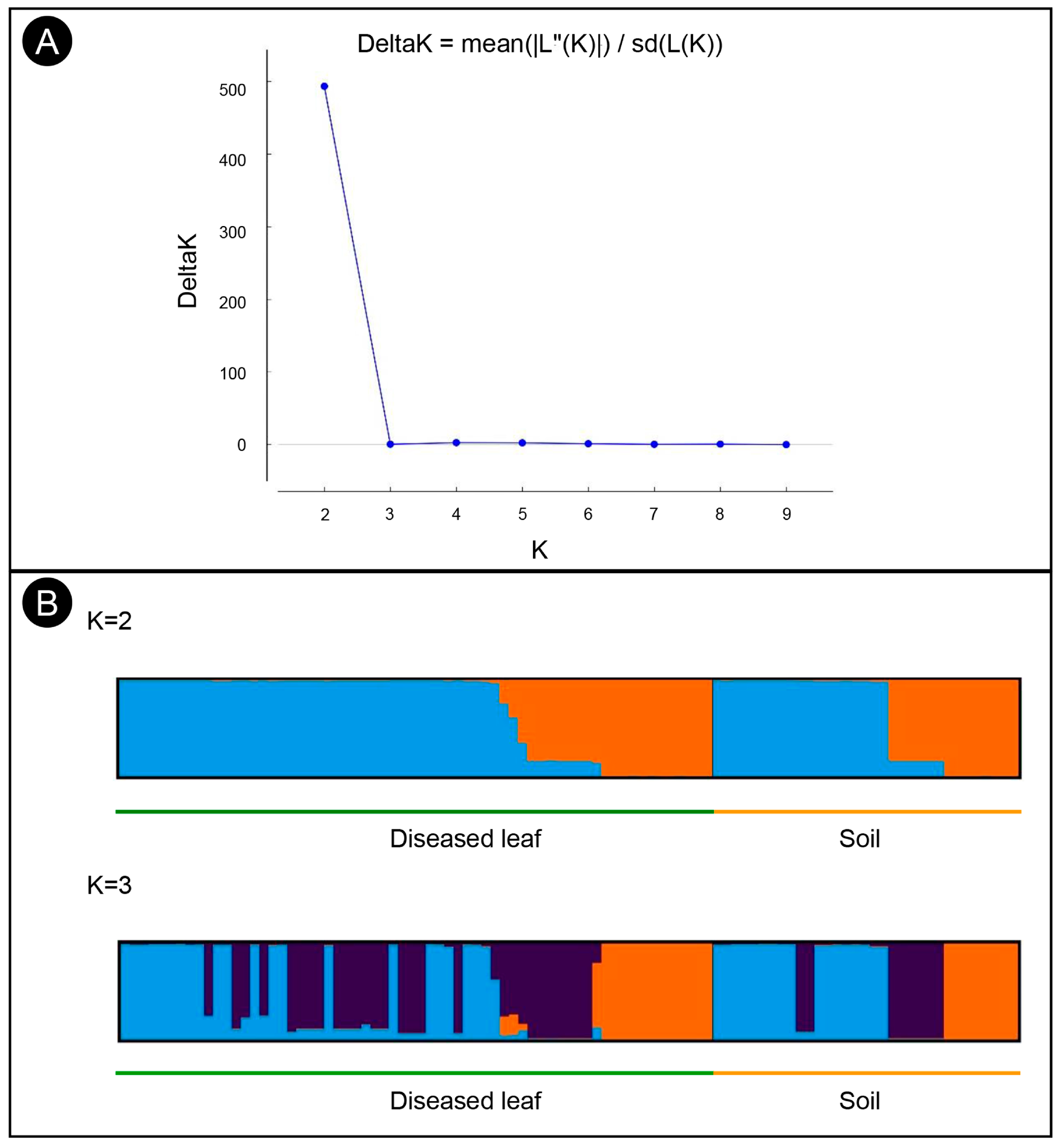
Structure analysis of the 97 isolates suggested that K = 2 was the best number of clusters for the investigated dataset (Figure 3A). A total of 62 isolates were assigned to cluster one, which included 43 isolates from the diseased leaf population and 19 isolates from the soil population. The remaining 35 isolates were assigned to cluster two, which included 21 isolates from the diseased leaf population and 14 isolates from the soil population (Figure 3B). An admixture between clusters one and two was observed within each population, and the admixture of isolates from the diseased leaf population was more heterogeneous than that from the soil population. The minimum spanning network (MSN) analysis showed that no clear clade was specifically related to either population (Figure 2). An analysis of the molecular variance suggests that there was no significant genetic differentiation between the diseased leaf and soil populations (p = 0.177). A higher genetic differentiation was observed within populations (99%), and a lower variation was observed between populations (1%) (Table 3).
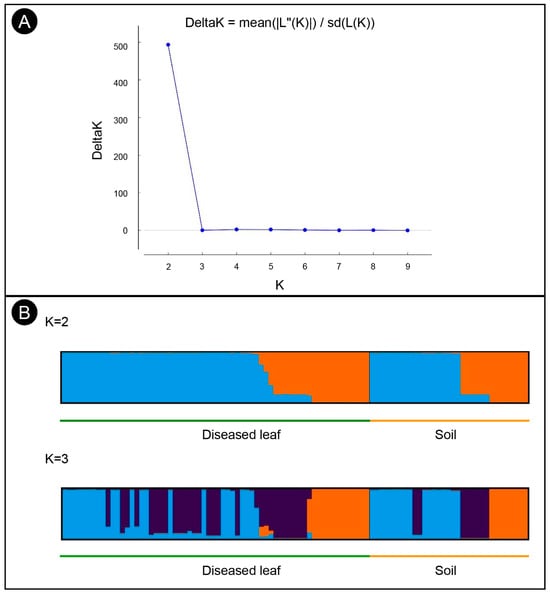
Figure 3.
Structure analyses of C. pseudoreteaudii isolates from diseased leaf and soil populations. (A) Each individual is displayed as a bar, which is divided into K colors, where K is the possible number of clusters. (B) The optimal number of genetic clusters (ΔK) = 2.

Table 3.
Analysis of molecular variance of C. pseudoreteaudii isolates from populations of diseased leaves and soil.
3.5. Population Reproduction Mode
The rBarD values of the diseased leaf and soil populations (population of diseased leaves: rBarD = 0.114, p = 0.001; population of soil: rBarD = 0.236, p = 0.001) were significantly greater than zero, suggesting that the asexual cycle or self-sterility represents the primary reproductive mode in diseased leaf and soil populations.
4. Discussion
The present study was undertaken to understand the genetic diversity and population structure of the Eucalyptus leaf blight pathogen C. pseudoreteaudii using polymorphic microsatellite markers. The genetic diversity of the isolates from diseased leaves and soil from the fungus collection site in Guangxi was high. The gene and genotype diversity of the C. pseudoreteaudii isolates from the diseased leaves were higher than those of the isolates from the soil. All of the genotypes detected in the soil were also found in the diseased leaves. No significant genetic differentiation existed between the diseased leaf and soil populations.
In the last decade, several studies on the population genetic diversity of Calonectria species have been conducted worldwide. These studies indicate the genetic diversity and population structure of the plant pathogens C. henricotiae, C. pauciramosa, and C. pseudonaviculata globally and of C. pteridis in Brazil [38,39,40,41]. Previous studies have suggested that these Calonectria spp. are clonally reproduced and that few genotypes are dominant in each Calonectria spp. [39,40,41,42]. This study indicates a high gene and genotype diversity and that multiple dominant genotypes are present in the C. pseudoreteaudii population at the fungus collection site in Guangxi, China. C. pseudoreteaudii in the investigated Eucalyptus plantation was not clonal, which increases the challenge of controlling the disease.
At the site at which C. pseudoreteaudii was isolated in this study, leaf blight caused by C. pseudoreteaudii was widely observed on the Eucalyptus trees throughout the plantation, whereas C. pseudoreteaudii was isolated only from a small proportion of the soil sampled under the diseased trees [10]. One of our aims in this study was to clarify the potential sources of C. pseudoreteaudii isolated from diseased leaves and soil. The genetic diversity, genetic structure, minimum spanning network, and molecular variance analyses of the diseased leaf and soil populations suggest that C. pseudoreteaudii in the soil was spread from the diseased leaves.
The results of previous studies consistently indicate that C. pseudoreteaudii is seldom isolated from soil in Eucalyptus plantations with leaf blight caused by this pathogen [4]. A relatively large number of C. pseudoreteaudii isolates were obtained from soils in a Eucalyptus plantation with leaf blight by Wu and Chen [10], which were collected from the Eucalyptus plantation a few days after rain. Some species of Calonectria are regarded as soil-borne fungi. These fungi can survive in soil for a long period of time because of their thick-walled microsclerotia [43]. Combining the results in this study, we hypothesized that C. pseudoreteaudii isolated from soil may drop from diseased leaves during the rainy season, and, therefore, these isolates are from pathogens on the diseased leaves and not soil-borne fungi. C. pseudoreteaudii in soil will easily die after the rainy season, especially during the dry season. It is challenging to find the appropriate season for soil sampling to obtain C. pseudoreteaudii. This hypothesis explains why C. pseudoreteaudii is seldom isolated from the soil in Eucalyptus plantations with leaf blight caused by this pathogen. Further research is needed to confirm this hypothesis.
The results of the standardized index of association in this study suggest that the asexual cycle or self-sterility was the primary reproductive mode in both the diseased leaf and soil populations. Previous studies have demonstrated that C. pseudoreteaudii is a heterothallic species [10,11]. The asexual cycle was likely the primary reproductive mode of C. pseudoreteaudii isolated in this study. Our previous study indicated that both mating types exist in C. pseudoreteaudii isolates from diseased leaves and soil [10]. The reproductive mode of C. pseudoreteaudii should be monitored regularly in the future. Sexual reproduction seems to drive potential recombination and could result in the appearance of more virulent genotypes of the pathogen. This will bring greater challenges to the disease control of such pathogens.
Calonectria pseudoreteaudii is a dominant species that causes Calonectria leaf blight on Eucalyptus trees in China. This species is widely distributed and causes disease in many Eucalyptus-planted regions in Fujian, Guangdong, Guangxi, and Hainan provinces [4,5,8,9,10,14,21]. Fungal populations with high levels of genetic variation have a greater evolutionary potential, which poses a greater “risk” to their hosts, and are likely to adapt more rapidly to resistant hosts than populations with low levels of genetic variation [44,45]. In this study, C. pseudoreteaudii was isolated from both diseased Eucalyptus leaves and soils, which indicates that C. pseudoreteaudii from both diseased leaves and soils should be considered in the disease control process. The high genetic diversity and spread of this pathogen between diseased leaves and soil may pose challenges in controlling the disease. C. pseudoreteaudii is common and easily obtained from diseased leaves of Eucalyptus plantations; however, it has been challenging to obtain C. pseudoreteaudii from the soil in these plantations. It is necessary to expand the sampling sites in China and neighboring countries and conduct a population biology study that will help us further clarify the genetic diversity, potential sources, and dispersal pathways of this important pathogen.
It is possible to screen for disease-tolerant Eucalyptus genotypes to reduce the adverse effects of leaf blight caused by Calonectria [8,10,46,47]. Fungal population with high levels of genetic variation are likely to have high variation in pathogenicity [48]. The results in this and previous studies suggested that diverse C. pseudoreteaudii isolates will be necessary for resistance screening to ensure the successful selection of Eucalyptus genotypes with durable disease resistance in China. Our previous research results indicated that differences in the tolerance to the tested C. pseudoreteaudii isolates existed among the seven tested E. urophylla × E. grandis genotypes [49]. It is necessary to test more Eucalyptus genetic materials to find C. pseudoreteaudii-tolerant Eucalyptus genotypes.
5. Conclusions
This study proved that the genetic diversity of C. pseudoreteaudii isolates from diseased leaves and soil from a fungus collection site in Guangxi was high, and that C. pseudoreteaudii in soil may spread from diseased leaves. The results in this study showed that C. pseudoreteaudii is an important pathogen causing Eucalyptus leaf blight in southern China. In the process of disease control and management of Eucalyptus leaf blight caused by Calonectria, C. pseudoreteaudii from both diseased leaves and soils in Eucalyptus plantations need to be carefully monitored. It is necessary to use diverse C. pseudoreteaudii isolates in the selection of disease-resistant Eucalyptus genotypes.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/microorganisms11112785/s1, Table S1. The 97 C. pseudoreteaudii isolates analyzed in this study and alleles scored at each locus based on the results obtained through sequencing. Figure S1. The genotype accumulation curve of the 64 C. pseudoreteaudii isolates from the diseased leaves that were genotyped with the 10 SSR primers. The x-axis shows the number of loci; the y-axis represents the number of multilocus genotypes (MLGs) observed in the dataset; and the red, dashed line shows 100% of the multilocus genotypes observed in this study. Figure S2. The genotype accumulation curve of the 33 C. pseudoreteaudii isolates from the soil that were genotyped with the 10 SSR primers. The x-axis shows the number of loci; the y-axis represents the number of multilocus genotypes (MLGs) observed in the dataset; and the red, dashed line shows 100% of the multilocus genotypes observed in this study.
Author Contributions
Conceptualization, S.C.; Formal analysis, W.W., W.L., F.L. and S.C.; Funding acquisition, S.C.; Investigation, S.C.; Methodology, W.W., W.L., F.L. and S.C.; Project administration, S.C.; Resources, W.W. and S.C.; Software, W.W., W.L. and F.L.; Supervision, S.C.; Writing—original draft, W.W.; Writing—review and editing, W.L., F.L. and S.C. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the National Key R&D Program of China (China-South Africa Forestry Joint Research Centre Project; project No. 2018YFE0120900), the National Ten-Thousand Talents Program (Project No. W03070115), and the Guangdong Top Young Talents Program in China (Project No. 20171172).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article and Supplementary Materials.
Acknowledgments
We thank QuanChao Wang and GuoQing Li (both at Research Institute of Fast-growing Trees, Chinese Academy of Forestry) for their assistance in collecting samples.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Alfenas, R.F.; Lombard, L.; Pereira, O.L.; Alfenas, A.C.; Crous, P.W. Diversity and potential impact of Calonectria species in Eucalyptus plantations in Brazil. Stud. Mycol. 2015, 80, 89–130. [Google Scholar] [CrossRef]
- Bose, R.; Banerjee, S.; Pandey, A.; Bhandari, M.S.; Barthwal, S.; Pandey, S. Calonectria leaf blight of Eucalyptus: A global review. Ann. Appl. Biol. 2023, 182, 6–28. [Google Scholar] [CrossRef]
- Li, W.W.; Chen, S.F.; Wingfield, M.J.; Duong, T.A. Calonectria queenslandica: Causal agent of Eucalyptus leaf blight in Southern China. Plant Dis. 2023, 107, 730–742. [Google Scholar] [CrossRef]
- Li, W.W.; Chen, S.F.; Wingfield, M.J.; Duong, T.A. Calonectria species associated with diseased leaves and soils in southern China Eucalyptus plantations. Phytopathol. Res. 2023, 5, 29. [Google Scholar] [CrossRef]
- Liang, X.Y.; Wang, Q.C.; Chen, S.F. Phylogeny, morphology, distribution, and pathogenicity of seven Calonectria species from leaf blighted Eucalyptus in HaiNan Island, China. Plant Dis. 2023, 107, 2579–2605. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.L.; Li, J.Q.; Wingfield, M.J.; Duong, T.A.; Wingfield, B.D.; Crous, P.W.; Chen, S.F. Reconsideration of species boundaries and proposed DNA barcodes for Calonectria. Stud. Mycol. 2020, 97, 100106. [Google Scholar] [CrossRef] [PubMed]
- Pham, N.; Barnes, I.; Chen, S.F.; Liu, F.F.; Dang, Q.; Pham, T.; Lombard, L.; Crous, P.; Wingfield, M.J. Ten new species of Calonectria from Indonesia and Vietnam. Mycologia 2019, 111, 78–102. [Google Scholar] [CrossRef]
- Wang, Q.C.; Chen, S.F. Calonectria pentaseptata causes severe leaf disease on cultivated Eucalyptus in Leizhou Peninsula of southern China. Plant Dis. 2020, 104, 493–509. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.Z.; Guo, W.S.; Ye, X.Z.; Huang, X.P.; Wu, Y.Z. Identification of Calonectria associated with Eucalyptus leaf blight in Fujian Province. J. Fujian Coll. For. 2013, 33, 176–182. [Google Scholar]
- Wu, W.X.; Chen, S.F. Species diversity, mating strategy and pathogenicity of Calonectria species from diseased leaves and soils in the Eucalyptus plantation in Southern China. J. Fungi 2021, 7, 73. [Google Scholar] [CrossRef]
- Li, J.Q.; Wingfield, B.D.; Wingfield, M.J.; Barnes, I.; Fourie, A.; Crous, P.W.; Chen, S.F. Mating genes in Calonectria and evidence for a heterothallic ancestral state. Persoonia 2020, 45, 163–176. [Google Scholar] [CrossRef]
- Lombard, L.; Zhou, X.D.; Crous, P.W.; Wingfield, B.D.; Wingfield, M.J. Calonectria species associated with cutting rot of Eucalyptus. Persoonia 2010, 24, 1–11. [Google Scholar] [CrossRef]
- Tarigan, M.; Pham, N.Q.; Jami, F.; Oliveira, L.S.; Saha, M.A.; Durán, A.; Wingfield, M.J. Calonectria species diversity on eucalypts in Indonesia. South. For. J. For. Sci. 2023, 85, 56–64. [Google Scholar] [CrossRef]
- Li, J.Q.; Wingfield, M.J.; Liu, Q.L.; Barnes, I.; Roux, J.; Lombard, L.; Crous, P.W.; Chen, S.F. Calonectria species isolated from Eucalyptus plantations and nurseries in South China. IMA Fungus 2017, 8, 259–286. [Google Scholar] [CrossRef] [PubMed]
- Bose, R.; Banerjee, S.; Negi, N.; Pandey, A.; Bhandari, M.S.; Pandey, S. Identification and pathogenicity of Calonectria pseudoreteaudii causing leaf blight of Eucalyptus––a new record for India. Physiol. Mol. Plant Pathol. 2022, 122, 101917. [Google Scholar] [CrossRef]
- Chen, C.; Liang, X.; Lin, Y.; Hsiang, T.; Xiang, M.M.; Zhang, Y. First report of leaf spot and stem blight on blueberry (Vaccinium corymbosum ‘Bluerain’) caused by Calonectria pseudoreteaudii in China. Plant Dis. 2023, 7, 1951. [Google Scholar] [CrossRef]
- Crous, P.W.; Shivas, R.G.; Wingfield, M.J.; Summerell, B.A.; Rossman, A.Y.; Alves, J.L.; Adams, G.C.; Barreto, R.W.; Bell, A.; Coutinho, M.L.; et al. Fungal Planet description sheets: 128–153. Persoonia 2012, 29, 146–201. [Google Scholar] [CrossRef]
- Jiang, G.Z.; Gao, F.; Yue, H.; He, X.Y. First report of fruit spot of Macadamia sp. caused by Calonectria pentaseptata in China. Plant Dis. 2020, 104, 575. [Google Scholar] [CrossRef]
- Jiang, Z.E.; Xie, J.; Wei, J.G.; Luo, J.; Wu, Y.J.; Luo, J.T.; Yang, X.H.; Yang, X.B. First report of husk black spot on Macadamia ternifolia caused by Calonectria pentaseptata in China. Plant Dis. 2020, 104, 1551. [Google Scholar] [CrossRef]
- Phanthavong, S.; Daly, A.; Weir, B.; Lee, D.; Park, D.; Balmas, V.; Burgess, L. First report of Calonectria pseudoreteaudii in Lao PDR associated with a leaf spot disease of Macadamia integrifolia. Australas. Plant Pathol. 2023, 52, 23–26. [Google Scholar] [CrossRef]
- Lombard, L.; Chen, S.F.; Mou, X.; Zhou, X.D.; Crous, P.W.; Wingfield, M.J. New species, hyper-diversity and potential importance of Calonectria spp. from Eucalyptus in South China. Stud. Mycol. 2015, 80, 151–188. [Google Scholar] [CrossRef] [PubMed]
- van Burik, J.A.H.; Schreckhise, R.W.; White, T.C.; Bowden, R.A.; Myerson, D. Comparison of six extraction techniques for isolation of DNA from filamentous fungi. Med. Mycol. 1998, 36, 299–303. [Google Scholar] [CrossRef] [PubMed]
- Li, W.W.; Liu, F.F.; Chen, S.F.; Wingfield, M.J.; Duong, T.A. High Genetic Diversity and Limited Regional Population Differentiation of Calonectria pseudoreteaudii Isolated from Diseased Eucalyptus Trees in Southern China; Research Institute of Fast-Growing Trees (RIFT): Zhanjiang, China; Chinese Academy of Forestry (CAF): Zhanjiang, China, 2023; manuscript in preparation. [Google Scholar]
- Kamvar, Z.N.; Tabima, J.F.; Grunwald, N.J. Poppr: An R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ 2014, 2, e281. [Google Scholar] [CrossRef] [PubMed]
- Dowling, M.E.; Bryson, P.K.; Boatwright, H.G.; Wilson, J.R.; Fan, Z.; Everhart, S.E.; Brannen, P.M.; Schnabel, G. Effect of fungicide applications on Monilinia fructicola population diversity and transposon movement. Phytopathology 2016, 106, 1504–1512. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GENALEX 6, genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Nielsen, R.; Tarpy, D.R.; Reeve, H.K. Estimating effective paternity number in social insects and the effective number of alleles in a population. Mol. Ecol. 2003, 12, 3157–3164. [Google Scholar] [CrossRef]
- Nei, M. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 1978, 89, 583–590. [Google Scholar] [CrossRef]
- Shannon, C.E. A mathematical theory of communication. ACM SIGMOBILE Mob. Comput. Commun. Rev. 2001, 5, 3–55. [Google Scholar] [CrossRef]
- Stoddart, J.A.; Taylor, J.F. Genotypic diversity: Estimation and prediction in samples. Genetics 1988, 118, 705–711. [Google Scholar] [CrossRef]
- Falush, D.; Stephens, M.; Pritchard, J.K. Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics 2003, 164, 1567–1587. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef] [PubMed]
- Earl, D.; von Holdt, B. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Kopelman, N.M.; Mayzel, J.; Jakobsson, M.; Rosenberg, N.A.; Mayrose, I. CLUMPAK: A program for identifying clustering modes and packaging population structure inferences across K. Mol. Ecol. Resour. 2015, 5, 1179–1191. [Google Scholar] [CrossRef] [PubMed]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5, genetic analysis in Excel. Population genetic software for teaching and research–an update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef] [PubMed]
- Agapow, P.M.; Burt, A. Indices of multilocus linkage disequilibrium. Mol. Ecol. Notes 2001, 1, 101102. [Google Scholar] [CrossRef]
- Grünwald, N.J.; Goodwin, S.B.; Milgroom, M.G.; Fry, W.E. Analysis of genotypic diversity data for populations of microorganisms. Phytopathology 2003, 93, 738–746. [Google Scholar] [CrossRef]
- Castroagudin, V.L.; Weiland, J.E.; Baysal-Gurel, F.; Cubeta, M.A.; Crouch, J.A. One clonal lineage of Calonectria pseudonaviculata is primarily responsible for the boxwood blight epidemic in the United States. Phytopathology 2020, 110, 1845–1853. [Google Scholar] [CrossRef]
- Freitas, R.G.; Alfenas, R.F.; Guimarães, L.M.S.; Badel, J.L.; Alfenas, A.C. Genetic diversity and aggressiveness of Calonectria pteridis in Eucalyptus spp. Plant Pathol. 2019, 68, 869–877. [Google Scholar] [CrossRef]
- LeBlanc, N.; Gehesquière, B.; Salgado-Salazar, C.; Heungens, K.; Crouch, J.A. Limited genetic diversity across pathogen populations responsible for the global emergence of boxwood blight identified using SSRs. Plant Pathol. 2019, 68, 861–868. [Google Scholar] [CrossRef]
- Li, J.Q.; Barnes, I.; Liu, F.F.; Wingfield, M.J.; Chen, S.F. Global genetic diversity and mating type distribution of Calonectria pauciramosa: An important wide-host-range plant pathogen. Plant Dis. 2021, 105, 1648–1656. [Google Scholar] [CrossRef]
- Wright, L.P.; Davis, A.J.; Wingfield, B.D.; Crous, P.W.; Brenneman, T.; Wingfield, M.J. Population structure of Cylindrocladium parasiticum infecting peanuts (Arachis hypogaea) in Georgia, USA. Eur. J. Plant Pathol. 2010, 127, 199–206. [Google Scholar] [CrossRef]
- Crous, P.W. Taxonomy and Pathology of Cylindrocladium (Calonectria) and Allied Genera; APS Press: St. Paul, MN, USA, 2002. [Google Scholar]
- McDonald, B.A.; McDermott, J.M. Population genetics of plant pathogenic fungi. Bioscience 1993, 43, 311–319. [Google Scholar] [CrossRef]
- McDonald, B.A.; Linde, C. Pathogen population genetics, evolutionary potential, and durable resistance. Annu. Rev. Phytopathol. 2002, 40, 349–379. [Google Scholar] [CrossRef] [PubMed]
- Alfenas, R.F.; Freitas, R.G.; Pereira, O.L.; Coutinho, M.M.; Zarpelon, T.G.; Cândido, T.S.; Alfenas, A.C. Screening of Corymbia and Eucalyptus species for resistance to Calonectria pteridis leaf blight. For. Pathol. 2016, 46, 76–81. [Google Scholar] [CrossRef]
- Rodas, C.A.; Lombard, L.; Gryzenhout, M.; Slipper, B.; Wingfield, M.J. Cylindrocladium blight of Eucalyptus grandis in Colombia. Australas. Plant Pathol. 2005, 34, 143–149. [Google Scholar] [CrossRef]
- Bai, Z.; Qin, Y.; Cao, K.; Du, J.; Han, Y.; Tan, Z.; Wu, G.; Tian, B.; Yang, Y.; Yu, Y.; et al. Genetic Diversity and Pathogenic Variation of the Rice False Smut Pathogen Ustilaginoidea virens from Different Rice Cultivars. Phytopathology 2023, 113, 549–558. [Google Scholar] [CrossRef]
- Wang, Q.C.; Chen, S.F. The resistances of eight Eucalyptus genotypes in southern China to Calonectria pentaseptata. Eucalypt Sci. Technol. 2020, 37, 1–9. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).