Identification, Characterization, and Production Optimization of 6-Methoxy-1H-Indole-2-Carboxylic Acid Antifungal Metabolite Produced by Bacillus toyonensis Isolate OQ071612
Abstract
:1. Introduction
2. Materials and Methods
2.1. Collection and Phylogenetic Analysis of the F1 Strain
2.2. Evaluation of the Antifungal Activity
2.3. Evaluation of the Fungicidal Activity
2.4. Active Metabolite Production
2.5. Testing the Intracellular and Extracellular Nature of the Antifungal Metabolites
2.6. Extraction and Purification
2.7. Physicochemical Properties of the Antifungal Metabolite(s)
2.8. Spectroscopic Analysis
2.9. Factors Affecting Antifungal Metabolite Production
2.10. Production Optimization Using RSM
2.11. Confirmation of the Model Used for Optimization
2.12. Statistical Analysis
3. Results
3.1. Antifungal Activity and the Identification of the F1 Isolate
3.2. Characterization of the Antifungal Metabolite
3.3. Solvent Extraction
3.4. Physicochemical Properties of the Antifungal Metabolite(s)
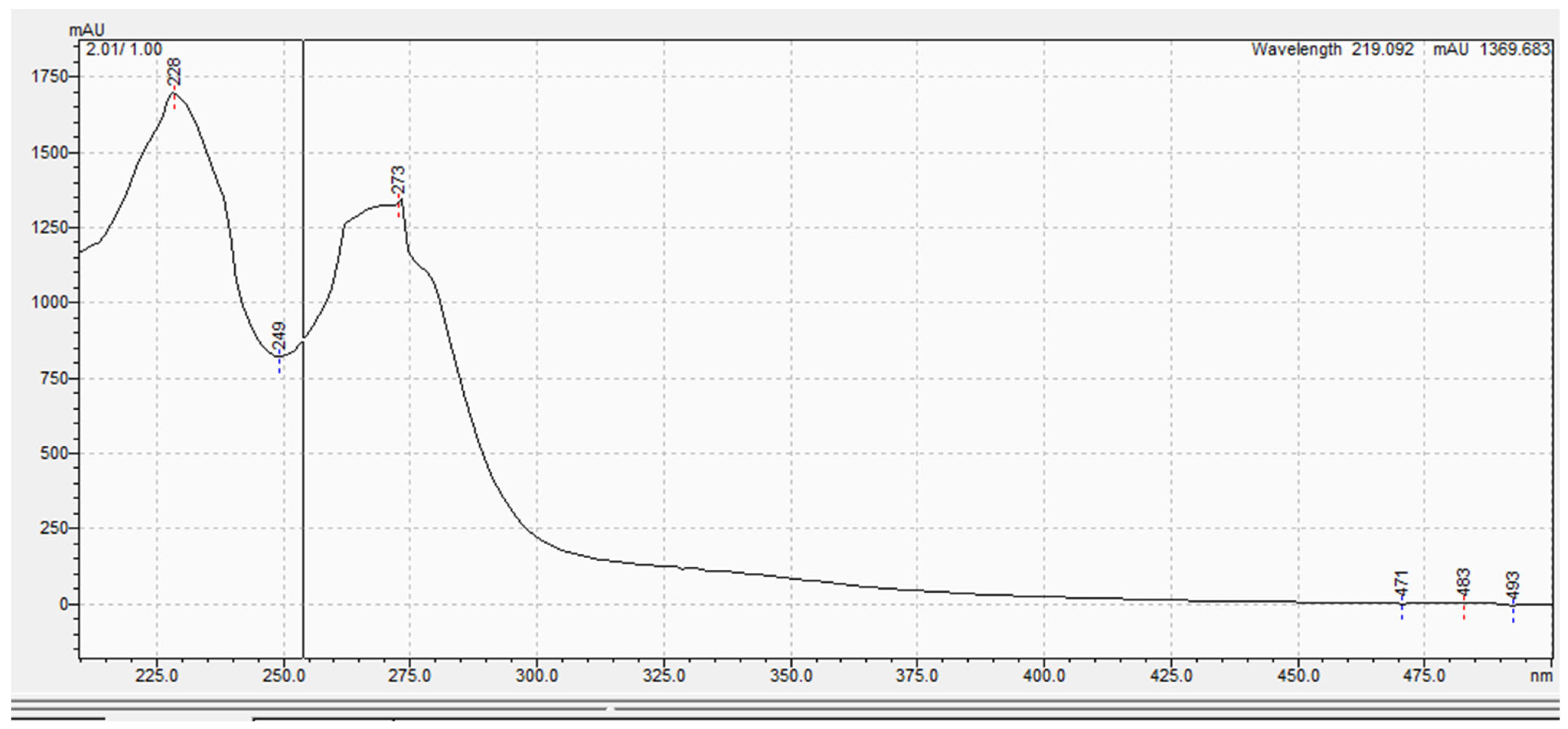
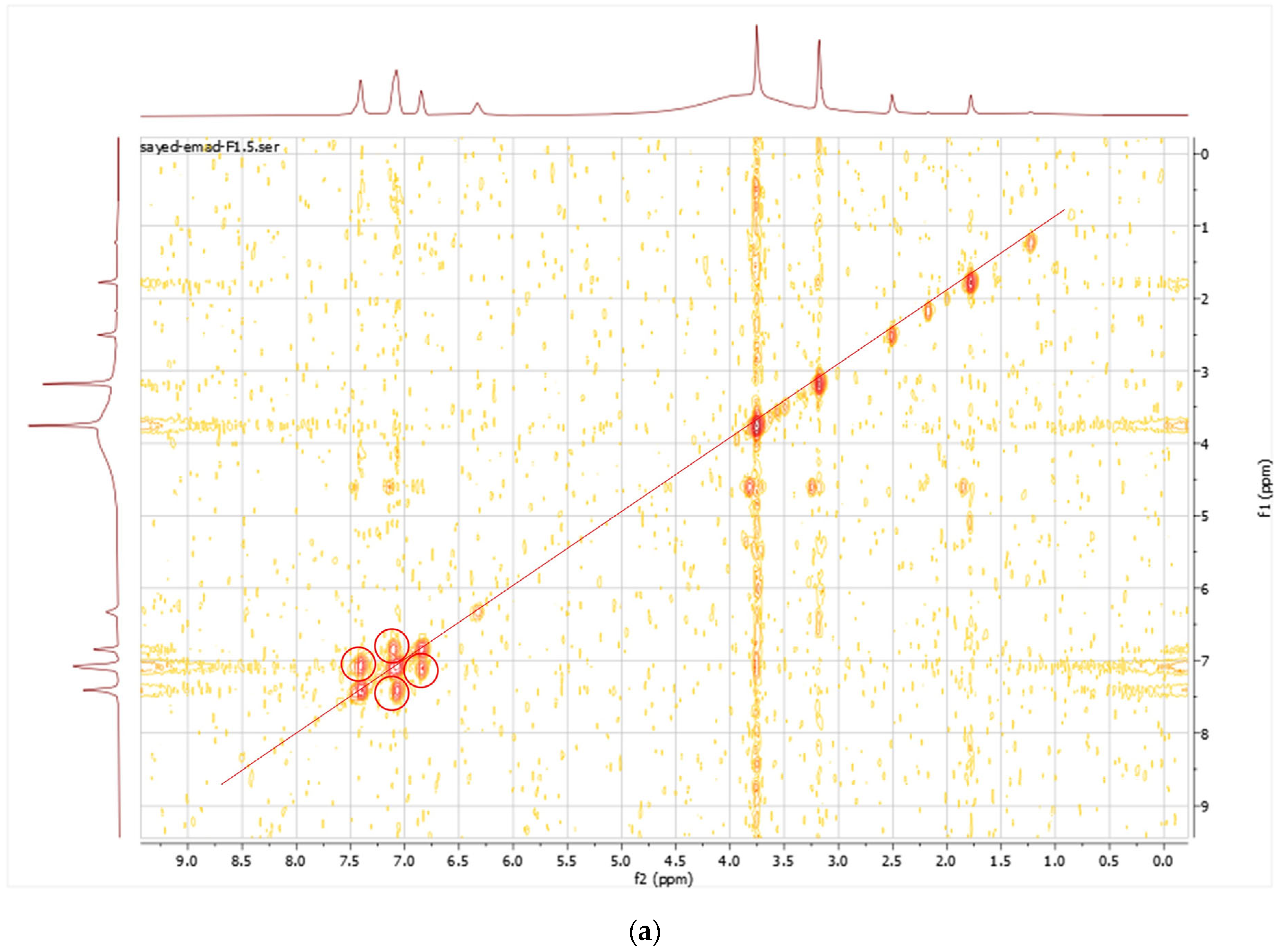
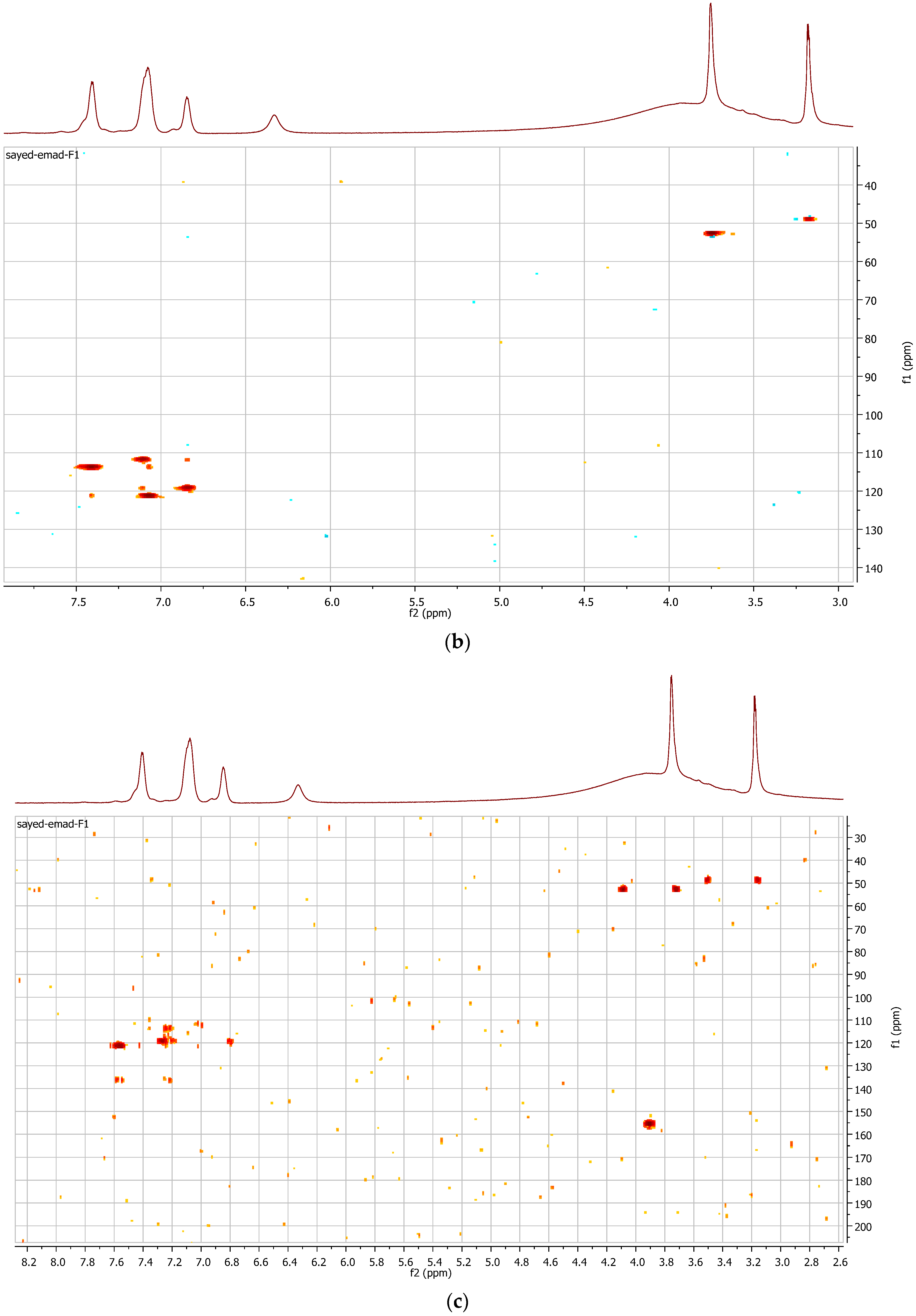
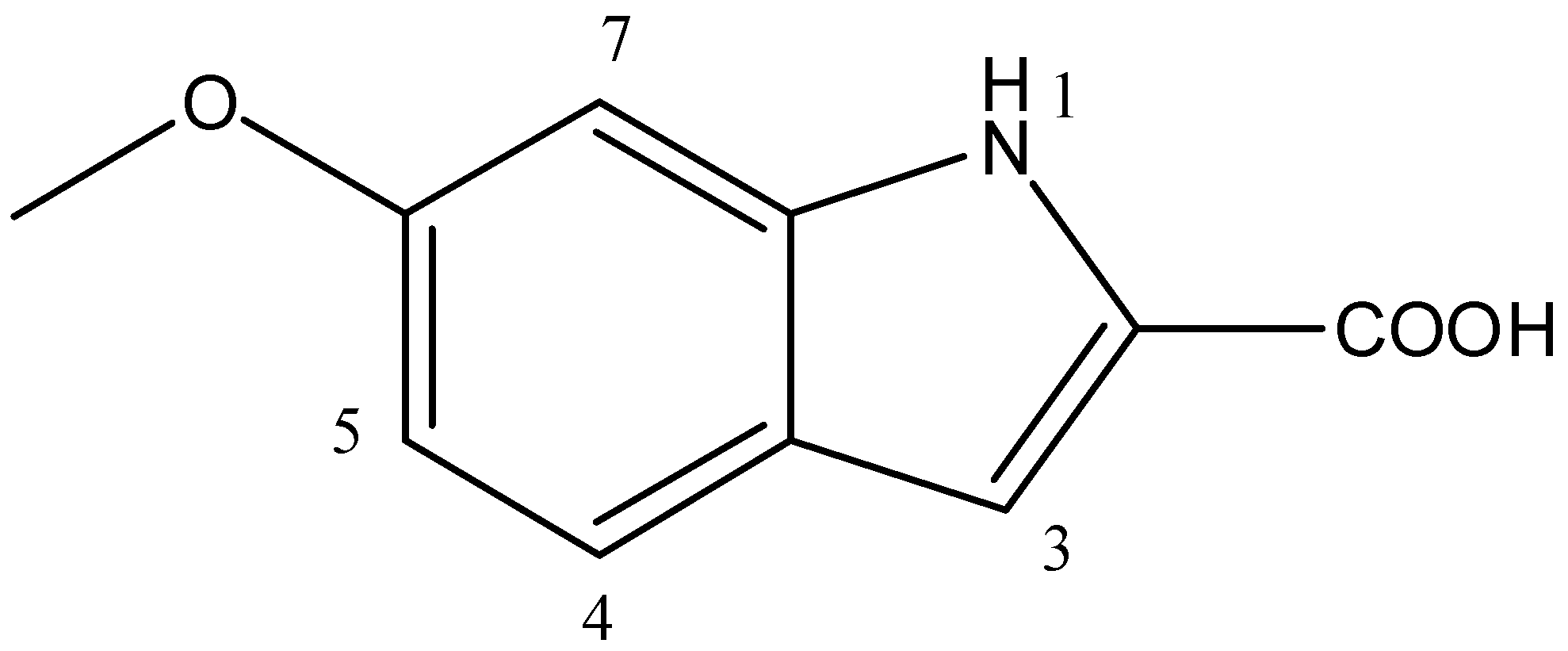
3.5. Spectroscopic Analysis
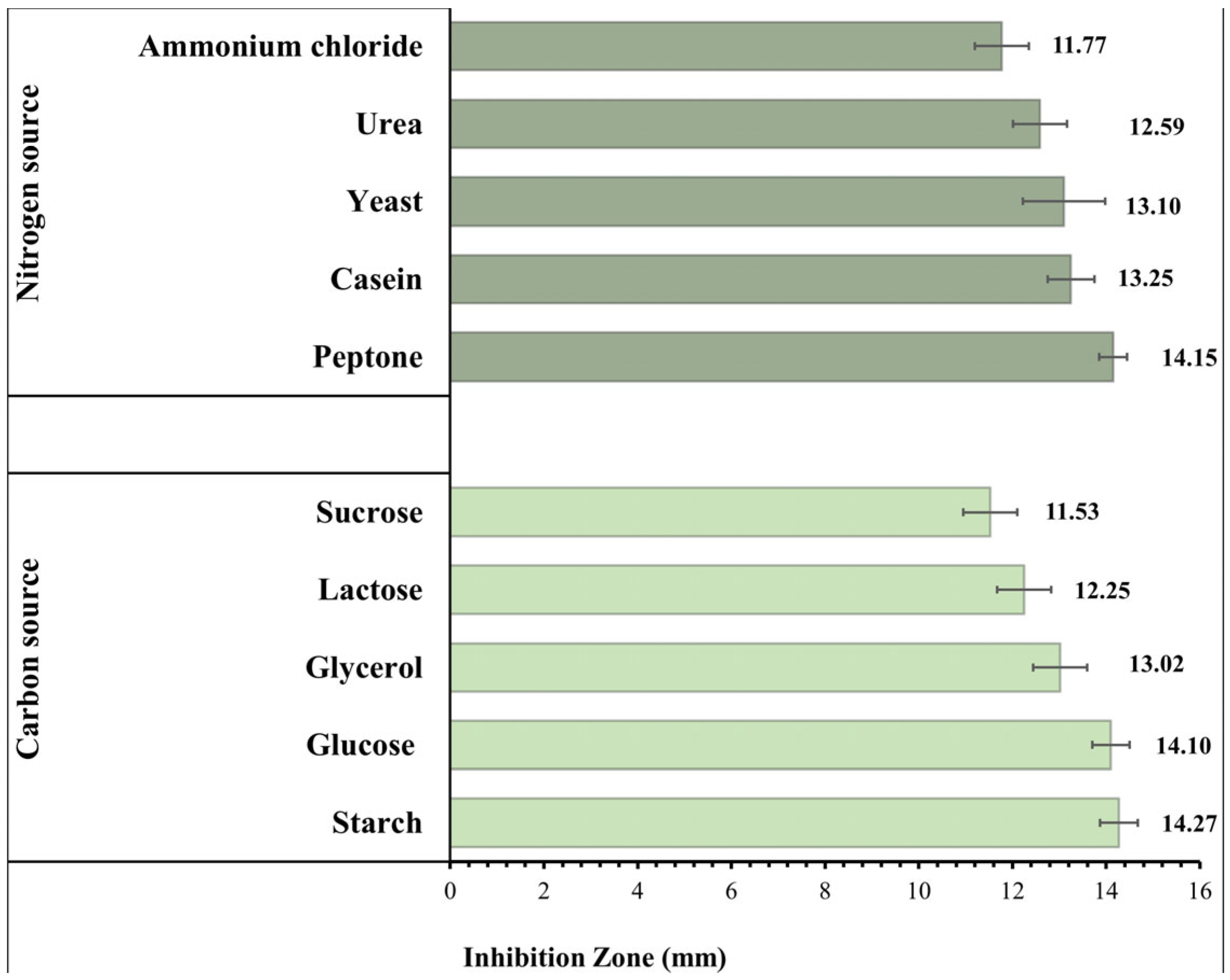
3.6. Effect of the Composition of the Culture Media
3.7. Optimization of the Bacterial Culture Conditions Using RSM
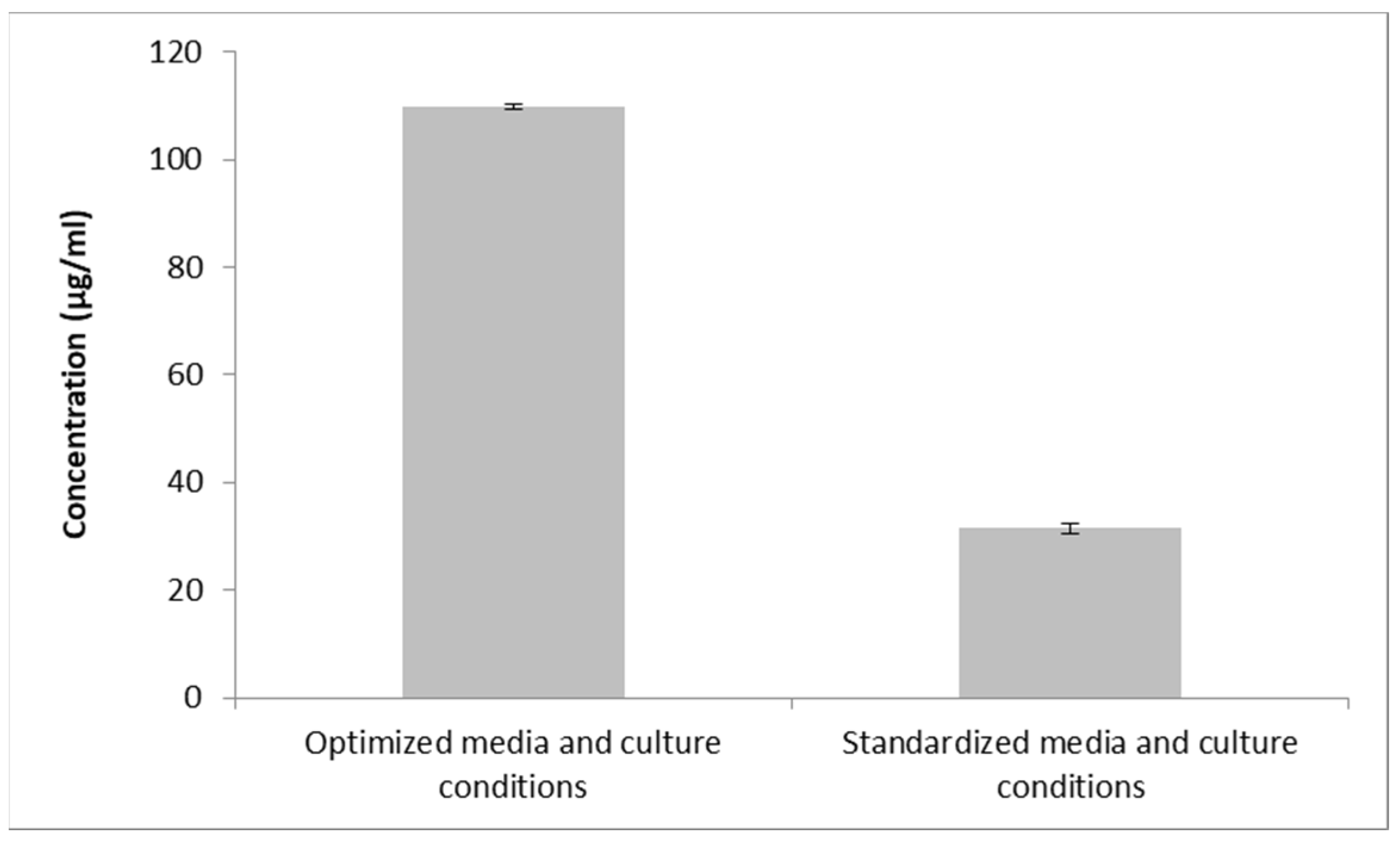
3.8. Confirmatory Experiment Using Optimal Conditions
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Crits-Christoph, A.; Diamond, S.; Butterfield, C.N.; Thomas, B.C.; Banfield, J.F. Novel soil bacteria possess diverse genes for secondary metabolite biosynthesis. Nature 2018, 558, 440–444. [Google Scholar] [CrossRef]
- Baazeem, A.; Almanea, A.; Manikandan, P.; Alorabi, M.; Vijayaraghavan, P.; Abdel-Hadi, A. In vitro antibacterial, antifungal, nematocidal and growth promoting activities of Trichoderma hamatum FB10 and its secondary metabolites. J. Fungi 2021, 7, 331. [Google Scholar] [CrossRef]
- Innes, A. The potential biocontrol agent Pseudomonas antimicrobica inhibits germination of conidia and outgrowth of Botrytis cinerea. Lett. Appl. Microbiol. 2001, 32, 346–348. [Google Scholar]
- Casadevall, A. Fungal diseases in the 21st century: The near and far horizons. Pathog. Immun. 2018, 3, 183. [Google Scholar] [CrossRef]
- Rodrigues, M.L.; Albuquerque, P.C. Searching for a change: The need for increased support for public health and research on fungal diseases. PLoS Negl. Trop. Dis. 2018, 12, e0006479. [Google Scholar] [CrossRef]
- Henriques, M.; Williams, D. Pathogenesis and Virulence of Candida albicans and Candida glabrata. Pathogens 2020, 9, 752. [Google Scholar] [CrossRef]
- Aoki, W.; Ueda, M. Characterization of antimicrobial peptides toward the development of novel antibiotics. Pharmaceuticals 2013, 6, 1055–1081. [Google Scholar] [CrossRef] [PubMed]
- Gangneux, J.-P.; Bougnoux, M.-E.; Dannaoui, E.; Cornet, M.; Zahar, J. Invasive fungal diseases during COVID-19: We should be prepared. J. De Mycol. Medicale 2020, 30, 100971. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Yu, Y.; Xu, J.; Shu, H.; Liu, H.; Wu, Y.; Zhang, L.; Yu, Z.; Fang, M.; Yu, T. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: A single-centered, retrospective, observational study. Lancet Respir. Med. 2020, 8, 475–481. [Google Scholar] [CrossRef]
- Chen, N.; Zhou, M.; Dong, X.; Qu, J.; Gong, F.; Han, Y.; Qiu, Y.; Wang, J.; Liu, Y.; Wei, Y. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study. Lancet 2020, 395, 507–513. [Google Scholar] [CrossRef] [PubMed]
- Dam, P.; Cardoso, M.H.; Mandal, S.; Franco, O.L.; Sağıroğlu, P.; Polat, O.A.; Kokoglu, K.; Mondal, R.; Mandal, A.K.; Ocsoy, I. Surge of mucormycosis during the COVID-19 pandemic. Travel Med. Infect. Dis. 2023, 52, 102557. [Google Scholar] [CrossRef] [PubMed]
- Zand, F.; Vakili, H.; Asmarian, N.; Masjedi, M.; Sabetian, G.; Nikandish, R.; Shafiee, E.; Tabatabaei Esfehani, A.; Azadi, F.; Sanaei Dashti, A. Unintended impact of COVID-19 pandemic on the rate of catheter related nosocomial infections and incidence of multiple drug resistance pathogens in three intensive care units not allocated to COVID-19 patients in a large teaching hospital. BMC Infect. Dis. 2023, 23, 11. [Google Scholar] [CrossRef]
- Szabo, B.G.; Lakatos, B.; Bobek, I.; Szabo, E.; Szlavik, J.; Vályi-Nagy, I. Invasive fungal infections among critically ill adult COVID-19 patients: First experiences from the national centre in Hungary. J. Med. Mycol. 2021, 31, 101198. [Google Scholar] [CrossRef] [PubMed]
- Vittoria, M.; Saggese, A.; Barletta, G.D.G.; Castaldi, S.; Isticato, R.; Baccigalupi, L.; Ricca, E. Sporulation efficiency and spore quality in a human intestinal isolate of Bacillus cereus. Res. Microbiol. 2023, 104030. [Google Scholar] [CrossRef]
- Vaca, J.; Ortiz, A.; Sansinenea, E. A study of bacteriocin like substances comparison produced by different species of Bacillus related to B. cereus group with specific antibacterial activity against foodborne pathogens. Arch. Microbiol. 2023, 205, 13. [Google Scholar] [CrossRef] [PubMed]
- Valizadeh, S.; Enayatizamir, N.; Ghomsheh, H.N.; Motamedi, H.; Moghadam, B.K. Characterization of the biosurfactant production and enzymatic potential of bacteria isolated from an oil-contaminated saline soil. Int. Microbiol. 2023, 26, 529–542. [Google Scholar] [CrossRef]
- Abdellatif, A.A.M.; Abd-Elrahman, T.M.A.; Sayed, M.A.E.; Ragab, A.A.E.-A.H.; Ibrahim, D.S.-E.S.-E. Isolation, Identification and Biocontrol Activity of Novel Chitinolytic Bacteria against Meloidogyne incognita Infecting Capsicum annuum L. Casp. J. Environ. Sci. 2023, 21, 49–68. [Google Scholar]
- Agustiani, R.D.; Oedjijono, O.; Rahmani, N.; Ekowati, N. Isolation and Characterization of Rhizospheric Bacteria Associated with Canna Plant for Production of Maltooligosaccharide Amylase. J. Trop. Biodivers. Biotechnol. 2023, 8, 78346. [Google Scholar] [CrossRef]
- Jiang, Y.-H.; Xin, W.-G.; Yang, L.-Y.; Ying, J.-P.; Zhao, Z.-S.; Lin, L.-B.; Li, X.-Z.; Zhang, Q.-L. A novel bacteriocin against Staphylococcus aureus from Lactobacillus paracasei isolated from Yunnan traditional fermented yogurt: Purification, antibacterial characterization, and antibiofilm activity. J. Dairy Sci. 2022, 105, 2094–2107. [Google Scholar] [CrossRef]
- Wang, J.; Xu, H.; Liu, S.; Song, B.; Liu, H.; Li, F.; Deng, S.; Wang, G.; Zeng, H.; Zeng, X. Toyoncin, a novel leaderless bacteriocin that is produced by Bacillus toyonensis XIN-YC13 and specifically targets B. cereus and Listeria monocytogenes. Appl. Environ. Microbiol. 2021, 87, e00185-21. [Google Scholar] [CrossRef]
- Sumiya, T.; Ishigaki, M.; Oh, K. Synthesis of imidazole and indole hybrid molecules and antifungal activity against rice blast. Int. J. Chem. Eng. Appl. 2017, 8, 233–236. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, J.-D.; Liu, C.-X.; Yuan, J.-H.; Wang, X.-J.; Xiang, W.-S. A new prenylated indole derivative from endophytic actinobacteria Streptomyces sp. neau-D50. Nat. Prod. Res. 2014, 28, 431–437. [Google Scholar] [CrossRef] [PubMed]
- Ali, G. Identification of volatile organic compounds produced by algae. Egypt. J. Phycol. 2004, 5, 71–81. [Google Scholar] [CrossRef]
- Gutierrez-Lugo, M.-T.; Woldemichael, G.M.; Singh, M.P.; Suarez, P.A.; Maiese, W.M.; Montenegro, G.; Timmermann, B.N. Isolation of three new naturally occurring compounds from the culture of Micromonospora sp. P1068. Nat. Prod. Res. 2005, 19, 645–652. [Google Scholar] [CrossRef]
- Steinmetz, H.; Mohr, K.I.; Zander, W.; Jansen, R.; Gerth, K.; Müller, R. Indiacens A and B: Prenyl indoles from the myxobacterium Sandaracinus amylolyticus. J. Nat. Prod. 2012, 75, 1803–1805. [Google Scholar] [CrossRef]
- Thirunavukkarasu, M.; Sawle, Y.; Lala, H. A comprehensive review on optimization of hybrid renewable energy systems using various optimization techniques. Renew. Sustain. Energy Rev. 2023, 176, 113192. [Google Scholar] [CrossRef]
- Chen, C.; Long, J.; Chen, W.; Liu, Z.; Guo, J. Modeling and prediction of spindle dynamic precision using the Kriging-based response surface method with a novel sampling strategy. Nonlinear Dyn. 2023, 111, 559–579. [Google Scholar] [CrossRef]
- El-Sayed, S.E.; El-Housseiny, G.S.; Abdelaziz, N.A.; El-Ansary, M.R.; Aboshanab, K.M. Optimized Production of the Allylamine Antifungal “Terbinafine” by Lysinibacillus Isolate MK212927 Using Response Surface Methodology. Infect. Drug Resist. 2020, 13, 3613–3626. [Google Scholar] [CrossRef]
- El-Housseiny, G.S.; Ibrahim, A.A.; Yassien, M.A.; Aboshanab, K.M. Production and statistical optimization of Paromomycin by Streptomyces rimosus NRRL 2455 in solid state fermentation. BMC Microbiol. 2021, 21, 34. [Google Scholar] [CrossRef]
- Ibrahim, A.A.; El-Housseiny, G.S.; Aboshanab, K.M.; Yassien, M.A.; Hassouna, N.A. Paromomycin production from Streptomyces rimosus NRRL 2455: Statistical optimization and new synergistic antibiotic combinations against multidrug resistant pathogens. BMC Microbiol. 2019, 19, 18. [Google Scholar] [CrossRef]
- El-Housseiny, G.S.; Aboshanab, K.M.; Aboulwafa, M.M.; Hassouna, N.A. Structural and Physicochemical Characterization of Rhamnolipids produced by Pseudomonas aeruginosa P6. AMB Express 2020, 10, 201. [Google Scholar] [CrossRef]
- Mansour, N.M.; Elkhatib, W.F.; Aboshanab, K.M.; Bahr, M.M.A. Inhibition of Clostridium difficile in Mice Using a Mixture of Potential Probiotic Strains Enterococcus faecalis NM815, E. faecalis NM915, and E. faecium NM1015: Novel Candidates to Control C. difficile Infection (CDI). Probiotics Antimicrob. Proteins 2018, 10, 511–522. [Google Scholar] [CrossRef] [PubMed]
- El-Sayed, S.E.; Abdelaziz, N.A.; El-Housseiny, G.S.; Aboshanab, K.M. Octadecyl 3-(3, 5-di-tert-butyl-4-hydroxyphenyl) propanoate, an antifungal metabolite of Alcaligenes faecalis strain MT332429 optimized through response surface methodology. Appl. Microbiol. Biotechnol. 2020, 104, 10755–10768. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Arribas, M.V.; Polo, M.C. Occurrence of lactic acid bacteria and biogenic amines in biologically aged wines. Food Microbiol. 2008, 25, 875–881. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Peela, S.; Kurada, V.B.; Terli, R. Studies on antagonistic marine actinomycetes from the Bay of Bengal. World J. Microbiol. Biotechnol. 2005, 21, 583–585. [Google Scholar] [CrossRef]
- Hossain, N.; Rahman, M. Antagonistic activity of antibiotic producing Streptomyces sp. against fish and human pathogenic bacteria. Braz. Arch. Biol. Technol. 2014, 57, 233–237. [Google Scholar] [CrossRef]
- Tiru, M.; Muleta, D.; Bercha, G.; Adugna, G. Antagonistic effect of rhizobacteria against coffee wilt disease caused by Gibberella xylarioides. Asian J. Plant Pathol. 2013, 7, 109–122. [Google Scholar] [CrossRef]
- Montealegre, J.R.; Reyes, R.; Pérez, L.M.; Herrera, R.; Silva, P.; Besoain, X. Selection of bioantagonistic bacteria to be used in biological control of Rhizoctonia solani in tomato. Electron. J. Biotechnol. 2003, 6, 115–127. [Google Scholar] [CrossRef]
- El-Sayed, S.E.; Abdelaziz, N.A.; Osman, H.-E.H.; El-Housseiny, G.S.; Aleissawy, A.E.; Aboshanab, K.M. Lysinibacillus Isolate MK212927: A Natural Producer of Allylamine Antifungal ‘Terbinafine’. Molecules 2021, 27, 201. [Google Scholar] [CrossRef]
- Baharlouei, A.; Sharifi-Sirchi, G.; Bonjar, G.S. Biological control of Sclerotinia sclerotiorum (oilseed rape isolate) by an effective antagonist Streptomyces. Afr. J. Biotechnol. 2011, 10, 5785–5794. [Google Scholar]
- Bonjar, G.S.; Farrokhi, P.R.; Aghighi, S.; Bonjar, L.S.; Aghelizadeh, A. Antifungal characterization of actinomycetes isolated from Kerman, Iran and their future prospects in biological control strategies in greenhouse and field conditions. Plant Pathol. J. 2005, 4, 78–84. [Google Scholar]
- Singh, R.K.; Kumar, D.P.; Solanki, M.K.; Singh, P.; Srivastva, A.K.; Kumar, S.; Kashyap, P.L.; Saxena, A.K.; Singhal, P.K.; Arora, D.K. Optimization of media components for chitinase production by chickpea rhizosphere associated Lysinibacillus fusiformis B-CM18. J. Basic Microbiol. 2013, 53, 451–460. [Google Scholar] [CrossRef] [PubMed]
- Pinu, F.R.; Villas-Boas, S.G. Extracellular Microbial Metabolomics: The State of the Art. Metabolites 2017, 7, 43. [Google Scholar] [CrossRef]
- Feliu, J.X.; Villaverde, A. An optimized ultrasonication protocol for bacterial cell disruption and recovery of β-galactosidase fusion proteins. Biotechnol. Tech. 1994, 8, 509–514. [Google Scholar] [CrossRef]
- Chawawisit, K.; Bhoopong, P.; Phupong, W.; Lertcanawanichakul, M. 2, 4-Di-tert-butylphenol, the bioactive compound produced by Streptomyces sp. KB1. J. Appl. Pharm. Sci. 2015, 5, 007–012. [Google Scholar] [CrossRef]
- Parthasarathi, S.; Sathya, S.; Bupesh, G.; Manikandan, M.; Kim, C.; Manikandan, T.; Balakrishnan, K. Isolation, Characterization and Extraction of antimicrobial compound from marine actinomycete Streptomyces hygroscopicus BDUS 49. Res. J. Biotechnol. Vol. 2012, 8, 3. [Google Scholar]
- Bhosale, H.; Kadam, T.; Mirajgave, R.; Holkar, S. Optimization and characterization of antifungal metabolite from a soil actinomycete Streptomyces indiaensis SRT1. Indian J. Biotechnol. 2018, 17, 261–271. [Google Scholar]
- Abdel-Nasser, A.; Hathout, A.S.; Badr, A.N.; Barakat, O.S.; Fathy, H.M. Extraction and characterization of bioactive secondary metabolites from lactic acid bacteria and evaluating their antifungal and antiaflatoxigenic activity. Biotechnol. Rep. 2023, 38, e00799. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.S.; Duraipandiyan, V.; Ignacimuthu, S. Isolation, screening and partial purification of antimicrobial antibiotics from soil Streptomyces sp. SCA 7. Kaohsiung J. Med. Sci. 2014, 30, 435–446. [Google Scholar] [CrossRef]
- Hefied, F.; Ahmed, Z.B.; Yousfi, M. Optimization of ultrasonic-assisted extraction of phenolic compounds and antioxidant activities From Pistacia atlantica Desf. galls using response surface methodology. J. Appl. Res. Med. Aromat. Plants 2023, 32, 100449. [Google Scholar] [CrossRef]
- Zhang, J.-T.; Wu, M.-F.; Ma, M.-H.; Zhao, L.; Zhu, J.-Y.; Nian, H.; Li, F.-L. Research on the wound healing effect of Shengji Huayu Formula ethanol extract-derived fractions in streptozotocin-induced diabetic ulcer rats. BMC Complement. Med. Ther. 2023, 23, 67. [Google Scholar] [CrossRef] [PubMed]
- Barbosa, D.; Gondim, C.; Silva-Henriques, M.; Soares, C.; Alves, D.; Santos, S.; Castro, R. Coriandrum sativum L. essential oil obtained from organic culture shows antifungal activity against planktonic and multi-biofilm Candida. Braz. J. Biol. 2023, 83, 1–9. [Google Scholar] [CrossRef]
- Meepagala, K.M.; Prabin, T. Antifungal Activity of Poncirus trifoliata Roots against Colletotrichum Species. Agric. Sci. 2023, 14, 346–355. [Google Scholar]
- Augustine, S.; Bhavsar, S.; Kapadnis, B. A non-polyene antifungal antibiotic from Streptomyces albidoflavus PU 23. J. Biosci. 2005, 30, 201–211. [Google Scholar] [CrossRef] [PubMed]
- Munimbazi, C.; Bullerman, L. Isolation and partial characterization of antifungal metabolites of Bacillus pumilus. J. Appl. Microbiol. 1998, 84, 959–968. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, J. Optimization of headspace solid-phase microextraction for analysis of ethyl carbamate in alcoholic beverages using a face-centered cube central composite design. Anal. Chim. Acta 2008, 627, 212–218. [Google Scholar] [CrossRef]
- Salazar, B.; Ortiz, A.; Keswani, C.; Minkina, T.; Mandzhieva, S.; Pratap Singh, S.; Rekadwad, B.; Borriss, R.; Jain, A.; Singh, H.B. Bacillus spp. as bio-factories for antifungal secondary metabolites: Innovation beyond whole organism formulations. Microbial. Ecol. 2022, 86, 1–24. [Google Scholar] [CrossRef]
- Kadjo, A.C.; Beugre, G.C.; Sess-Tchotch, D.-A.; Kedjebo, K.B.D.; Mounjouenpou, P.; Durand, N.; Fontana, A.; Guehi, S.T. Screening of Anti-fungal Bacillus Strains and Influence of their Application on Cocoa Beans Fermentation and Final Bean Quality. J. Adv. Microbiol. 2023, 23, 8–17. [Google Scholar] [CrossRef]
- Lopes, R.; Cerdeira, L.; Tavares, G.S.; Ruiz, J.C.; Blom, J.; Horácio, E.C.; Mantovani, H.C.; Queiroz, M.V.d. Genome analysis reveals insights of the endophytic Bacillus toyonensis BAC3151 as a potentially novel agent for biocontrol of plant pathogens. World J. Microbiol. Biotechnol. 2017, 33, 185. [Google Scholar] [CrossRef]
- Agamennone, V.; van Straalen, J.; Brouwer, A.; de Boer, T.E.; Hensbergen, P.J.; Zaagman, N.; Braster, M.; van Straalen, N.M.; Roelofs, D.; Janssens, T.K. Genome annotation and antimicrobial properties of Bacillus toyonensis VU-DES 13, isolated from the Folsomia candida gut. Entomol. Exp. Appl. 2019, 167, 269–285. [Google Scholar] [CrossRef]
- Sauka, D.H.; Peralta, C.; Pérez, M.P.; Onco, M.I.; Fiodor, A.; Caballero, J.; Caballero, P.; Berry, C.; Del Valle, E.E.; Palma, L. Bacillus toyonensis biovar Thuringiensis: A novel entomopathogen with insecticidal activity against lepidopteran and coleopteran pests. Biol. Control 2022, 167, 104838. [Google Scholar] [CrossRef]
- Balcerek, M.; Szmigiel-Bakalarz, K.; Lewańska, M.; Günther, D.; Oeckler, O.; Malik, M.; Morzyk-Ociepa, B. Experimental and computational study on dimers of 5-halo-1H-indole-2-carboxylic acids and their microbiological activity. J. Mol. Struct. 2023, 1274, 134492. [Google Scholar] [CrossRef]
- Sa-Uth, C.; Rattanasena, P.; Chandrapatya, A.; Bussaman, P. Modification of Medium Composition for Enhancing the Production of Antifungal Activity from Xenorhabdus stockiae PB09 by Using Response Surface Methodology. Int. J. Microbiol. 2018, 2018, 3965851. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Ahluwalia, V.; Saran, S.; Kumar, J.; Patel, A.K.; Singhania, R.R. Recent developments on solid-state fermentation for production of microbial secondary metabolites: Challenges and solutions. Bioresour. Technol. 2021, 323, 124566. [Google Scholar] [CrossRef] [PubMed]
- Costa, E.; Teixidó, N.; Usall, J.; Atarés, E.; Viñas, I. The effect of nitrogen and carbon sources on growth of the biocontrol agent Pantoea agglomerans strain CPA-2. Lett. Appl. Microbiol. 2002, 35, 117–120. [Google Scholar] [CrossRef]
- Singh, C.; Parmar, R.S.; Jadon, P.; Kumar, A. Optimization of cultural conditions for production of antifungal bioactive metabolites by Streptomyces spp. isolated from soil. Int. J. Curr. Microbiol. Appl. Sci. 2017, 6, 386–396. [Google Scholar] [CrossRef]
- Shakeel, Q.; Lyu, A.; Zhang, J.; Wu, M.; Chen, S.; Chen, W.; Li, G.; Yang, L. Optimization of the cultural medium and conditions for production of antifungal substances by Streptomyces platensis 3-10 and evaluation of its efficacy in suppression of clubroot disease (Plasmodiophora brassicae) of oilseed rape. Biol. Control. 2016, 101, 59–68. [Google Scholar] [CrossRef]
- Jacob, J.; Rajendran, R.U.; Priya, S.H.; Purushothaman, J.; Saraswathy Amma, D.K.B.N. Enhanced antibacterial metabolite production through the application of statistical methodologies by a Streptomyces nogalater NIIST A30 isolated from Western Ghats forest soil. PLoS ONE 2017, 12, e0175919. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Lan, X.; Jia, R.; Hu, L.; Wang, Y. Response Surface Methodology (RSM) Mediated Optimization of Medium Components for Mycelial Growth and Metabolites Production of Streptomyces alfalfae XN-04. Microorganisms 2022, 10, 1854. [Google Scholar] [CrossRef]
- Abhini, K.; Rajan, A.B.; Fathimathu Zuhara, K.; Sebastian, D. Response surface methodological optimization of l-asparaginase production from the medicinal plant endophyte Acinetobacter baumannii ZAS1. J. Genet. Eng. Biotechnol. 2022, 20, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Jose, P.A.; Jebakumar, S.R.D. Phylogenetic appraisal of antagonistic, slow growing actinomycetes isolated from hypersaline inland solar salterns at Sambhar salt Lake, India. Front. Microbiol. 2013, 4, 190. [Google Scholar] [CrossRef]
- Hii, L.S.; Rosfarizan, M.; Ling, T.C.; Ariff, A.B. Statistical optimization of pullulanase production by Raoultella planticola DSMZ 4617 using sago starch as carbon and peptone as nitrogen sources. Food Bioprocess Technol. 2012, 5, 729–737. [Google Scholar] [CrossRef]
- Chattopadhyay, D.; Sen, S. Optimisation of cultural conditions for antifungal antibiotic accumulation by Streptomyces rochei G164. Hindustan Antibiot. Bull. 1997, 39, 64–71. [Google Scholar]
- Kanini, G.S.; Katsifas, E.A.; Savvides, A.L.; Karagouni, A.D. Streptomyces rochei ACTA1551, an indigenous Greek isolate studied as a potential biocontrol agent against Fusarium oxysporum f. sp. lycopersici. BioMed Res. Int. 2013, 2013, 387230. [Google Scholar] [CrossRef]
- Souagui, Y.; Tritsch, D.; Grosdemange-Billiard, C.; Kecha, M. Optimization of antifungal production by an alkaliphilic and halotolerant actinomycete, Streptomyces sp. SY-BS5, using response surface methodology. J. Mycol. Medicale 2015, 25, 108–115. [Google Scholar] [CrossRef]
- Pandian, S.R.K.; Deepak, V.; Sivasubramaniam, S.D.; Nellaiah, H.; Sundar, K. Optimization and purification of anticancer enzyme L-glutaminase from Alcaligenes faecalis KLU102. Biologia 2014, 69, 1644–1651. [Google Scholar] [CrossRef]
- Demirel, M.; Kayan, B. Application of response surface methodology and central composite design for the optimization of textile dye degradation by wet air oxidation. Int. J. Ind. Chem. 2012, 3, 24. [Google Scholar] [CrossRef]
- Mourabet, M.; El Rhilassi, A.; El Boujaady, H.; Bennani-Ziatni, M.; Taitai, A. Use of response surface methodology for optimization of fluoride adsorption in an aqueous solution by Brushite. Arab. J. Chem. 2017, 10, S3292–S3302. [Google Scholar] [CrossRef]
- Kasiri, M.B.; Modirshahla, N.; Mansouri, H. Decolorization of organic dye solution by ozonation; Optimization with response surface methodology. Int. J. Ind. Chem. 2013, 4, 3. [Google Scholar] [CrossRef]
- Chen, X.-C.; Bai, J.-X.; Cao, J.-M.; Li, Z.-J.; Xiong, J.; Zhang, L.; Hong, Y.; Ying, H.-J. Medium optimization for the production of cyclic adenosine 3′, 5′-monophosphate by Microbacterium sp. no. 205 using response surface methodology. Bioresour. Technol. 2009, 100, 919–924. [Google Scholar] [CrossRef] [PubMed]
- El-Housseiny, G.S.; Aboulwafa, M.M.; Aboshanab, K.A.; Hassouna, N.A.H. Optimization of Rhamnolipid Production by P. aeruginosa Isolate P6. J. Surfactants Deterg. 2016, 19, 943–955. [Google Scholar] [CrossRef]



| Factor | Symbol | Level | ||
|---|---|---|---|---|
| −1 | 0 | +1 | ||
| Starch (g/L) | A | 1 | 3 | 5 |
| Peptone (g/L) | B | 1 | 3 | 5 |
| Temperature (°C) | C | 30 | 35 | 40 |
| pH | D | 6 | 7 | 8 |
| Agitation rate (rpm) | E | 150 | 225 | 300 |
| Solvent | Extraction Yield (mg/mL) | Mean Inhibition Zones (mm) ± SD | |
|---|---|---|---|
| C. albicans | A. niger | ||
| Ethyl acetate | 0.28 | 14.4 ± 0.32 | 14.1 ± 0.58 |
| Chloroform | 0.26 | 14.11 ± 0.16 | 14.0 ± 0 |
| n-hexane Dichloromethane | 0.22 0.19 | 14.0 ± 0 13.2 ± 0.58 | 13.9 ± 0.72 13.0 ± 0 |
| Acetone | 0.15 | 12.43 ± 0.52 | 12.1 ± 0.15 |
| Diethyl ether | 0.11 | 11.3 ± 0.42 | 11.4 ± 0.33 |
| n-butanol(n-butyl alcohol) | - | - | - |
| Ethanol | - | - | - |
| Methanol | - | - | - |
| Pooled Fractions (PFs) | Ratio of Chloroform: Ethyl Acetate | Elutes Recovered | Retardation Factor (RF) | Dry Weight of Each PF (mg) | Mean Inhibition Zones (mm) ± SD | |
|---|---|---|---|---|---|---|
| C. albicans | A. niger | |||||
| 1 | Chloroform 100% | 1–4 | 0.87 | 0.63 | - | - |
| 2 | 95:5 | 5–12 | 0.32 | 0.25 | - | - |
| 3 | 90:10 | 13–19 | 0.44 | 0.71 | 11.0 ± 0 | - |
| 4 | 85:15 | 20–27 | 0.57 | 0.34 | 11.3 ± 0.14 | 11.0 ± 0 |
| 5 | 80:20 | 28–44 | 0.23 | 0.65 | 11.22 ± 0.28 | 11.3 ± 0.50 |
| 6 | 75:25 | 45–54 | 0.85 | 0.11 | 12.33 ± 0.61 | 12.11 ± 0.52 |
| 7 | 70:30 | 55–62 | 0.79 | 0.42 | 12.5 ± 0.12 | 12.26 ± 0.64 |
| 8 | 65:35 | 63–72 | 0.65 | 0.22 | 12.58 ± 0.21 | 11.6 ± 0.0.58 |
| 9 | 60:40 | 73–75 | 0.52 | 0.64 | 13.34 ± 0.64 | 13.12 ± 0.18 |
| 10 | 55:45 | 76–83 | 0.89 | 0.89 | 13.76 ± 0.30 | 12.3 ± 0.43 |
| 11 | 50:50 | 84–86 | 0.18 | 0.90 | 13.6 ± 0.48 | 13.1 ± 0.11 |
| 12 | 45:55 | 87–90 | 0.38 | 0.40 | 13.3 ± 0.23 | 12.76 ± 0.68 |
| 13 | 40:60 | 91–96 | 0.76 | 0.56 | 13.76 ± 0.12 | 13.4 ± 0.71 |
| 14 | 35:65 | 97–101 | 0.41 | 0.71 | 14.3 ± 0.64 | 14.0 ± 0 |
| 15 | 30:70 | 102–108 | 0.58 | 0.65 | 14.7 ± 0.56 | 14.3 ± 0.33 |
| 16 | 25:75 | 109–114 | 0.62 | 0.79 | 14.7 ± 0.16 | 14.6 ± 0.18 |
| 17 | 20:80 | 115–120 | 0.11 | 0.40 | 15.6 ± 0.44 | 15.4 ± 0.35 |
| 18 | 15:85 | 121–126 | 0.21 | 0.81 | 15.8 ± 0.72 | 15.6 ± 0.62 |
| 19 | 10:90 | 127–134 | 0.25 | 0.21 | 15.4 ± 0.14 | 15.1 ± 0.27 |
| 20 | 5:95 | 135–139 | 0.97 | 0.23 | 11.3 ± 0.33 | 11.1 ± 0.12 |
| 21 | 2.5:97.5 | 140–142 | 0.70 | 0.49 | 12.76 ± 0.54 | 12.4 ± 0.15 |
| 22 | Ethylacetate | 143–145 | 0.94 | 0.36 | 13.6 ± 0.32 | 13.3 ± 0.26 |
| Position | δH (MeOD, 400 MHz, J in Hz) | δC Obtained from, COSY, HMBC and HSQC Spectra (MeOD, 100 MHz) |
|---|---|---|
| 1 | (NH) 6.3 (s) | - |
| 2 | - | 124.1 (C) |
| 3 | 7.10 (s) | 111.75 (CH) |
| 4 | 7.41 (m) | 114 (CH) |
| 5 | 6.85 (m) | 119.6 (CH) |
| 6 | 156 (C) | |
| 7 | 7.08 (brs) | 121.6 |
| 6-OCH3 | 3.76 (s) | 53.5 (C) |
| Run | A: Starch | B: Peptone | C: Temperature | D: pH | E: Agitation Rate | Observed Inhibition Zone (mm) | Predicted Inhibition Zone (mm) |
|---|---|---|---|---|---|---|---|
| 1 | 5 | 3 | 35 | 7 | 225 | 18.6 | 18.09 |
| 2 | 3 | 3 | 30 | 7 | 225 | 13.76 | 14.46 |
| 3 | 1 | 3 | 35 | 7 | 225 | 11.76 | 11.43 |
| 4 | 1 | 5 | 40 | 8 | 150 | 12.3 | 12.59 |
| 5 | 5 | 5 | 40 | 6 | 150 | 20.3 | 20.01 |
| 6 | 5 | 5 | 30 | 6 | 300 | 19 | 18.94 |
| 7 | 3 | 3 | 35 | 7 | 150 | 14.76 | 14.99 |
| 8 | 5 | 1 | 40 | 8 | 150 | 17.3 | 17.25 |
| 9 | 1 | 1 | 30 | 6 | 150 | 11.3 | 10.73 |
| 10 | 5 | 1 | 40 | 6 | 300 | 17.7 | 17.54 |
| 11 | 3 | 5 | 35 | 7 | 225 | 15.76 | 15.77 |
| 12 | 1 | 5 | 40 | 6 | 300 | 12.76 | 12.89 |
| 13 | 3 | 3 | 40 | 7 | 225 | 15.3 | 15.07 |
| 14 | 5 | 1 | 30 | 8 | 300 | 16.3 | 16.17 |
| 15 | 5 | 5 | 30 | 8 | 150 | 19.3 | 18.65 |
| 16 | 3 | 3 | 35 | 6 | 225 | 15 | 15.14 |
| 17 | 3 | 3 | 35 | 7 | 300 | 14 | 14.53 |
| 18 | 3 | 3 | 35 | 8 | 225 | 13.3 | 14.38 |
| 19 | 3 | 1 | 35 | 7 | 225 | 13 | 13.76 |
| 20 | 1 | 1 | 40 | 8 | 300 | 11 | 10.12 |
| 21 | 1 | 5 | 30 | 8 | 300 | 12 | 11.52 |
| 22 | 3 | 3 | 35 | 7 | 225 | 14.3 | 14.76 |
| Source | Sum of Squares | df | Mean Square | F-Value | p-Value | |
|---|---|---|---|---|---|---|
| Model | 0.0117 | 5 | 0.0023 | 183.58 | <0.0001 | significant |
| A-Starch source | 0.0106 | 1 | 0.0106 | 827.52 | <0.0001 | |
| B-peptone source | 0.0009 | 1 | 0.0009 | 67.96 | <0.0001 | |
| C-Temperature | 0.0001 | 1 | 0.0001 | 6.93 | 0.0181 | |
| D-pH | 0.0001 | 1 | 0.0001 | 8.62 | 0.0097 | |
| E-Agitation rate | 0.0000 | 1 | 0.0000 | 2.05 | 0.1718 | |
| Residual | 0.0002 | 16 | 0.0000 | |||
| Corrected Total | 0.0119 | 21 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
El-Sayed, S.E.; Abdelaziz, N.A.; Ali, A.A.; Alshahrani, M.Y.; Aboshanab, K.M.; El-Housseiny, G.S. Identification, Characterization, and Production Optimization of 6-Methoxy-1H-Indole-2-Carboxylic Acid Antifungal Metabolite Produced by Bacillus toyonensis Isolate OQ071612. Microorganisms 2023, 11, 2835. https://doi.org/10.3390/microorganisms11122835
El-Sayed SE, Abdelaziz NA, Ali AA, Alshahrani MY, Aboshanab KM, El-Housseiny GS. Identification, Characterization, and Production Optimization of 6-Methoxy-1H-Indole-2-Carboxylic Acid Antifungal Metabolite Produced by Bacillus toyonensis Isolate OQ071612. Microorganisms. 2023; 11(12):2835. https://doi.org/10.3390/microorganisms11122835
Chicago/Turabian StyleEl-Sayed, Sayed E., Neveen A. Abdelaziz, Amer Al Ali, Mohammad Y. Alshahrani, Khaled M. Aboshanab, and Ghadir S. El-Housseiny. 2023. "Identification, Characterization, and Production Optimization of 6-Methoxy-1H-Indole-2-Carboxylic Acid Antifungal Metabolite Produced by Bacillus toyonensis Isolate OQ071612" Microorganisms 11, no. 12: 2835. https://doi.org/10.3390/microorganisms11122835
APA StyleEl-Sayed, S. E., Abdelaziz, N. A., Ali, A. A., Alshahrani, M. Y., Aboshanab, K. M., & El-Housseiny, G. S. (2023). Identification, Characterization, and Production Optimization of 6-Methoxy-1H-Indole-2-Carboxylic Acid Antifungal Metabolite Produced by Bacillus toyonensis Isolate OQ071612. Microorganisms, 11(12), 2835. https://doi.org/10.3390/microorganisms11122835








