Isolation and Identification of Microorganisms and Antibiotic Resistance Microorganisms from Beehives Located in Palm, Corn and Longan Plantations, Thailand
Abstract
:1. Introduction
2. Materials and Methods
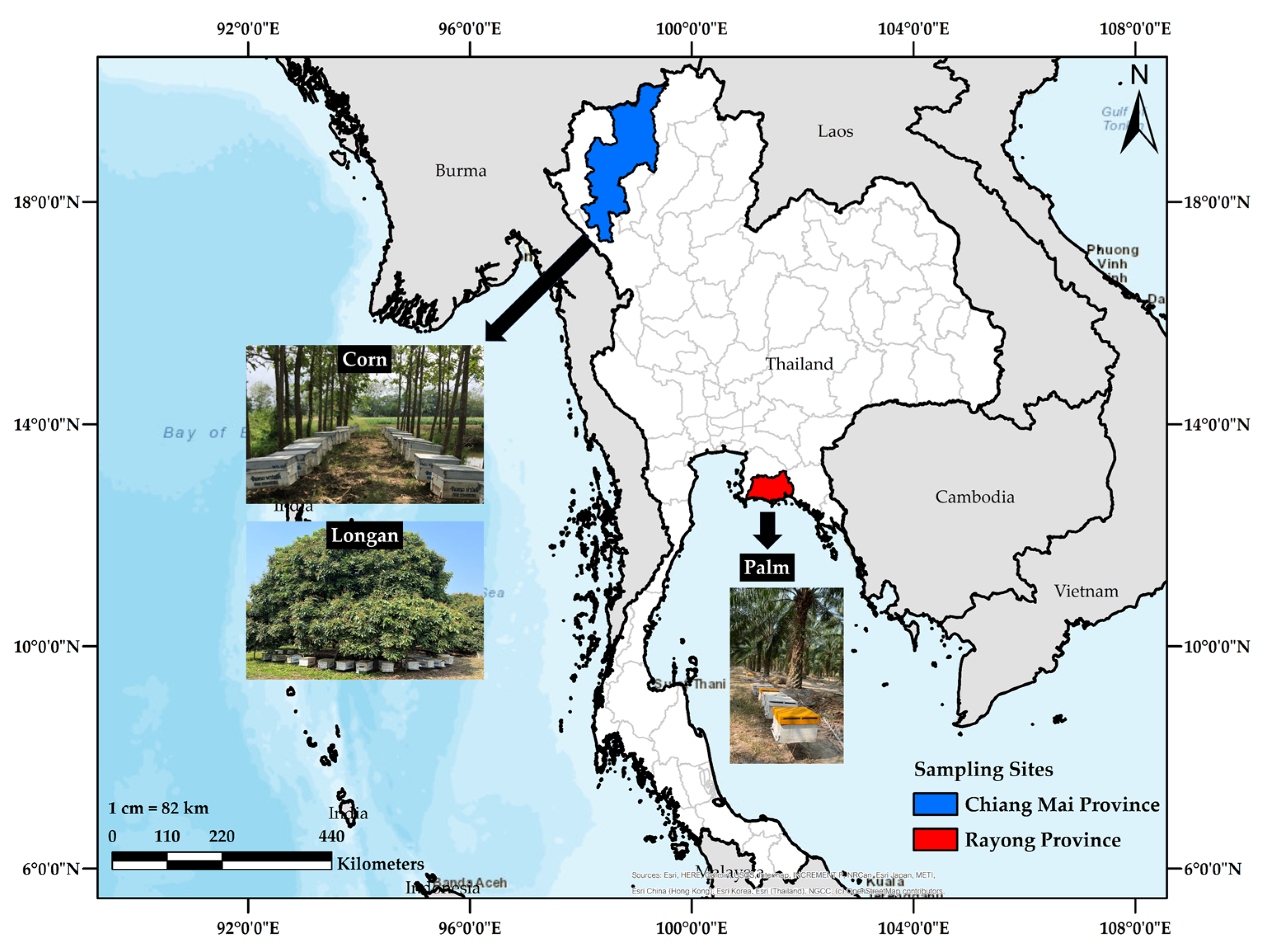
2.1. Sample Collection
2.2. Microbial Culture and Identification
2.3. Antimicrobial Susceptibility Test
2.4. Data Analysis
3. Results
3.1. Microbial Culture and Identification
3.2. Antimicrobial Drug Susceptibility Test
4. Discussion
4.1. Bacterial Identification
4.2. Antimicrobial Drug Susceptibility Test
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Serwecińska, L. Antimicrobials and Antibiotic-Resistant Bacteria: A Risk to the Environment and to Public Health. Water 2020, 12, 3313. [Google Scholar] [CrossRef]
- Dadgostar, P. Antimicrobial Resistance: Implications and Costs. Infect. Drug Resist. 2019, 12, 3903–3910. [Google Scholar] [CrossRef]
- Prestinaci, F.; Pezzotti, P.; Pantosti, A. Antimicrobial resistance: A global multifaceted phenomenon. Pathog. Glob. Health. 2015, 109, 309–318. [Google Scholar] [CrossRef]
- World Health Organization. Sixty-Eighth World Health Assembly; World Health Organization: Geneva, Switzerland, 2015. [Google Scholar]
- World Health Organization. Global Action Plan on Antimicrobial Resistance; WHO Document Production Services: Geneva, Switzerland, 2015. [Google Scholar]
- Khalifa, S.A.M.; Elshafiey, E.H.; Shetaia, A.A.; El-Wahed, A.A.A.; Algethami, A.F.; Musharraf, S.G.; AlAjmi, M.F.; Zhao, C.; Masry, S.H.D.; Abdel-Daim, M.M.; et al. Overview of Bee Pollination and Its Economic Value for Crop Production. Insects 2021, 12, 688. [Google Scholar] [CrossRef]
- Anderson, K.E.; Sheehan, T.H.; Mott, B.M.; Maes, P.; Snyder, L.; Schwan, M.R.; Walton, A.; Jones, B.M.; Corby-Harris, V. Microbial ecology of the hive and pollination landscape: Bacterial associates from floral nectar, the alimentary tract and stored food of honey bees (Apis mellifera). PLoS ONE 2013, 8, e83125. [Google Scholar] [CrossRef]
- Sinacori, M.; Francesca, N.; Alfonzo, A.; Cruciata, M.; Sannino, C.; Settanni, L.; Moschetti, G. Cultivable microorganisms associated with honeys of different geographical and botanical origin. Food Microbiol. 2014, 38, 284–294. [Google Scholar] [CrossRef]
- Santorelli, L.A.; Wilkinson, T.; Abdulmalik, R.; Rai, Y.; Creevey, C.J.; Huws, S.; Gutierrez-Merino, J. Beehives possess their own distinct microbiomes. Environ. Microbiome 2023, 18, 1. [Google Scholar] [CrossRef]
- Anderson, K.E.; Carroll, M.J.; Sheehan TI, M.; Mott, B.M.; Maes, P.; Corby-Harris, V. Hive-stored pollen of honey bees: Many lines of evidence are consistent with pollen preservation, not nutrient conversion. Mol. Ecol. 2014, 23, 5904–5917. [Google Scholar] [CrossRef]
- Corby-Harris, V.; Maes, P.; Anderson, K.E. The bacterial communities associated with honey bee (Apis mellifera) foragers. PLoS ONE 2014, 9, e95056. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute. M100 Performance Standards for Antimicrobial Susceptibility Testing, 32nd ed.; Clinical and Laboratory Standards Institute: Wayne, NY, USA, 2022. [Google Scholar]
- Clinical and Laboratory Standards Institute. M45 Methods for Antimicrobial Dilution and Disk Susceptibility Testing of Infrequently Isolated or Fastidious Bacteria, 3rd ed.; Clinical and Laboratory Standards Institute: Wayne, NY, USA, 2015. [Google Scholar]
- Aarestrup, F.M.; Seyfarth, A.M.; Emborg, H.D.; Pedersen, K.; Hendriksen, R.S.; Bager, F. Effect of abolishment of the use of antimicrobial agents for growth promotion on occurrence of antimicrobial resistance in fecal enterococci from food animals in Denmark. Antimicrob. Agents Chemother. 2001, 45, 2054–2059. [Google Scholar] [CrossRef]
- Ahmadi, A.; Khezri, A.; Nørstebø, H.; Ahmad, R. A culture-, amplification-independent, and rapid method for identification of pathogens and antibiotic resistance profile in bovine mastitis milk. Front. Microbiol. 2023, 13, 1104701. [Google Scholar] [CrossRef]
- Lorenzo, J.M.; Munekata, P.E.; Dominguez, R.; Pateiro, M.; Saraiva, J.A.; Franco, D. Main Groups of Microorganisms of Relevance for Food Safety and Stability: General Aspects and Overall Description. Innov. Technol. Food Preserv. 2018, 13, 53–107. [Google Scholar]
- Nguyen, A.T.; Tallent, S.M. Screening food for Bacillus cereus toxins using whole genome sequencing. Food Microbiol. 2019, 78, 164–170. [Google Scholar] [CrossRef]
- Hölzel, C.S.; Tetens, J.L.; Schwaiger, K. Unraveling the Role of Vegetables in Spreading Antimicrobial-Resistant Bacteria: A Need for Quantitative Risk Assessment. Foodborne Pathog. Dis. 2018, 15, 671–688. [Google Scholar] [CrossRef]
- Martha, G. Identification and roles of non-pathogenic microflora associated with honey bees. FEMS Microbiol. Lett. 1997, 155, 1–10. [Google Scholar]
- de Almeida, C.V.; Taddei, A.; Amedei, A. The controversial role of Enterococcus faecalis in colorectal cancer. Ther. Adv. Gastroenterol. 2018, 11, 1756284818783606. [Google Scholar] [CrossRef]
- Ryan, K.J.; Ray, C.G. Sherris Medical Microbiology, 4th ed.; McGraw Hill: New York, NY, USA, 2004; pp. 294–295. [Google Scholar]
- Murray, B.E. The life and times of the Enterococcus. Clin. Microbiol. Rev. 1990, 3, 46–65. [Google Scholar] [CrossRef]
- Hidron, A.I.; Edwards, J.R.; Patel, J.; Horan, T.C.; Sievert, D.M.; Pollock, D.A.; Fridkin, S.K. NHSN annual update: Antimicrobial-resistant pathogens associated with healthcare-associated infections: Annual summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2006–2007. Infect. Control Hosp. Epidemiol. 2008, 29, 996–1011. [Google Scholar] [CrossRef]
- Jacquemyn, H.; Lenaerts, M.; Brys, R.; Willems, K.; Honnay, O.; Lievens, B. Among-population variation in microbial community structure in the floral nectar of the bee-pollinated forest herb Pulmonaria officinalis L. PLoS ONE 2013, 8, e56917. [Google Scholar] [CrossRef]
- Madigan, M.; Martinko, J. Brock Biomlogy of Microorganisms, 11th ed.; Prentice Hall: New York, NY, USA, 2005; ISBN 978-0-13-144329-7. [Google Scholar]
- Sandot, A.; Grall, N.; Rodier, T.; Bunel, V.; Godet, C.; Weisenburger, G.; Tran-Dinh, A.; Montravers, P.; Mordant, P.; Castier, Y.; et al. Risk of Bronchial Complications After Lung Transplantation With Respiratory Corynebacteria. Results from a Monocenter Retrospective Cohort Study. Transpl. Int. 2023, 36, 10942. [Google Scholar] [CrossRef]
- Doughari, H.J.; Ndakidemi, P.A.; Human, I.S.; Benade, S. The ecology, biology and pathogenesis of Acinetobacter spp.: An overview. Microbes Environ. 2011, 26, 101–112. [Google Scholar] [CrossRef]
- Layla, S.A.; Mayyada, F.D. Prevalence and antibiotic susceptibility patterns of Pantoea spp. isolated form clinical and environmental sources in Iraq. Int. J. ChemTech Res. 2016, 9, 430–437. [Google Scholar]
- O’Mahony, M.M.; Dobson, A.D.; Barnes, J.D.; Singleton, I. The use of ozone in the remediation of polycyclic aromatic hydrocarbon contaminated soil. Chemosphere 2006, 63, 307–314. [Google Scholar] [CrossRef]
- Valenstein, P.; Bardy, G.H.; Cox, C.C.; Zwadyk, P. Pseudomonas alcaligenes endocarditis. Am. J. Clin. Pathol. 1983, 79, 245–247. [Google Scholar] [CrossRef]
- Suzuki, M.; Suzuki, S.; Matsui, M.; Hiraki, Y.; Kawano, F.; Shibayama, K. Genome Sequence of a Strain of the Human Pathogenic Bacterium Pseudomonas alcaligenes That Caused Bloodstream Infection. Genome Announc. 2013, 1, e00919-13. [Google Scholar] [CrossRef]
- Clark, L.L.; Dajc, J.J.; McLean, C.H.; Bartell, J.G.; Stroman, D.W. Pseudomonas otitidis sp. nov., isolated from patients with otic infections. Int. J. Syst. Evol. Microbiol. 2006, 56, 709–714. [Google Scholar] [CrossRef]
- Tsuchida, S.; Umemura, H.; Nakayama, T. Current Status of Matrix-Assisted Laser Desorption/Ionization-Time-of-Flight Mass Spectrometry (MALDI-TOF MS) in Clinical Diagnostic Microbiology. Molecules 2020, 25, 4775. [Google Scholar] [CrossRef]
- Grenga, L.; Pible, O.; Armengaud, J. Pathogen proteotyping: A rapidly developing application of mass spectrometry to address clinical concerns. Clin. Mass Spectrom. 2019, 14 Pt A, 9–17. [Google Scholar] [CrossRef]
- Church, D.L.; Cerutti, L.; Gürtler, A.; Griener, T.; Zelazny, A.; Emler, S. Performance and Application of 16S rRNA Gene Cycle Sequencing for Routine Identification of Bacteria in the Clinical Microbiology Laboratory. Clin. Microbiol. Rev. 2020, 33, e00053-19. [Google Scholar] [CrossRef]
- Ozma, M.A.; Abbasi, A.; Asgharzadeh, M.; Pagliano, P.; Guarino, A.; Köse, Ş.; Samadi Kafil, H. Antibiotic therapy for pan-drug-resistant infections. Infez. Med. 2022, 30, 525–531. [Google Scholar]
- Parmanik, A.; Das, S.; Kar, B.; Bose, A.; Dwivedi, G.R.; Pandey, M.M. Current Treatment Strategies Against Multidrug-Resistant Bacteria: A Review. Curr. Microbiol. 2022, 79, 388. [Google Scholar] [CrossRef]
- Chalalai, R.; Srisamai, V.; Suporn, T.; Phitsanu, T. Species Diversity and Antimicrobial Susceptibility Properties of Staphylococcus Isolated from Broiler Feces in Selected Farms, Thailand. J. Public Health 2017, 47, 44–55. [Google Scholar]
- Kirby-Bauer Disk Susceptibility Test. LibreTextTM. Available online: https://bio.libretexts.org/ (accessed on 3 October 2023).
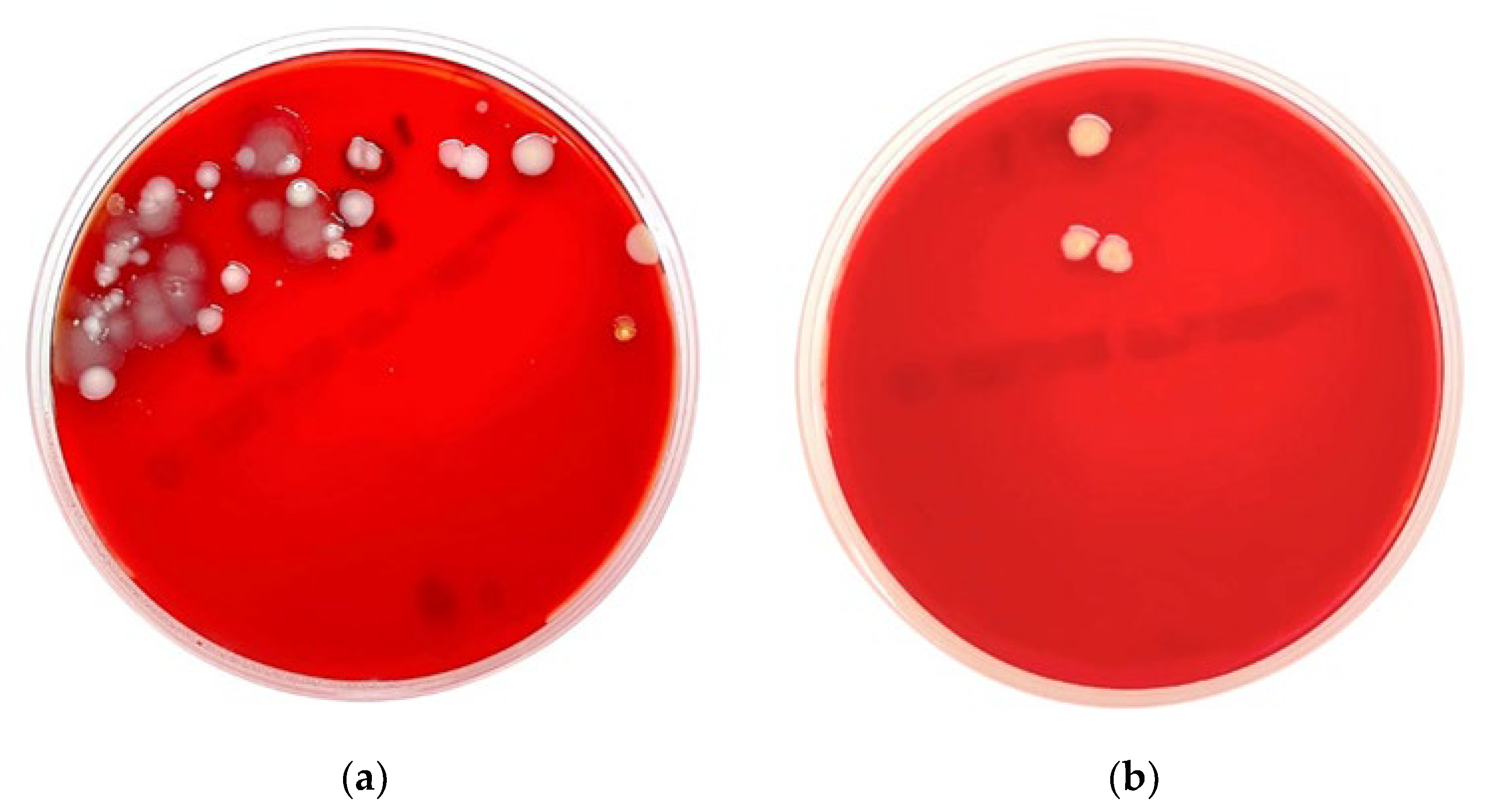

| Swab No. | Plantation | Gram Stain Morphology | Microbial Identification | |
|---|---|---|---|---|
| Biochemical Tests | MALDI-TOF MS (VITEK® MS, bioMerieux, Marcy-l’Étoile, France) | |||
| 1 | Palm | Not performed | Yeast | Candida orthopsilosis |
| 2 | Palm | Gram-positive cocci | Coagulase-negative Staphylococcus spp. | Staphylococcus sciuri |
| Gram-positive cocci | Coagulase-negative Staphylococcus spp. | Unidentified | ||
| 3 | Palm | Gram-positive bacilli | Coryneform bacteria | Unidentified |
| 4 | Palm | Gram-positive cocci | Coagulase-negative Staphylococcus spp. | Staphylococcus sciuri |
| 5 | Palm | No growth | No growth | Not performed |
| 6 | Palm | Gram-positive bacilli | Coryneform bacteria | Unidentified |
| Gram-negative bacilli | Pseudomonas spp. | Pseudomonas otitidis | ||
| Gram-negative bacilli | Non-fermentative Gram-negative bacilli | Acinetobacter junii | ||
| 7 | Palm | Gram-positive cocci | Coagulase-negative Staphylococcus spp. | Staphylococcus gallinarum |
| Gram-positive cocci | Coagulase-negative Staphylococcus spp. | Staphylococcus sciuri | ||
| Gram-negative bacilli | Pantoea spp. | Pantoea dispersa | ||
| 8 | Palm | Gram-positive bacilli with spore | Bacillus spp. | Bacillus altitudinis/pumilus |
| Gram-positive bacilli | Bacillus spp. | Bacillus megaterium | ||
| Not conducted | Yeast | Candida guilliermondis + Candida permentati | ||
| 9 | Palm | Gram-positive cocci | Coagulase-negative Staphylococcus spp. | Staphylococcus sciuri |
| Gram-positive cocci | Coagulase-negative Staphylococcus spp. | Staphylococcus gallinarum | ||
| Gram-negative bacilli | Pantoea spp. | Pantoea ananatis | ||
| 10 | Palm | Gram-positive bacilli | Coryneform bacteria | Exiguobacterium acetylicum |
| Gram-negative bacilli | Pantoea spp. | Unidentified | ||
| Gram-negative bacilli | Non-fermentative Gram negative bacilli | Acinetobacter junii | ||
| 11 | Longan | Gram-positive bacilli | Coryneform bacteria | Unidentified |
| Gram-negative bacilli | Pantoea spp. | Pantoea dispersa | ||
| 12 | Corn | Gram-positive bacilli | Coryneform bacteria | Unidentified |
| 13 | Corn | Gram-positive bacilli | Bacillus spp. | Bacillus megaterium |
| Gram-positive bacilli | Coryneform bacteria | Unidentified | ||
| 14 | Corn | Gram-positive bacilli | Bacillus spp. | Unidentified |
| Gram-positive bacilli | Bacillus spp. | Bacillus cereus gr. | ||
| 15 | Corn | Gram-positive bacilli | Bacillus spp. | Bacillus cereus gr. |
| Gram-negative cocco-bacilli | Unidentified Gram-negative bacteria | Unidentified | ||
| 16 | Corn | Gram-negative cocco-bacilli | Non-fermentative Gram-negative bacilli | Acinetobacter schindleri |
| Gram-negative cocco-bacilli | Non-fermentative Gram-negative bacilli | Acinetobacter radioresistens | ||
| Gram-negative bacilli | Pseudomonas spp. | Pseudomonas alcaligenes | ||
| 17 | Corn | Gram-positive bacilli | Coryneform bacteria | Unidentified |
| Gram-negative cocco-bacilli | Non-fermentative Gram-negative bacilli | Acinetobacter lwoffii | ||
| Gram-positive bacilli | Bacillus spp. | Bacillus flexus | ||
| Gram-positive cocco-bacilli | Enterococcus faecalis | Enterococcus faecalis | ||
| Gram-negative bacilli | Non-fermentative Gram-negative bacilli | Chryseobacterium gleum | ||
| Plantation/No. of Colony Source (n = 37) | Type of Microorganisms | ||
|---|---|---|---|
| Gram-Positive [59.5% (22/37)] | Gram-Negative [35.1% (13/37)] | Fungus (Yeast) [5.4% (2/37)] | |
| Palm/ (n = 20) | Gram-positive [60% (12/20)] Staphylococcus sciuri [33.3% [4/12)] Staphylococcus gallinarum [16.7% (2/12)] Bacillus altitudinis/pumilus [8.3% (1/12)] Bacillus megaterium [8.3% (1/12)] Exiguobacterium acetylicum [8.3% (1/12)] Unidentified Coagulase-negative Staphylococcus spp. [8.3% (1/12)] Unidentified Coryneform bacteria [16.7% (2/12)] | Gram-negative [30% (6/20)] Acinetobacter junii [33.3% (2/6)] Pantoea ananatis [16.7% (1/6)] Pantoea dispersa [16.7% (1/6)] Pseudomonas otitidis [16.7% (1/6)] Unidentified Pantoea spp. [16.7% (1/6)] | Fungus (Yeast) (10% (2/20)] Candida orthopsilosis [50% (1/2)] Candida guilliermondis + Candida permentati [50% (1/2)] |
| Longan/ (n = 2) | Gram-positive [50% (1/2)] Unidentified Coryneform bacteria [100% (1/1)] | Gram-negative [50% (1/2)] Pantoea dispersa [100% (1/1)] | Not Found |
| Corn/ (n = 15) | Gram-positive [60% (9/15)] Bacillus cereus gr. [22.2% (2/9)] Bacillus megaterium [11% (1/9)] Bacillus flexus [11% (1/9)] Enterococcus faecalis [11% (1/9)] Unidentified Bacillus spp. [11% (1/9)] Unidentified Coryneform bacteria [33.3% (3/9)] | Gram-negative [40% (6/15)] Acinetobacter schindleri [16.7% (1/6)] Acinetobacter radioresistens [16.7% (1/6)] Pseudomonas alcaligenes [16.7% (1/6)] Acinetobacter lwoffii [16.7% (1/6)] Chryseobacterium gleum [16.7% (1/6)] Unidentified Gram-negative bacteria [16.7% (1/6)] | Not Found |
| Swab No. | Plantation-Based Swab Source | Microorganism | Antibiotic Inhibition Zone Size (mm) 1 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| VA30 2 | CX30 3 | CIP5 4 | LEV5 4 | CAZ30 5 | CTX30 5 | IMI10 6 | MEM10 6 | |||
| 2 | Palm | Staphylococcus sciuri | 21 | 23 (R) | ND | ND | ND | ND | ND | ND |
| Unidentified Coagulase negative Staphylococcus spp. | 19 | 22 (R) | ND | ND | ND | ND | ND | ND | ||
| 4 | Palm | Staphylococcus sciuri | 20 | 24 (R) | ND | ND | ND | ND | ND | ND |
| 6 | Palm | Pseudomonas otitidis | ND | ND | 38 | 32 | 18 | 18 | 19 | 19 |
| Acinetobacter junii | ND | ND | 34 (S) | 34 (S) | 32 (S) | 32 (S) | 40 (S) | 34 (S) | ||
| 7 | Palm | Staphylococcus gallinarum | 19 | 21 (R) | ND | ND | ND | ND | ND | ND |
| Staphylococcus sciuri | 19 | 20 (R) | ND | ND | ND | ND | ND | ND | ||
| Pantoea dispersa | ND | ND | 40 | 36 | 34 | 30 | 30 | 34 | ||
| 9 | Palm | Staphylococcus sciuri | 18 | 22 (R) | ND | ND | ND | ND | ND | ND |
| Staphylococcus gallinarum | 19 | 23 (R) | ND | ND | ND | ND | ND | ND | ||
| Pantoea ananatis | ND | ND | 36 | 36 | 28 | 34 | 30 | 32 | ||
| 10 | Palm | Acinetobacter junii | ND | ND | 34 (S) | 32 (S) | 28 (S) | 30 (S) | 26 (S) | 30 (S) |
| Unidentified Pantoea spp. | ND | ND | 22 | 20 | 21 | 21 | 34 | 22 | ||
| 11 | Longan | Pantoea dispersa | ND | ND | 38 | 32 | 26 | 28 | 26 | 30 |
| 15 | Corn | Unidentified Gram-negative bacteria | ND | ND | 44 | 40 | 34 | 32 | 32 | 42 |
| 16 | Corn | Acinetobacter schindleri | ND | ND | 26 (S) | 26 (S) | 16 (R) | 20 (R) | 30 (S) | 28 (S) |
| Acinetobacter radioresistens | ND | ND | 26 (S) | 26 (S) | 23 (S) | 25 (S) | 28 (S) | 32 (S) | ||
| Pseudomonas alcaligenes | ND | ND | 36 | 28 | 11 | 8 | 23 | 14 | ||
| 17 | Corn | Acinetobacter lwoffii | ND | ND | 28 (S) | 28 (S) | 19 (S) | 24 (S) | 36 (S) | 30 (S) |
| Enterococcus faecalis | 20 (S) | ND | ND | ND | ND | ND | ND | ND | ||
| Chryseobacterium gleum | ND | ND | 24 | 25 | 19 | 0 | 14 | 0 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dokuta, S.; Yadoung, S.; Jeeno, P.; Hongjaisee, S.; Khamnoi, P.; Manochomphu, S.; Danmek, K.; Maitip, J.; Chuttong, B.; Hongsibsong, S. Isolation and Identification of Microorganisms and Antibiotic Resistance Microorganisms from Beehives Located in Palm, Corn and Longan Plantations, Thailand. Microorganisms 2023, 11, 2855. https://doi.org/10.3390/microorganisms11122855
Dokuta S, Yadoung S, Jeeno P, Hongjaisee S, Khamnoi P, Manochomphu S, Danmek K, Maitip J, Chuttong B, Hongsibsong S. Isolation and Identification of Microorganisms and Antibiotic Resistance Microorganisms from Beehives Located in Palm, Corn and Longan Plantations, Thailand. Microorganisms. 2023; 11(12):2855. https://doi.org/10.3390/microorganisms11122855
Chicago/Turabian StyleDokuta, Sirikwan, Sumed Yadoung, Peerapong Jeeno, Sayamon Hongjaisee, Phadungkiat Khamnoi, Sirinya Manochomphu, Khanchai Danmek, Jakkrawut Maitip, Bajaree Chuttong, and Surat Hongsibsong. 2023. "Isolation and Identification of Microorganisms and Antibiotic Resistance Microorganisms from Beehives Located in Palm, Corn and Longan Plantations, Thailand" Microorganisms 11, no. 12: 2855. https://doi.org/10.3390/microorganisms11122855
APA StyleDokuta, S., Yadoung, S., Jeeno, P., Hongjaisee, S., Khamnoi, P., Manochomphu, S., Danmek, K., Maitip, J., Chuttong, B., & Hongsibsong, S. (2023). Isolation and Identification of Microorganisms and Antibiotic Resistance Microorganisms from Beehives Located in Palm, Corn and Longan Plantations, Thailand. Microorganisms, 11(12), 2855. https://doi.org/10.3390/microorganisms11122855






