One Step Closer to Enigmatic USCα Methanotrophs: Isolation of a Methylocapsa-like Bacterium from a Subarctic Soil
Abstract
:1. Introduction
2. Materials and Methods
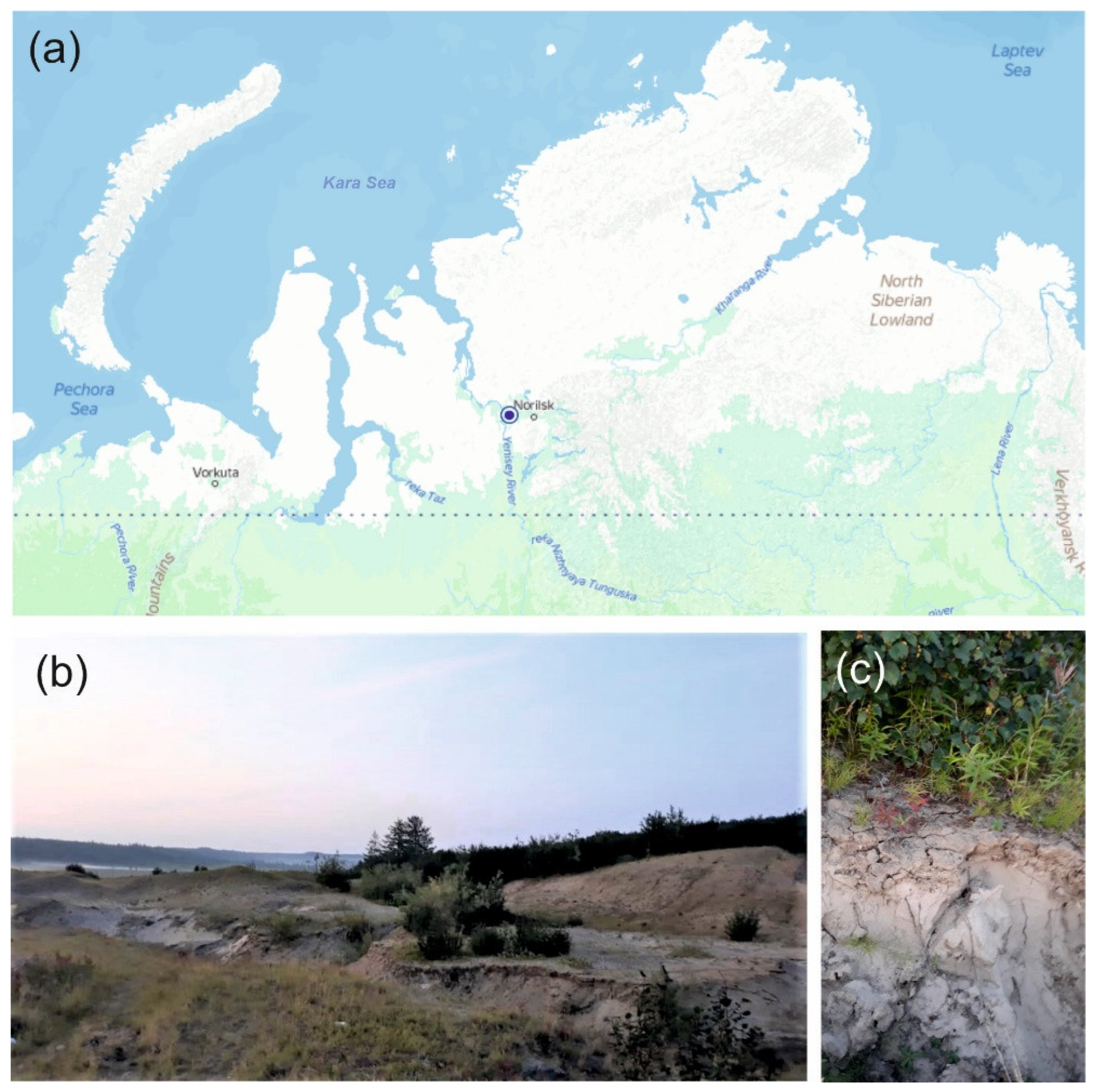
2.1. Sampling Site
2.2. Soil DNA Extraction and High-Throughput Sequencing of 16S rRNA Genes
2.3. Enrichment and Isolation of Methane-Oxidizing Bacteria
2.4. Microscopic Studies
2.5. Growth Experiments
2.6. Genome Sequencing and Annotation
2.7. Phylogenomic Analysis and Genome-Encoded Features
2.8. Global Distribution Analysis
2.9. Sequence Accession Numbers
3. Results
3.1. Prokaryote Diversity and Methanotrophic Bacteria Identified in a Subarctic Soil
3.2. Isolation and Identification of Strain D3K7
3.3. Growth Characteristics
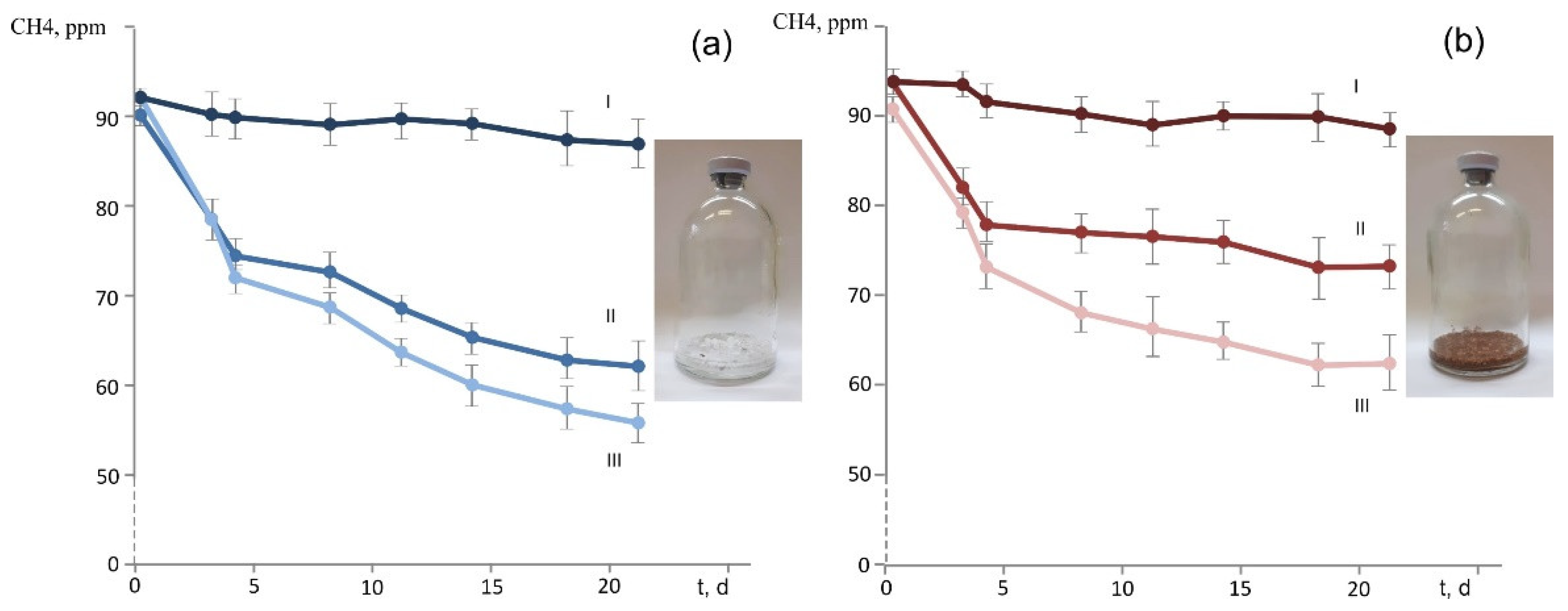
3.4. Oxidation of CH4 Provided in Low Concentrations
3.5. Genome Sequencing and Phylogenomic Placement
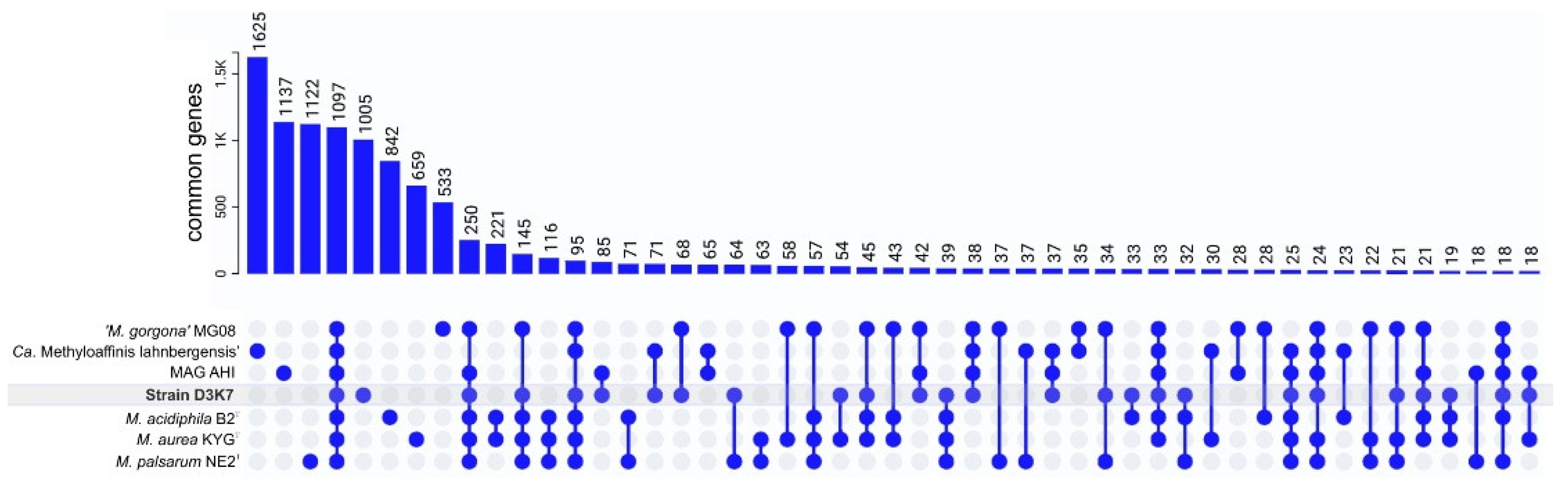
3.6. Pangenome Analysis, Shared and Unique Functional Characteristics
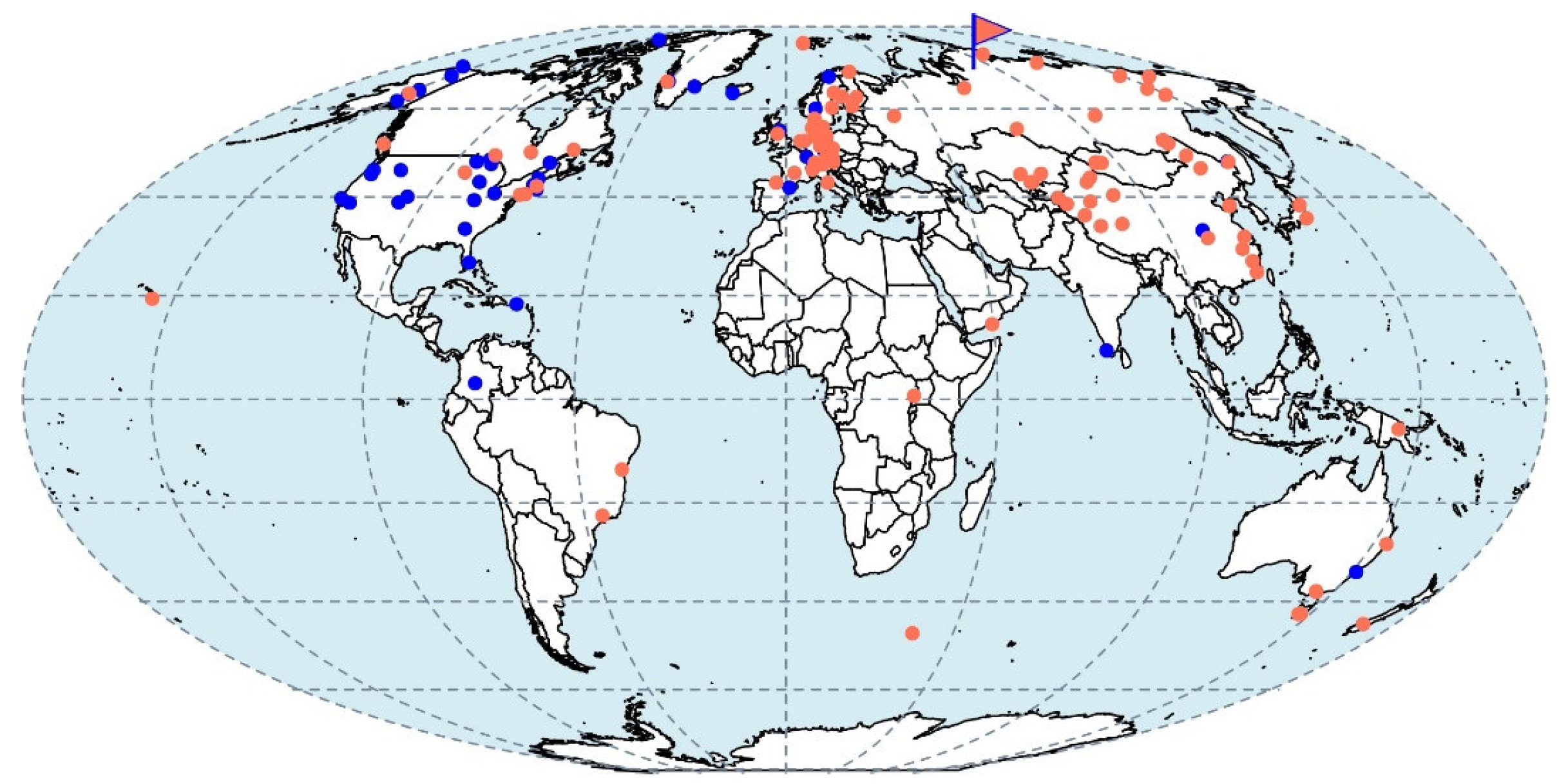
3.7. Biogeography of Strain D3K7-like Methanotrophs
4. Discussion
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dlugokencky, E.J.; Nisbet, E.G.; Fisher, R.; Lowry, D. Global atmospheric methane: Budget, changes and dangers. Philos. Trans. A Math. Phys. Eng. Sci. 2011, 369, 2058–2072. [Google Scholar] [CrossRef] [PubMed]
- Graham, D.E.; Wallenstein, M.D.; Vishnivetskaya, T.A.; Waldrop, M.P.; Phelps, T.J.; Pfiffner, S.M.; Onstott, T.C.; Whyte, L.G.; Rivkina, E.M.; Gilichinsky, D.A.; et al. Microbes in thawing permafrost: The unknown variable in the climate change equation. ISME J. 2012, 6, 709–712. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, D.M.; Koven, C.D.; Swenson, S.C.; Riley, W.J.; Slater, A.G. Permafrost thaw and resulting soil moisture changes regulate projected high-latitude CO2 and CH4 emissions. Environ. Res. Lett. 2015, 10, 094011. [Google Scholar] [CrossRef]
- Schuur, E.A.; McGuire, A.D.; Schädel, C.; Grosse, G.; Harden, J.W.; Hayes, D.J.; Hugelius, G.; Koven, C.D.; Kuhry, P.; Lawrence, D.M.; et al. Climate change and the permafrost carbon feedback. Nature 2015, 520, 171–179. [Google Scholar] [CrossRef]
- Oh, Y.; Zhuang, Q.; Liu, L.; Welp, L.R.; Lau, M.C.Y.; Onstott, T.C.; Medvigy, D.; Bruhwiler, L.; Dlugokencky, E.J.; Hugelius, G.; et al. Reduced net methane emissions due to microbial methane oxidation in a warmer Arctic. Nat. Clim. Chang. 2020, 10, 317–321. [Google Scholar] [CrossRef]
- Voigt, C.; Virkkala, A.-M.; Gosselin, G.H.; Bennett, K.A.; Black, T.A.; Detto, M.; Chevrier-Dion, C.; Guggenberger, G.; Hashmi, W.; Kohl, L.; et al. Arctic soil methane sink increases with drier conditions and higher ecosystem respiration. Nat. Clim. Chang. 2023, 13, 1095–1104. [Google Scholar] [CrossRef]
- Whalen, S.C.; Reeburgh, W.S. Consumption of atmospheric methane by tundra soils. Nature 1990, 346, 160–162. [Google Scholar] [CrossRef]
- Emmerton, C.A.; St. Louis, V.L.; Lehnherr, I.; Humphreys, E.R.; Rydz, E.; Kosolofski, H.R. The net exchange of methane with high Arctic landscapes during the summer growing season. Biogeosci. Discuss. 2014, 11, 1673–1706. [Google Scholar] [CrossRef]
- Martineau, C.; Pan, Y.; Bodrossy, L.; Yergeau, E.; Whyte, L.G.; Greer, C.W. Atmospheric methane oxidizers are present and active in Canadian high Arctic soils. FEMS Microbiol. Ecol. 2014, 89, 257–269. [Google Scholar] [CrossRef] [PubMed]
- Jørgensen, C.J.; Johansen, K.M.L.; Westergaard-Nielsen, A.; Elberling, B. Net regional methane sink in High Arctic soils of northeast Greenland. Nat. Geosci. 2015, 8, 20–23. [Google Scholar] [CrossRef]
- Lau, M.C.; Stackhouse, B.T.; Layton, A.C.; Chauhan, A.; Vishnivetskaya, T.A.; Chourey, K.; Ronholm, J.; Mykytczuk, N.C.S.; Bennett, P.C.; Lamarche-Gagnon, G.; et al. An active atmospheric methane sink in high Arctic mineral cryosols. ISME J. 2015, 9, 1880–1891. [Google Scholar] [CrossRef]
- Hanson, R.S.; Hanson, T.E. Methanotrophic bacteria. Microbiol. Rev. 1996, 60, 439–471. [Google Scholar] [CrossRef]
- Trotsenko, Y.A.; Murrell, J.C. Metabolic aspects of aerobic obligate methanotrophy. Adv. Appl. Microbiol. 2008, 63, 183–229. [Google Scholar] [CrossRef]
- Chistoserdova, L.; Lidstrom, M.E. Aerobic methylotrophic prokaryotes. Prokaryotes 2006, 2, 618. [Google Scholar]
- Dedysh, S.N.; Knief, C. Diversity and phylogeny of described aerobic methanotrophs. In Methane Biocatalysis: Paving the Way to Sustainability; Springer: Cham, Switzerland, 2018; pp. 17–42. [Google Scholar] [CrossRef]
- Khmelenina, V.N.; But, S.Y.; Rozova, O.N.; Trotsenko, Y.A. Metabolic Features of Aerobic Methanotrophs: News and Views. Curr. Issues Mol. Biol. 2019, 33, 85–100. [Google Scholar] [CrossRef]
- Knief, C. Diversity and habitat preferences of cultivated and uncultivated aerobic methanotrophic bacteria evaluated based on pmoA as molecular marker. Front. Microbiol. 2015, 6, 1346. [Google Scholar] [CrossRef] [PubMed]
- Singleton, C.M.; McCalley, C.K.; Woodcroft, B.J.; Boyd, J.A.; Evans, P.N.; Hodgkins, S.B.; Chanton, J.P.; Frolking, S.; Crill, P.M.; Saleska, S.R.; et al. Methanotrophy across a natural permafrost thaw environment. ISME J. 2018, 10, 2544–2558. [Google Scholar] [CrossRef] [PubMed]
- Belova, S.E.; Danilova, O.V.; Ivanova, A.A.; Merkel, A.Y.; Dedysh, S.N. Methane-oxidizing communities in Lichen-dominated forested tundra are composed exclusively of high-affinity USCα methanotrophs. Microorganisms 2020, 8, 2047. [Google Scholar] [CrossRef] [PubMed]
- Knief, C.; Lipski, A.; Dunfield, P.F. Diversity and activity of methanotrophic bacteria in different upland soils. Appl. Environ. Microbiol. 2003, 69, 6703–6714. [Google Scholar] [CrossRef] [PubMed]
- Holmes, A.J.; Roslev, P.; McDonald, I.R.; Iversen, N.; Henriksen, K.; Murrell, J.C. Characterization of methanotrophic bacterial populations in soils showing atmospheric methane uptake. Appl. Environ. Microbiol. 1999, 65, 3312–3318. [Google Scholar] [CrossRef]
- Dedysh, S.N.; Horz, H.P.; Dunfield, P.F.; Liesack, W. A novel pmoA lineage represented by the acidophilic methanotrophic bacterium Methylocapsa acidiphila correction of acidophila B2. Arch. Microbiol. 2001, 177, 117–121. [Google Scholar] [CrossRef] [PubMed]
- Dedysh, S.N.; Khmelenina, V.N.; Suzina, N.E.; Trotsenko, Y.A.; Semrau, J.D.; Liesack, W.; Tiedje, J.M. Methylocapsa acidiphila gen. nov., sp. nov., a novel methane-oxidizing and dinitrogen-fixing acidophilic bacterium from Sphagnum bog. Int. J. Syst. Evol. Microbiol. 2002, 52, 251–261. [Google Scholar] [CrossRef] [PubMed]
- Henckel, T.; Jäckel, U.; Schnell, S.; Conrad, R. Molecular analyses of novel methanotrophic communities in forest soil that oxidize atmospheric methane. Appl. Environ. Microbiol. 2000, 66, 1801–1808. [Google Scholar] [CrossRef] [PubMed]
- Degelmann, D.M.; Borken, W.; Drake, H.L.; Kolb, S. Different atmospheric methane-oxidizing communities in European beech and Norway spruce soils. Appl. Environ. Microbiol. 2010, 76, 3228–3235. [Google Scholar] [CrossRef] [PubMed]
- Sabrekov, A.F.; Danilova, O.V.; Terentieva, I.E.; Ivanova, A.A.; Belova, S.E.; Litti, Y.V.; Glagolev, M.V.; Dedysh, S.N. Atmospheric methane consumption and methanotroph communities in West Siberian boreal upland forest ecosystems. Forests 2021, 12, 1738. [Google Scholar] [CrossRef]
- Pratscher, J.; Vollmers, J.; Wiegand, S.; Dumont, M.G.; Kaster, A.K. Unravelling the identity, metabolic potential and global biogeography of the atmospheric methane-oxidizing Upland Soil Cluster α. Environ. Microbiol. 2018, 20, 1016–1029. [Google Scholar] [CrossRef]
- Tveit, A.T.; Hestnes, A.G.; Robinson, S.L.; Schintlmeister, A.; Dedysh, S.N.; Jehmlich, N.; von Bergen, M.; Herbold, C.; Wagner, M.; Richter, A.; et al. Widespread soil bacterium that oxidizes atmospheric methane. Proc. Natl. Acad. Sci. USA 2019, 23, 8515–8524. [Google Scholar] [CrossRef]
- Frey, B.; Rime, T.; Phillips, M.; Stierli, B.; Hajdas, I.; Widmer, F.; Hartmann, M. Microbial diversity in European alpine permafrost and active layers. FEMS Microbiol. Ecol. 2016, 92, fiw018. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Subramanian, S.; Faith, J.J.; Gevers, D.; Gordon, J.I.; Knight, R.; Mills, D.A.; Caporaso, J.G. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods 2013, 10, 57–59. [Google Scholar] [CrossRef]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Dedysh, S.N.; Didriksen, A.; Danilova, O.V.; Belova, S.E.; Liebner, S.; Svenning, M.M. Methylocapsa palsarum sp. nov., a methanotroph isolated from a subArctic discontinuous permafrost ecosystem. Int. J. Syst. Evol. Microbiol. 2015, 65, 3618–3624. [Google Scholar] [CrossRef]
- Dedysh, S.N.; Beletsky, A.V.; Ivanova, A.A.; Kulichevskaya, I.S.; Suzina, N.E.; Philippov, D.A.; Rakitin, A.L.; Mardanov, A.V.; Ravin, N.V. Wide distribution of Phycisphaera-like planctomycetes from WD2101 soil group in peatlands and genome analysis of the first cultivated representative. Environ. Microbiol. 2021, 23, 1510–1526. [Google Scholar] [CrossRef]
- Wilson, K. Preparation of genomic DNA from bacteria. Curr. Protoc. Mol. Biol. 2001, 56, 2–4. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing 454 reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial 456 genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment 463 tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. 466 BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; O’Neill, K.R.; Haft, D.H.; DiCuccio, M.; Chetvernin, V.; Badretdin, A.; Coulouris, G.; Chitsaz, F.; Derbyshire, M.K.; Durkin, A.S.; et al. 469 RefSeq: Expanding the Prokaryotic Genome Annotation Pipeline reach with protein family 470 model curation. Nucleic Acids Res. 2021, 49, D1020–D1028. [Google Scholar] [CrossRef]
- Parks, D.H.; Chuvochina, M.; Chaumeil, P.A.; Rinke, C.; Mussig, A.J.; Hugenholtz, P. A complete domain-to-species taxonomy for Bacteria and Archaea. Nat. Biotechnol. 2020, 38, 1079–1086. [Google Scholar] [CrossRef] [PubMed]
- Chaumeil, P.A.; Mussig, A.J.; Hugenholtz, P.; Parks, D.H. GTDB-Tk: A toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics 2020, 36, 1925–1927. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Page, A.J.; Cummins, C.A.; Hunt, M.; Wong, V.K.; Reuter, S.; Holden, M.T.G.; Fookes, M.; Falush, D.; Keane, J.A.; Parkhill, J. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef]
- Lagkouvardos, I.; Joseph, D.; Kapfhammer, M.; Giritli, S.; Horn, M.; Haller, D.; Clavel, T. IMNGS: A comprehensive open resource of processed 16S rRNA microbial profiles for ecology and diversity studies. Sci. Rep. 2016, 6, 33721. [Google Scholar] [CrossRef]
- Belova, S.E.; Kulichevskaya, I.S.; Bodelier, P.L.E.; Dedysh, S.N. Methylocystis bryophila sp. nov., a facultatively methanotrophic bacterium from acidic Sphagnum peat, and emended description of the genus Methylocystis (ex Whittenbury et al. 1970) Bowman et al. 1993. Int. J. Syst. Evol. Microbiol. 2013, 63, 1096–1104. [Google Scholar] [CrossRef] [PubMed]
- Dunfield, P.F.; Belova, S.E.; Vorob’ev, A.V.; Cornish, S.L.; Dedysh, S.N. Methylocapsa aurea sp. nov., a facultative methanotroph possessing a particulate methane monooxygenase, and emended description of the genus Methylocapsa. Int. J. Syst. Evol. Microbiol. 2010, 60, 2659–2664. [Google Scholar] [CrossRef]
- Rusley, C.; Onstott, T.C.; Vishnivetskaya, T.A.; Layton, A.; Chauhan, A.; Pfiffner, S.M.; Whyte, L.G.; Lau, M.C.Y. Metagenome-Assembled Genome of USCα AHI, a Potential High-Affinity Methanotroph from Axel Heiberg Island, Canadian High Arctic. Microbiol. Resour. Announc. 2019, 8, e01178-19. [Google Scholar] [CrossRef]
- Ji, M.; Greening, C.; Vanwonterghem, I.; Carere, C.R.; Bay, S.K.; Steen, J.A.; Montgomery, K.; Lines, T.; Beardall, J.; van Dorst, J.; et al. Atmospheric trace gases support primary production in Antarctic desert surface soil. Nature 2017, 552, 400–403. [Google Scholar] [CrossRef]
- Bale, N.J.; Rijpstra, W.I.C.; Sahonero-Canavesi, D.X.; Oshkin, I.Y.; Belova, S.E.; Dedysh, S.N.; Sinninghe Damsté, J.S. Fatty Acid and Hopanoid Adaption to Cold in the Methanotroph Methylovulum psychrotolerans. Front. Microbiol. 2019, 10, 589. [Google Scholar] [CrossRef] [PubMed]
- Mirzoyan, S.; Romero-Pareja, P.M.; Coello, M.D.; Trchounian, A.; Trchounian, K. Evidence for hydrogenase-4 catalyzed biohydrogen production in Escherichia coli. Int. J. Hydrogen Energy 2017, 42, 21697–21703. [Google Scholar] [CrossRef]
- Tabita, F.R.; Satagopan, S.; Hanson, T.E.; Kreel, N.E.; Scott, S.S. Distinct form I, II, III, and IV Rubisco proteins from the three kingdoms of life provide clues about Rubisco evolution and structure/function relationships. J. Exp. Bot. 2008, 59, 1515–1524. [Google Scholar] [CrossRef]
- Hanson, T.E.; Tabita, F.R. A ribulose-1,5-bisphosphate carboxylase/oxygenase (RubisCO)-like protein from Chlorobium tepidum that is involved with sulfur metabolism and the response to oxidative stress. Proc. Natl. Acad. Sci. USA 2001, 98, 4397–4402. [Google Scholar] [CrossRef] [PubMed]
- Imker, H.J.; Singh, J.; Warlick, B.P.; Tabita, F.R.; Gerlt, J.A. Mechanistic diversity in the RuBisCO superfamily: A novel isomerization reaction catalyzed by the RuBisCO-like protein from Rhodospirillum rubrum. Biochemistry 2008, 47, 11171–11173. [Google Scholar] [CrossRef]
- Pratscher, J.; Dumont, M.G.; Conrad, R. Assimilation of acetate by the putative atmospheric methane oxidizers belonging to the USCα clade. Environ. Microbiol. 2011, 13, 2692–2701. [Google Scholar] [CrossRef]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Danilova, O.V.; Oshkin, I.Y.; Belova, S.E.; Miroshnikov, K.K.; Ivanova, A.A.; Dedysh, S.N. One Step Closer to Enigmatic USCα Methanotrophs: Isolation of a Methylocapsa-like Bacterium from a Subarctic Soil. Microorganisms 2023, 11, 2800. https://doi.org/10.3390/microorganisms11112800
Danilova OV, Oshkin IY, Belova SE, Miroshnikov KK, Ivanova AA, Dedysh SN. One Step Closer to Enigmatic USCα Methanotrophs: Isolation of a Methylocapsa-like Bacterium from a Subarctic Soil. Microorganisms. 2023; 11(11):2800. https://doi.org/10.3390/microorganisms11112800
Chicago/Turabian StyleDanilova, Olga V., Igor Y. Oshkin, Svetlana E. Belova, Kirill K. Miroshnikov, Anastasia A. Ivanova, and Svetlana N. Dedysh. 2023. "One Step Closer to Enigmatic USCα Methanotrophs: Isolation of a Methylocapsa-like Bacterium from a Subarctic Soil" Microorganisms 11, no. 11: 2800. https://doi.org/10.3390/microorganisms11112800





