Optimization of Siderophore Production in Three Marine Bacterial Isolates along with Their Heavy-Metal Chelation and Seed Germination Potential Determination
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Isolation and Screening of Siderophore-Producing Marine Bacteria
2.3. Characterization of Siderophore-Producing Marine Bacteria
2.4. Growth Curve Estimation
2.5. Siderophore Production and Estimation
2.6. Effect of Production Parameters on Siderophore Production
2.7. Heavy-Metal Chelation
2.8. Seed Germination
2.9. Statistical Analyses
3. Results
3.1. Identification and Characterization of Siderophore-Producing Marine Bacterial Isolates
3.2. Production and Estimation of Siderophores
3.3. Effect of Process Parameters on Siderophore Production
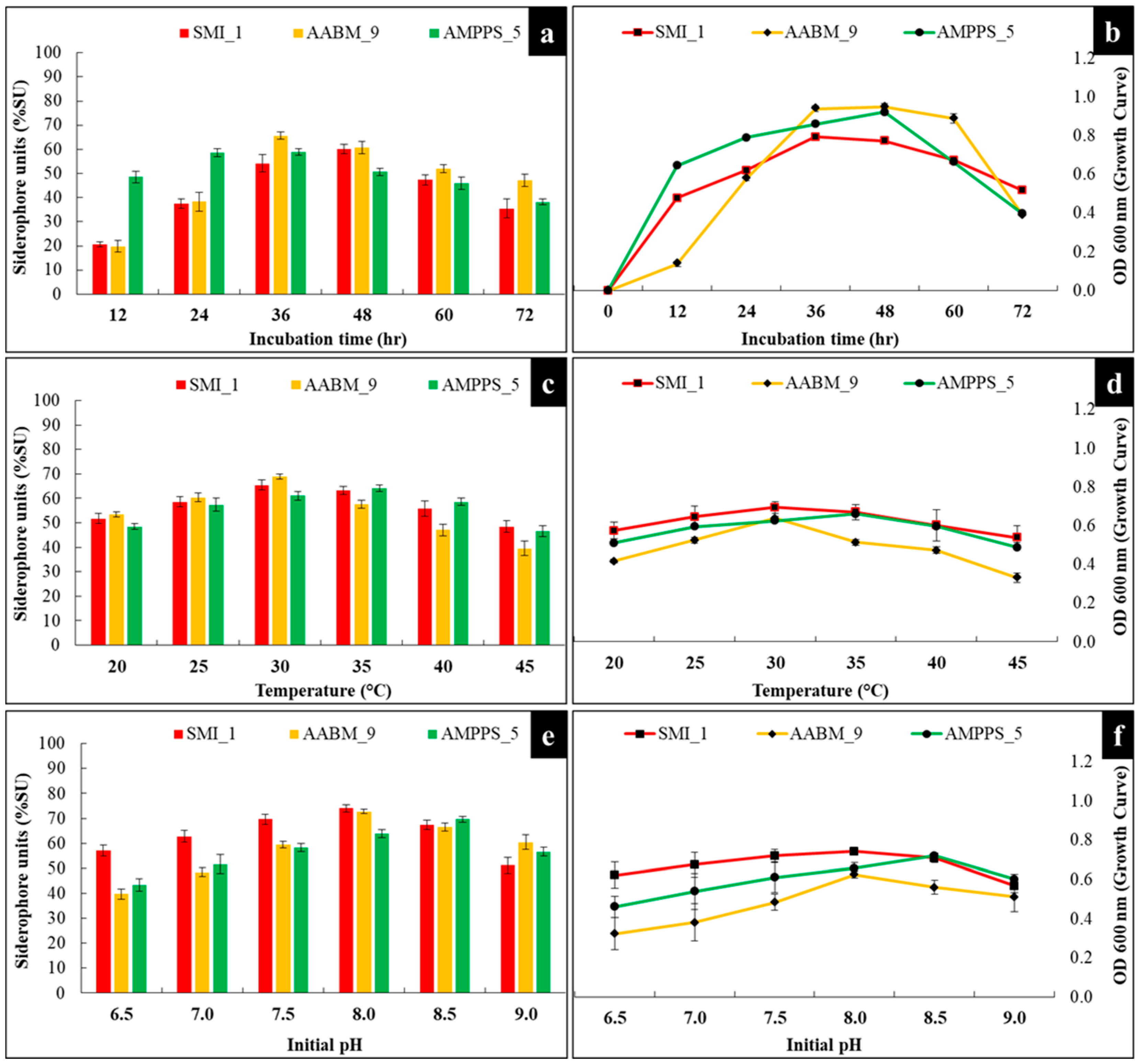
3.3.1. Incubation Time
3.3.2. Temperature
3.3.3. Initial pH
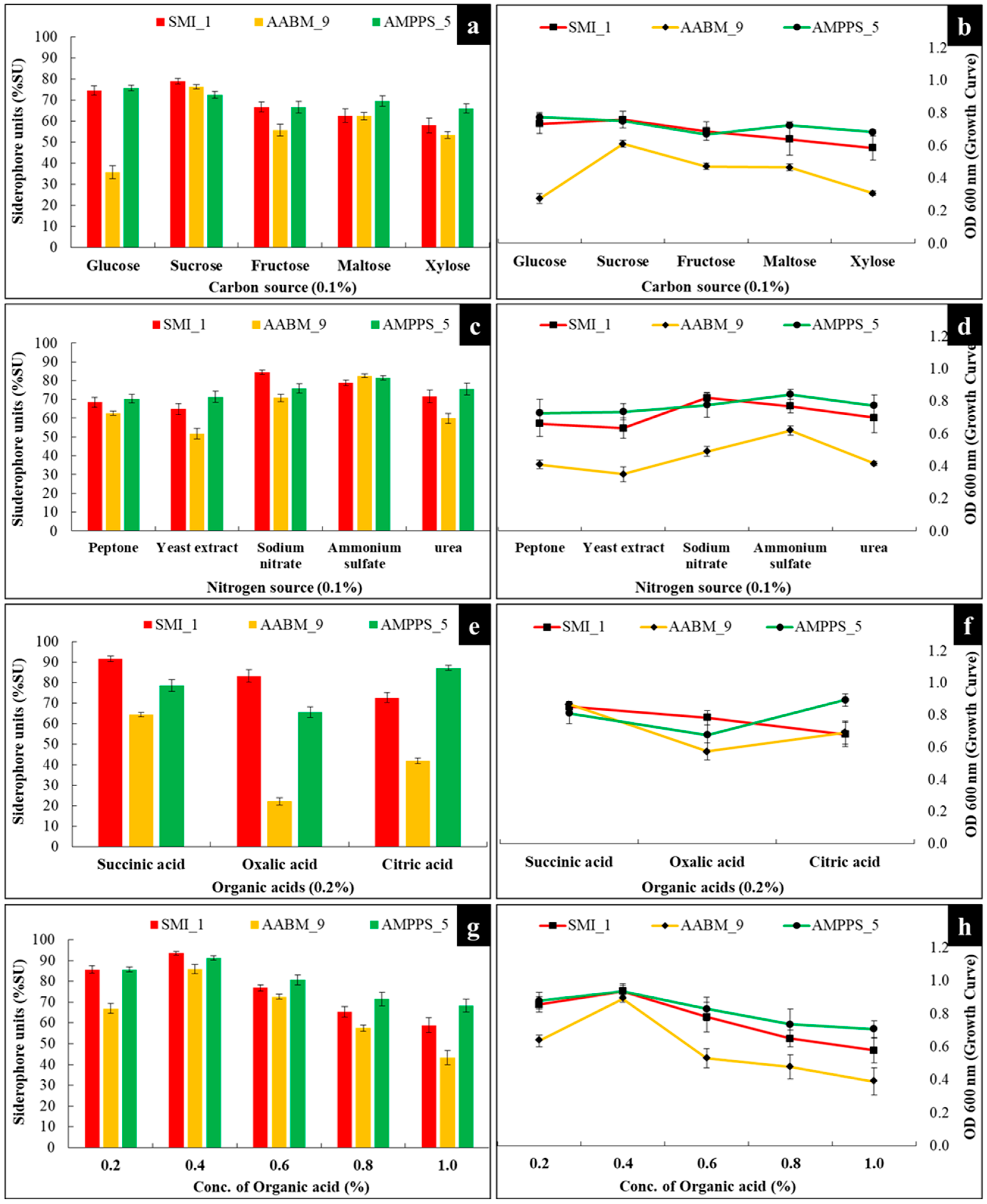
3.3.4. Carbon Source
3.3.5. Nitrogen Source
3.3.6. Organic Acids
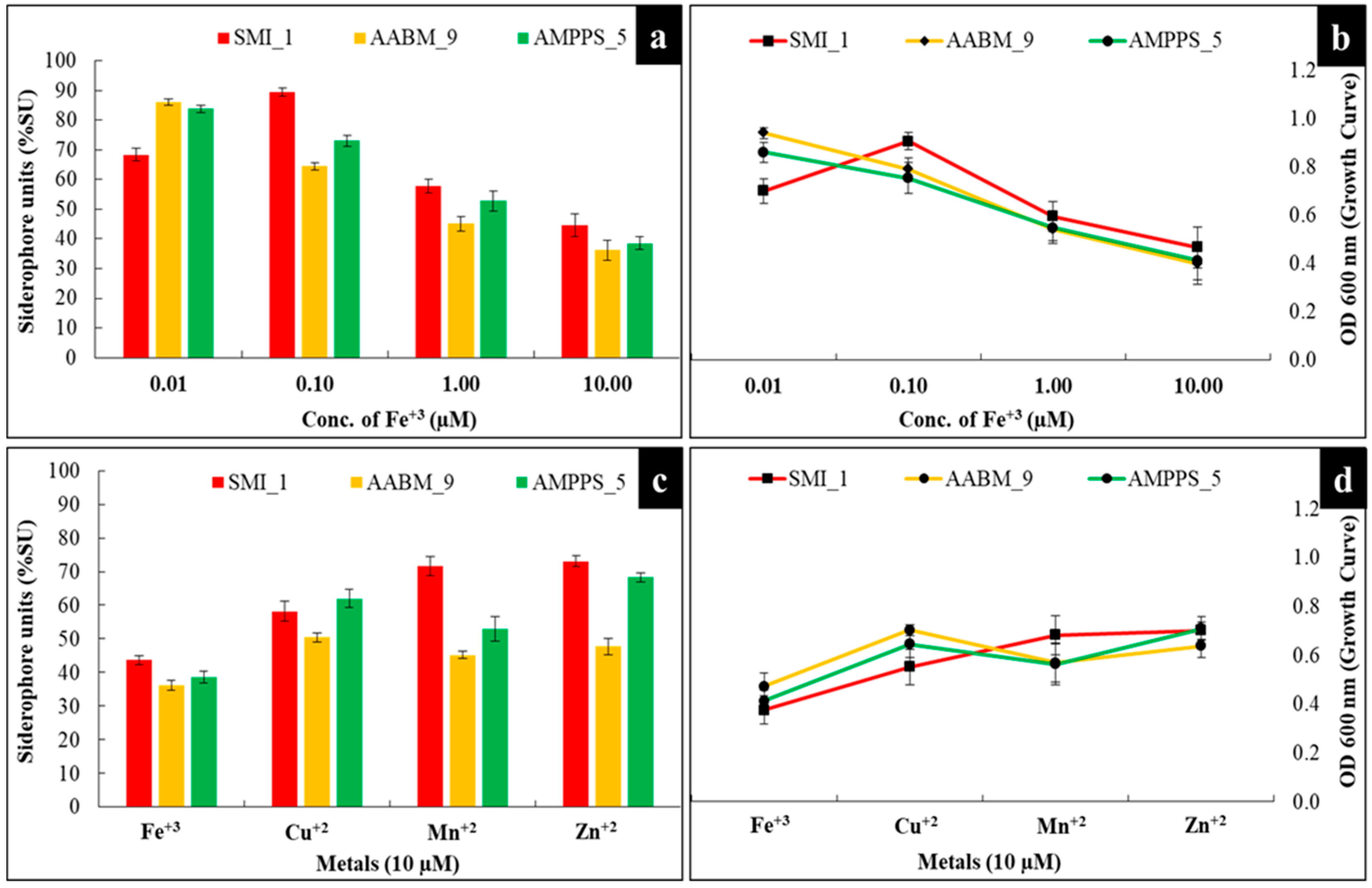
3.3.7. Concentration of Iron (Fe+3)
3.3.8. Different Metal Ions
3.4. Heavy-Metal Chelation
3.5. Seed Germination
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yue, Z.; Chen, Y.; Hao, Y.; Wang, C.; Zhang, Z.; Chen, C.; Liu, H.; Liu, Y.; Li, L.; Sun, Z. Bacillus sp. WR12 Alleviates Iron Deficiency in Wheat via Enhancing Siderophore- and Phenol-Mediated Iron Acquisition in Roots. Plant Soil 2022, 471, 247–260. [Google Scholar] [CrossRef]
- Teta, R.; Esposito, G.; Kundu, K.; Stornaiuolo, M.; Scarpato, S.; Pollio, A.; Costantino, V. A Glimpse at Siderophores Production by Anabaena Flos-aquae UTEX 1444. Mar. Drugs 2022, 20, 0256. [Google Scholar] [CrossRef]
- Sun, Y.; Wu, J.; Shang, X.; Xue, L.; Ji, G.; Chang, S.; Niu, J.; Emaneghemi, B. Screening of Siderophore-Producing Bacteria and Their Effects on Promoting the Growth of Plants. Curr. Microbiol. 2022, 79, 150. [Google Scholar] [CrossRef] [PubMed]
- Soares, E.V. Perspective on the Biotechnological Production of Bacterial Siderophores and Their Use. Appl. Microbiol. Biotechnol. 2022, 106, 3985–4004. [Google Scholar] [CrossRef] [PubMed]
- Sultana, S.; Alam, S.; Karim, M.M. Screening of Siderophore-Producing Salt-Tolerant Rhizobacteria Suitable for Supporting Plant Growth in Saline Soils with Iron Limitation. J. Agric. Food Res. 2021, 4, 100150. [Google Scholar] [CrossRef]
- Sheng, M.; Jia, H.; Zhang, G.; Zeng, L.; Zhang, T.; Long, Y.; Lan, J.; Hu, Z.; Zeng, Z.; Wang, B.; et al. Siderophore Production by Rhizosphere Biological Control Bacteria Brevibacillus Brevis GZDF3 of Pinellia Ternata and Its Antifungal Effects on Candida Albicans. J. Microbiol. Biotechnol. 2020, 30, 689–699. [Google Scholar] [CrossRef] [PubMed]
- Kalyan, V.K.; Meena, S.; Karthikeyan, S.; Jawahar, D. Isolation, Screening, Characterization, and Optimization of Bacterium Isolated from Calcareous Soils for Siderophore Production. Arch. Microbiol. 2022, 204, 721. [Google Scholar] [CrossRef]
- Lemare, M.; Puja, H.; David, S.R.; Mathieu, S.; Ihiawakrim, D.; Geoffroy, V.A.; Rigouin, C. Engineering Siderophore Production in Pseudomonas to Improve Asbestos Weathering. Microb. Biotechnol. 2022, 15, 2351–2363. [Google Scholar] [CrossRef]
- Manck, L.E.; Park, J.; Tully, B.J.; Poire, A.M.; Bundy, R.M.; Dupont, C.L.; Barbeau, K.A. Petrobactin, a Siderophore Produced by Alteromonas, Mediates Community Iron Acquisition in the Global Ocean. ISME J. 2022, 16, 358–369. [Google Scholar] [CrossRef]
- Shen, Q.; Dai, G.; Ravichandran, V.; Liu, Y.; Zhong, L.; Sui, H.; Ren, X.; Jiao, N.; Zhang, Y.; Zhou, H.; et al. Saccharochelins A-H, Cytotoxic Amphiphilic Siderophores from the Rare Marine Actinomycete Saccharothrix sp. D09. J. Nat. Prod. 2021, 84, 2149–2156. [Google Scholar] [CrossRef]
- Butler, A.; Theisen, R.M. Iron(III)–Siderophore Coordination Chemistry: Reactivity of Marine Siderophores. Coord. Chem. Rev. 2010, 254, 288–296. [Google Scholar] [CrossRef] [PubMed]
- Cavas, L.; Kirkiz, I. Characterization of Siderophores from Escherichia Coli Strains through Genome Mining Tools: An AntiSMASH Study. AMB Express 2022, 12, 74. [Google Scholar] [CrossRef] [PubMed]
- Nithyapriya, S.; Lalitha, S.; Sayyed, R.Z.; Reddy, M.S.; Dailin, D.J.; El Enshasy, H.A.; Luh Suriani, N.; Herlambang, S. Production, Purification, and Characterization of Bacillibactin Siderophore of Bacillus subtilis and Its Application for Improvement in Plant Growth and Oil Content in Sesame. Sustainability 2021, 13, 5394. [Google Scholar] [CrossRef]
- Galica, T.; Borbone, N.; Mareš, J.; Kust, A.; Caso, A.; Esposito, G.; Hájek, J.; Reháková, K.; Urajová, P.; Costantino, V.; et al. Cyanochelins, an Overlooked Class of Widely Distributed. Appl. Environ. Microbiol. 2021, 87, e0312820. [Google Scholar] [CrossRef] [PubMed]
- Moore, L.E.; Heller, M.I.; Barbeau, K.A.; Moffett, J.W.; Bundy, R.M. Organic Complexation of Iron by Strong Ligands and Siderophores in the Eastern Tropical North Pacific Oxygen Deficient Zone. Mar. Chem. 2021, 236, 104021. [Google Scholar] [CrossRef]
- Barakat, H.; Qureshi, K.A.; Alsohim, A.S.; Rehan, M. The Purified Siderophore from Streptomyces Tricolor HM10 Accelerates Recovery from Iron-Deficiency-Induced Anemia in Rats. Molecules 2022, 27, 4010. [Google Scholar] [CrossRef]
- Park, J.; Durham, B.P.; Key, R.S.; Groussman, R.D.; Pinedo-Gonzalez, P.; Hawco, N.J.; John, S.G.; Carlson, M.C.G.; Lindell, D.; Juranek, L.; et al. Siderophore Production and Utilization by Microbes in the North Pacific Ocean. bioRxiv 2022. [Google Scholar] [CrossRef]
- Katumba, G.L.; Tran, H.; Henderson, J.P. The Yersinia High-Pathogenicity Island Encodes a Siderophore-Dependent Copper Response System in Uropathogenic Escherichia Coli. MBio 2022, 13, e0239121. [Google Scholar] [CrossRef]
- Wang, W.; Qiu, Z.; Tan, H.; Cao, L. Siderophore Production by Actinobacteria. BioMetals 2014, 27, 623–631. [Google Scholar] [CrossRef]
- Cruz-Morales, P.; Ramos-Aboites, H.E.; Licona-Cassani, C.; Selem-Mójica, N.; Mejía-Ponce, P.M.; Souza-Saldívar, V.; Barona-Gómez, F. Actinobacteria Phylogenomics, Selective Isolation from an Iron Oligotrophic Environment and Siderophore Functional Characterization, Unveil New Desferrioxamine Traits. FEMS Microbiol. Ecol. 2017, 93, fix086. [Google Scholar] [CrossRef]
- Dimkpa, C.O.; Merten, D.; Svatoš, A.; Büchel, G.; Kothe, E. Siderophores Mediate Reduced and Increased Uptake of Cadmium by Streptomyces Tendae F4 and Sunflower (Helianthus Annuus), Respectively. J. Appl. Microbiol. 2009, 107, 1687–1696. [Google Scholar] [CrossRef]
- Asad, S.A.; Farooq, M.; Afzal, A.; West, H. Integrated Phytobial Heavy Metal Remediation Strategies for a Sustainable Clean Environment—A Review. Chemosphere 2019, 217, 925–941. [Google Scholar] [CrossRef]
- Role of Microbial Communities for Sustainability; Seneviratne, G., Zavahir, J.S., Eds.; Microorganisms for Sustainability; Springer: Singapore, 2021; Volume 29, ISBN 978-981-15-9911-8. [Google Scholar]
- Tamariz-Angeles, C.; Huamán, G.D.; Palacios-Robles, E.; Olivera-Gonzales, P.; Castañeda-Barreto, A. Characterization of Siderophore-Producing Microorganisms Associated to Plants from High-Andean Heavy Metal Polluted Soil from Callejón de Huaylas (Ancash, Perú). Microbiol. Res. 2021, 250, 126811. [Google Scholar] [CrossRef] [PubMed]
- Timofeeva, A.M.; Galyamova, M.R.; Sedykh, S.E. Bacterial Siderophores: Classification, Biosynthesis, Perspectives of Use in Agriculture. Plants 2022, 11, 3065. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, E.; Holmström, S.J.M. Siderophores in Environmental Research: Roles and Applications. Microb. Biotechnol. 2014, 7, 196–208. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.M.; Jeon, C.O.; Kim, C.J. Bacillus taeanensis sp. Nov., a Halophilic Gram-Positive Bacterium from a Solar Saltern in Korea. Int. J. Syst. Evol. Microbiol. 2006, 56, 2903–2908. [Google Scholar] [CrossRef][Green Version]
- Kumari, S.; Kiran, S.; Kumari, S.; Kumar, P.; Singh, A. Optimization of Siderophore Production by Bacillus subtilis DR2 and Its Effect on Growth of Coriandrum Sativum. Res. Sq. 2021, 48, 467–475. [Google Scholar] [CrossRef]
- Clarridge, J.E. Impact of 16S RRNA Gene Sequence Analysis for Identification of Bacteria on Clinical Microbiology and Infectious Diseases. Clin. Microbiol. Rev. 2004, 17, 840–862. [Google Scholar] [CrossRef] [PubMed]
- Darby, A.C.; Chandler, S.M.; Welburn, S.C.; Douglas, A.E. Aphid-Symbiotic Bacteria Cultured in Insect Cell Lines. Appl. Environ. Microbiol. 2005, 71, 4833–4839. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Manwar, A.V.; Khandelwal, S.R.; Chaudhari, B.L.; Meyer, J.M.; Chincholkar, S.B. Siderophore Production by a Marine Pseudomonas Aeruginosa and Its Antagonistic Action against Phytopathogenic Fungi. Appl. Biochem. Biotechnol.-Part A Enzym. Eng. Biotechnol. 2004, 118, 243–251. [Google Scholar] [CrossRef]
- Payne, S.M. Detection, Isolation, and Characterization of Siderophores. Methods Enzymol. 1994, 235, 329–344. [Google Scholar]
- Rashmi, V.; ShylajaNaciyar, M.; Rajalakshmi, R.; D’Souza, S.F.; Prabaharan, D.; Uma, L. Siderophore Mediated Uranium Sequestration by Marine Cyanobacterium Synechococcus Elongatus BDU 130911. Bioresour. Technol. 2013, 130, 204–210. [Google Scholar] [CrossRef] [PubMed]
- Arnow, B.Y.L.E. Hydrochloric Acid and Enough Distilled Water to Make a Volume of 1 Liter. Preserve under Toluene. 531. Comp. A J. Comp. Educ. 1937, 531–537. [Google Scholar]
- Wang, Y.; Huang, W.; Li, Y.; Yu, F.; Penttinen, P. Isolation, Characterization, and Evaluation of a High-Siderophore-Yielding Bacterium from Heavy Metal–Contaminated Soil. Environ. Sci. Pollut. Res. 2022, 29, 3888–3899. [Google Scholar] [CrossRef] [PubMed]
- Patel, P.R.; Shaikh, S.S.; Sayyed, R.Z. Modified Chrome Azurol S Method for Detection and Estimation of Siderophores Having Affinity for Metal Ions Other than Iron. Environ. Sustain. 2018, 1, 81–87. [Google Scholar] [CrossRef]
- Kumar, P.; Thakur, S.; Dhingra, G.K.; Singh, A.; Pal, M.K.; Harshvardhan, K.; Dubey, R.C.; Maheshwari, D.K. Inoculation of Siderophore Producing Rhizobacteria and Their Consortium for Growth Enhancement of Wheat Plant. Biocatal. Agric. Biotechnol. 2018, 15, 264–269. [Google Scholar] [CrossRef]
- Mishra, D.; Chitara, M.K.; Negi, S.; Pal singh, J.; Kumar, R.; Chaturvedi, P. Biosynthesis of Zinc Oxide Nanoparticles via Leaf Extracts of Catharanthus roseus (L.) G. Don and Their Application in Improving Seed Germination Potential and Seedling Vigor of Eleusine coracana (L.) Gaertn. Adv. Agric. 2023, 2023, 7412714. [Google Scholar] [CrossRef]
- Lynne, A.M.; Haarmann, D.; Louden, B.C. Use of Blue Agar CAS Assay for Siderophore Detection. J. Microbiol. Biol. Educ. 2011, 12, 51–53. [Google Scholar] [CrossRef]
- Murakami, C.; Tanaka, A.R.; Sato, Y.; Kimura, Y.; Morimoto, K. Easy Detection of Siderophore Production in Diluted Growth Media Using an Improved CAS Reagent. J. Microbiol. Methods 2021, 189, 106310. [Google Scholar] [CrossRef]
- Santos, S.; Neto, I.F.F.; Machado, M.D.; Soares, H.M.V.M.; Soares, E.V. Siderophore Production by Bacillus megaterium: Effect of Growth Phase and Cultural Conditions. Appl. Biochem. Biotechnol. 2014, 172, 549–560. [Google Scholar] [CrossRef] [PubMed]
- Sasirekha, B.; Srividya, S. Siderophore Production by Pseudomonas Aeruginosa FP6, a Biocontrol Strain for Rhizoctonia Solani and Colletotrichum Gloeosporioides Causing Diseases in Chilli. Agric. Nat. Resour. 2016, 50, 250–256. [Google Scholar] [CrossRef]
- Sinha, A.K.; Parli Venkateswaran, B.; Tripathy, S.C.; Sarkar, A.; Prabhakaran, S. Effects of Growth Conditions on Siderophore Producing Bacteria and Siderophore Production from Indian Ocean Sector of Southern Ocean. J. Basic Microbiol. 2019, 59, 412–424. [Google Scholar] [CrossRef] [PubMed]
- Patel, P.R.; Shaikh, S.S.; Sayyed, R. Process Optimization for Siderophore Production and Evaluation of Bioefficacy and Root Colonizing Potential of Alcaligenes sp. In Advances in Bio and Medico Sciences; Excel India Publishers: New Delhi, India, 2016; pp. 145–152. [Google Scholar]
- Yu, S.; Teng, C.; Bai, X.; Liang, J.; Song, T.; Dong, L.; Jin, Y.; Qu, J. Optimization of Siderophore Production by Bacillus sp. PZ-1 and Its Potential Enhancement of Phytoextration of PB from Soil. J. Microbiol. Biotechnol. 2017, 27, 1500. [Google Scholar] [CrossRef]
- Tailor, A.J.; Joshi, B.H. Characterization and Optimization of Siderophore Production from Pseudomonas Fluorescens Strain Isolated from Sugarcane Rhizosphere. J. Environ. Res. Dev. 2012, 6, 688–694. [Google Scholar]
- Venkat Kumar, S.; Menon, S.; Agarwal, H.; Gopalakrishnan, D. Characterization and Optimization of Bacterium Isolated from Soil Samples for the Production of Siderophores. Resour. Technol. 2017, 3, 434–439. [Google Scholar] [CrossRef]
- Fazary, A.E.; Al-Shihri, A.S.; Alfaifi, M.Y.; Saleh, K.A.; Alshehri, M.A.; Elbehairi, S.E.I.; Ju, Y.H. Microbial Production of Four Biodegradable Siderophores under Submerged Fermentation. Int. J. Biol. Macromol. 2016, 88, 527–541. [Google Scholar] [CrossRef]
- Shaikh, S.S.; Wani, S.J.; Sayyed, R.Z. Statistical-Based Optimization and Scale-up of Siderophore Production Process on Laboratory Bioreactor. 3 Biotech 2016, 6, 69. [Google Scholar] [CrossRef]
- Colombowala, A.; Aruna, K. Studies on Optimization of Siderophore Production By Pseudomonas Aeruginosa Azar 11 Isolated From Aquatic Soil and Its Antibacterial Activity. Int. J. Pharm. Biol. Sci. 2018, 8, 714–731. [Google Scholar]
- Sarvepalli, M.; Korrapati, N. Statistical Optimisation of Process Parameters Involved in Siderophore Production of Marine Bacterial Isolate Marinobacter sp. SVU_3. Biomass Convers. Biorefinery 2023, 1–12. [Google Scholar] [CrossRef]
- Singh, P.; Khan, A.; Kumar, R.; Kumar, R.; Singh, V.K.; Srivastava, A. Recent Developments in Siderotyping: Procedure and Application. World J. Microbiol. Biotechnol. 2020, 36, 178. [Google Scholar] [CrossRef]
- Butler, A. Marine Microbial Siderophores: Reactivity and Structural Diversity. Chemist 2022, 94, 1–16. [Google Scholar]
- Liu, R.; Huang, Z.; Dong, C.; Shao, Z. Lottiidibacillus patelloidae gen. nov., sp. nov., Isolated from the Intestinal Tract of a Marine Limpet and Reclassification of Bacillus taeanensis as Maribacillus taeanensis gen. nov., comb. nov. Antonie Van Leeuwenhoek 2019, 112, 797–807. [Google Scholar] [CrossRef]
- Palleroni, N.J.; Doudoroff, M.; Stanier, R.Y.; Solánes, R.E.; Mandel, M. Taxonomy of the Aerobic Pseudomonads: The Properties of the Pseudomonas Stutzeri Group. J. Gen. Microbiol. 1970, 60, 215–231. [Google Scholar] [CrossRef] [PubMed]
- Yen, K.M.; Karl, M.R. Identification of a New Gene, TmoF, in the Pseudomonas Mendocina KR1 Gene Cluster Encoding Toluene-4-Monooxygenase. J. Bacteriol. 1992, 174, 7253–7261. [Google Scholar] [CrossRef]
- Guo, W.; Song, C.; Kong, M.; Geng, W.; Wang, Y.; Wang, S. Simultaneous Production and Characterization of Medium-Chain-Length Polyhydroxyalkanoates and Alginate Oligosaccharides by Pseudomonas Mendocina NK-01. Appl. Microbiol. Biotechnol. 2011, 92, 791–801. [Google Scholar] [CrossRef] [PubMed]
- Wilson, M.K.; Abergel, R.J.; Raymond, K.N.; Arceneaux, J.E.L.; Byers, B.R. Siderophores of Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis. Biochem. Biophys. Res. Commun. 2006, 348, 320–325. [Google Scholar] [CrossRef]
- Wu, Q.; Throckmorton, K.; Maity, M.; Chevrette, M.G.; Braun, D.R.; Rajski, S.R.; Currie, C.R.; Thomas, M.G.; Bugni, T.S. Bacillibactins e and F from a Marine Sponge-Associated Bacillus sp. J. Nat. Prod. 2021, 84, 136–141. [Google Scholar] [CrossRef]
- Mokracka, J.; Koczura, R.; Kaznowski, A. Yersiniabactin and Other Siderophores Produced by Clinical Isolates of Enterobacter Spp. and Citrobacter Spp. FEMS Immunol. Med. Microbiol. 2004, 40, 51–55. [Google Scholar] [CrossRef]
- Hussein, K.A.; Joo, J.H. Stimulation, Purification, and Chemical Characterization of Siderophores Produced by the Rhizospheric Bacterial Strain Pseudomonas Putida. Rhizosphere 2017, 4, 16–21. [Google Scholar] [CrossRef]
- Kaplan, A.R.; Wuest, W.M. Promiscuous Pseudomonas: Uptake of Non-Endogenous Ligands for Iron Acquisition. Tetrahedron Lett. 2021, 75, 153204. [Google Scholar] [CrossRef]
- Vindeirinho, J.M.; Soares, H.M.V.M.; Soares, E.V. Modulation of Siderophore Production by Pseudomonas Fluorescens Through the Manipulation of the Culture Medium Composition. Appl. Biochem. Biotechnol. 2021, 193, 607–618. [Google Scholar] [CrossRef]
- Balado, M.; Lages, M.A.; Fuentes-Monteverde, J.C.; Martínez-Matamoros, D.; Rodríguez, J.; Jiménez, C.; Lemos, M.L. The Siderophore Piscibactin Is a Relevant Virulence Factor for Vibrio Anguillarum Favored at Low Temperatures. Front. Microbiol. 2018, 9, 1766. [Google Scholar] [CrossRef]
- Aziz, O.A.A.; Helal, G.A.; Galal, Y.G.M.; Kader, A.; Rofaida, S. Fungal Siderophores Production in Vitro as Affected by Some Abiotic Factors. Int. J. Curr. Microbiol. Appl. Sci. 2016, 5, 210–222. [Google Scholar] [CrossRef]
- Singh, A.; Mishra, A.K. Influence of Various Levels of Iron and Other Abiotic Factors on Siderophorogenesis in Paddy Field Cyanobacterium Anabaena Oryzae. Appl. Biochem. Biotechnol. 2015, 176, 372–386. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.B.; Kanao, T.; Hedrich, S. Redox Transformations of Iron at Extremely Low PH: Fundamental and Applied Aspects. Front. Microbiol. 2012, 3, 96. [Google Scholar] [CrossRef] [PubMed]
- Murugappan, R.M.; Aravinth, A.; Rajaroobia, R.; Karthikeyan, M.; Alamelu, M.R. Optimization of MM9 Medium Constituents for Enhancement of Siderophoregenesis in Marine Pseudomonas Putida Using Response Surface Methodology. Indian J. Microbiol. 2012, 52, 433–441. [Google Scholar] [CrossRef] [PubMed]
- Prabhu, G.N.; Bindu, P. Optimization of Process Parameters for Siderophore Production under Solid State Fermentation Using Polystyrene Beads as Inert Support. J. Sci. Ind. Res. 2016, 75, 621–625. [Google Scholar]
- Valdebenito, M.; Crumbliss, A.L.; Winkelmann, G.; Hantke, K. Environmental Factors Influence the Production of Enterobactin, Salmochelin, Aerobactin, and Yersiniabactin in Escherichia Coli Strain Nissle 1917. Int. J. Med. Microbiol. 2006, 296, 513–520. [Google Scholar] [CrossRef]
- Virpiranta, H.; Banasik, M.; Taskila, S.; Leiviskä, T.; Halttu, M.; Sotaniemi, V.H.; Tanskanen, J. Isolation of Efficient Metal-Binding Bacteria from Boreal Peat Soils and Development of Microbial Biosorbents for Improved Nickel Scavenging. Water 2020, 12, 2000. [Google Scholar] [CrossRef]
- Sayyed, R.Z.; Seifi, S.; Patel, P.R.; Shaikh, S.S.; Jadhav, H.P.; Enshasy, H. El Siderophore Production in Groundnut Rhizosphere Isolate, Achromobacter sp. RZS2 Influenced by Physicochemical Factors and Metal Ions. Environ. Sustain. 2019, 2, 117–124. [Google Scholar] [CrossRef]
- Ferreira, C.M.H.; Vilas-Boas, Â.; Sousa, C.A.; Soares, H.M.V.M.; Soares, E.V. Comparison of Five Bacterial Strains Producing Siderophores with Ability to Chelate Iron under Alkaline Conditions. AMB Express 2019, 9, 78. [Google Scholar] [CrossRef] [PubMed]
- Gaonkar, T.; Bhosle, S. Effect of Metals on a Siderophore Producing Bacterial Isolate and Its Implications on Microbial Assisted Bioremediation of Metal Contaminated Soils. Chemosphere 2013, 93, 1835–1843. [Google Scholar] [CrossRef] [PubMed]
- Johnstone, T.C.; Nolan, E.M. Beyond Iron: Non-Classical Biological Functions of Bacterial Siderophores. Dalt. Trans. 2015, 44, 6320–6339. [Google Scholar] [CrossRef] [PubMed]
- Hussein, K.A.; Joo, J.H. Zinc Ions Affect Siderophore Production by Fungi Isolated from the Panax Ginseng Rhizosphere. J. Microbiol. Biotechnol. 2019, 29, 105–113. [Google Scholar] [CrossRef]
- Schijf, J.; Christenson, E.A.; Potter, K.J. Different Binding Modes of Cu and Pb vs. Cd, Ni, and Zn with the Trihydroxamate Siderophore Desferrioxamine B at Seawater Ionic Strength. Mar. Chem. 2015, 173, 40–51. [Google Scholar] [CrossRef]
- Wiche, O.; Tischler, D.; Fauser, C.; Lodemann, J.; Heilmeier, H. Effects of Citric Acid and the Siderophore Desferrioxamine B (DFO-B) on the Mobility of Germanium and Rare Earth Elements in Soil and Uptake in Phalaris Arundinacea. Int. J. Phytoremediat. 2017, 19, 746–754. [Google Scholar] [CrossRef]
- Retamal-Morales, G.; Mehnert, M.; Schwabe, R.; Tischler, D.; Zapata, C.; Chávez, R.; Schlömann, M.; Levicán, G. Detection of Arsenic-Binding Siderophores in Arsenic-Tolerating Actinobacteria by a Modified CAS Assay. Ecotoxicol. Environ. Saf. 2018, 157, 176–181. [Google Scholar] [CrossRef]
- Yi, Y.; Hou, Z.; Shi, Y.; Zhang, C.; Zhu, L.; Sun, X.; Zhang, R.; Wang, Z. Pseudomonas Fluorescens RB5 as a Biocontrol Strain for Controlling Wheat Sheath Blight Caused by Rhizoctonia Cerealis. Agronomy 2023, 13, 1986. [Google Scholar] [CrossRef]
- Roslan, M.A.M.; Zulkifli, N.N.; Sobri, Z.M.; Zuan, A.T.K.; Cheak, S.C.; Rahman, N.A.A. Seed Biopriming with P- A Nd K-Solubilizing Enterobacter hormaechei sp. Improves the Early Vegetative Growth and the P and K Uptake of Okra (Abelmoschus esculentus) Seedling. PLoS ONE 2020, 15, e0232860. [Google Scholar] [CrossRef]





| Characteristic(s) | Marine Bacterial Isolate(s) | ||
|---|---|---|---|
| SMI_1 | AABM_9 | AMPPS_5 | |
| Source | Marine sediment | Marine sediment | Marine water |
| Cell morphology | Gram-positive, rod-shaped | Gram-negative, rod-shaped | Gram-negative, rod-shaped |
| Colony morphology | Circular, cream, white, smooth edged, and slightly raised | Circular, smooth edged, convex, and light-yellow color | Smooth, flat, non-wrinkled, and pale brownish yellow |
| Hydrolysis of gelatin | Negative | Negative | Positive |
| Oxidase | Positive | Negative | Positive |
| Catalase | Positive | Positive | Positive |
| Closest relatives in NCBI GenBank | Bacillus taeanensis | Enterobacter sp. | Pseudomonas mendocina |
| Time | Temperature | Initial pH | Carbon Source | Nitrogen Source | Organic Acids | Conc. of Organic Acid | Conc. of Fe+3 | Different Metals | SMI_1 |
|---|---|---|---|---|---|---|---|---|---|
| (h) | (°C) | (0.1%) | (0.1%) | (0.2%) | (%) | (µM) | (10 µM) | (%SU) | |
| 12 | 28 | 7 | - | - | Succinic acid | 0.2 | - | - | 20.56 ± 1.09 |
| 24 | 37.62 ± 1.92 | ||||||||
| 36 | 54.17 ± 3.61 | ||||||||
| 48 | 60.17 ± 1.94 | ||||||||
| 60 | 47.38 ± 2.05 | ||||||||
| 72 | 35.54 ± 3.87 | ||||||||
| 48 | 20 | 7 | - | - | Succinic acid | 0.2 | - | - | 51.73 ± 2.03 |
| 25 | 58.52 ± 2.10 | ||||||||
| 30 | 65.45 ± 2.05 | ||||||||
| 35 | 63.19 ± 1.71 | ||||||||
| 40 | 55.91 ± 3.15 | ||||||||
| 45 | 48.51 ± 2.47 | ||||||||
| 48 | 30 | 6.5 | - | - | Succinic acid | 0.2 | - | - | 57.13 ± 2.18 |
| 7.0 | 62.83 ± 2.37 | ||||||||
| 7.5 | 69.66 ± 1.98 | ||||||||
| 8.0 | 74.04 ± 1.39 | ||||||||
| 8.5 | 67.42 ± 2.02 | ||||||||
| 9.0 | 51.20 ± 3.29 | ||||||||
| 48 | 30 | 8.0 | Glucose | - | Succinic acid | 0.2 | - | - | 74.40 ± 2.23 |
| Sucrose | 78.91 ± 1.38 | ||||||||
| Fructose | 66.68 ± 2.45 | ||||||||
| Maltose | 62.60 ± 3.15 | ||||||||
| Xylose | 57.97 ± 3.53 | ||||||||
| 48 | 30 | 8.0 | Sucrose | Peptone | Succinic acid | 0.2 | - | - | 68.48 ± 2.66 |
| Yeast extract | 64.87 ± 2.92 | ||||||||
| Sodium nitrate | 84.53 ± 1.17 | ||||||||
| Ammonium sulfate | 78.69 ± 1.45 | ||||||||
| Urea | 71.43 ± 3.43 | ||||||||
| 48 | 30 | 8.0 | Sucrose | Sodium nitrate | Succinic acid | 0.2 | - | - | 91.74 ± 1.38 |
| Oxalic acid | 83.35 ± 3.04 | ||||||||
| Citric acid | 72.64 ± 2.40 | ||||||||
| 48 | 30 | 8.0 | Sucrose | Sodium nitrate | Succinic acid | 0.2 | - | 85.58 ± 1.79 | |
| 0.4 | 93.57 ± 0.91 | ||||||||
| 0.6 | 76.81 ± 1.57 | ||||||||
| 0.8 | 65.33 ± 2.45 | ||||||||
| 1.0 | 58.91 ± 3.43 | ||||||||
| 48 | 30 | 8.0 | Sucrose | Sodium nitrate | Succinic acid | 0.4 | 0.01 | - | 68.35 ± 2.20 |
| 0.10 | 89.45 ± 1.34 | ||||||||
| 1.00 | 57.85 ± 2.40 | ||||||||
| 10.0 | 44.62 ± 3.81 | ||||||||
| 48 | 30 | 8.0 | Sucrose | Sodium nitrate | Succinic acid | 0.4 | - | Fe+3 | 43.68 ± 1.36 |
| Cu+2 | 58.21 ± 2.94 | ||||||||
| Mn+2 | 71.70 ± 2.83 | ||||||||
| Zn+2 | 73.09 ± 1.68 |
| Time | Temperature | Initial pH | Carbon Source | Nitrogen Source | Organic Acids | Conc. of Organic Acid | Conc. of Fe+3 | Different Metals | AABM_9 |
|---|---|---|---|---|---|---|---|---|---|
| (h) | (°C) | (0.1%) | (0.1%) | (0.2%) | (%) | (µM) | (10 µM) | (%SU) | |
| 12 | 28 | 7 | - | - | Succinic acid | 0.2 | - | - | 19.86 ± 2.32 |
| 24 | 38.31 ± 4.04 | ||||||||
| 36 | 65.68 ± 1.43 | ||||||||
| 48 | 60.81 ± 2.57 | ||||||||
| 60 | 52.10 ± 1.68 | ||||||||
| 72 | 47.23 ± 2.56 | ||||||||
| 36 | 20 | 7 | - | - | Succinic acid | 0.2 | - | - | 53.38 ± 1.11 |
| 25 | 60.36 ± 1.78 | ||||||||
| 30 | 68.88 ± 1.01 | ||||||||
| 35 | 57.59 ± 1.67 | ||||||||
| 40 | 47.08 ± 2.43 | ||||||||
| 45 | 39.57 ± 2.97 | ||||||||
| 36 | 30 | 6.5 | - | - | Succinic acid | 0.2 | - | - | 39.76 ± 2.12 |
| 7.0 | 48.48 ± 1.79 | ||||||||
| 7.5 | 59.45 ± 1.25 | ||||||||
| 8.0 | 72.76 ± 0.79 | ||||||||
| 8.5 | 66.57 ± 1.52 | ||||||||
| 9.0 | 60.50 ± 2.85 | ||||||||
| 36 | 30 | 8.0 | Glucose | - | Succinic acid | 0.2 | - | - | 35.77 ± 3.16 |
| Sucrose | 76.40 ± 1.06 | ||||||||
| Fructose | 55.68 ± 2.76 | ||||||||
| Maltose | 62.35 ± 1.67 | ||||||||
| Xylose | 53.22 ± 1.63 | ||||||||
| 36 | 30 | 8.0 | Sucrose | Peptone | Succinic acid | 0.2 | - | - | 62.57 ± 1.05 |
| Yeast extract | 51.69 ± 2.82 | ||||||||
| Sodium nitrate | 70.82 ± 1.98 | ||||||||
| Ammonium sulfate | 82.54 ± 1.10 | ||||||||
| Urea | 59.92 ± 2.62 | ||||||||
| 36 | 30 | 8.0 | Sucrose | Ammonium sulfate | Succinic acid | 0.2 | - | - | 64.49 ± 1.01 |
| Oxalic acid | 22.11 ± 1.83 | ||||||||
| Citric acid | 41.94 ± 1.47 | ||||||||
| 36 | 30 | 8.0 | Sucrose | Ammonium sulfate | Succinic acid | 0.2 | - | - | 66.91 ± 2.40 |
| 0.4 | 85.90 ± 2.32 | ||||||||
| 0.6 | 72.58 ± 1.27 | ||||||||
| 0.8 | 57.46 ± 1.53 | ||||||||
| 1.0 | 43.29 ± 3.36 | ||||||||
| 36 | 30 | 8.0 | Sucrose | Ammonium sulfate | Succinic acid | 0.4 | 0.01 | - | 86.14 ± 1.08 |
| 0.10 | 64.54 ± 1.23 | ||||||||
| 1.00 | 45.19 ± 2.47 | ||||||||
| 10.0 | 36.23 ± 3.45 | ||||||||
| 36 | 30 | 8.0 | Sucrose | Ammonium sulfate | Succinic acid | 0.4 | - | Fe+3 | 36.23 ± 1.45 |
| Cu+2 | 50.40 ± 1.33 | ||||||||
| Mn+2 | 45.20 ± 1.05 | ||||||||
| Zn+2 | 47.78 ± 2.43 |
| Time | Temperature | Initial pH | Carbon Source | Nitrogen Source | Organic Acids | Conc. of Organic Acid | Conc. of Fe+3 | Different Metals | AMPPS_5 |
|---|---|---|---|---|---|---|---|---|---|
| (h) | (°C) | (0.1%) | (0.1%) | (0.2%) | (%) | (µM) | (10 µM) | (%SU) | |
| 12 | 28 | 7 | - | - | Succinic acid | 0.2 | - | - | 48.54 ± 2.52 |
| 24 | 58.58 ± 1.57 | ||||||||
| 36 | 58.86 ± 1.30 | ||||||||
| 48 | 50.68 ± 1.43 | ||||||||
| 60 | 45.94 ± 2.64 | ||||||||
| 72 | 38.27 ± 1.25 | ||||||||
| 36 | 20 | 7 | - | - | Succinic acid | 0.2 | - | - | 48.50 ± 1.28 |
| 25 | 57.37 ± 2.68 | ||||||||
| 30 | 61.08 ± 1.72 | ||||||||
| 35 | 64.05 ± 1.30 | ||||||||
| 40 | 58.64 ± 1.56 | ||||||||
| 45 | 46.60 ± 2.34 | ||||||||
| 36 | 35 | 6.5 | - | - | Succinic acid | 0.2 | - | - | 43.39 ± 2.44 |
| 7.0 | 51.66 ± 3.83 | ||||||||
| 7.5 | 58.36 ± 1.54 | ||||||||
| 8.0 | 63.90 ± 1.61 | ||||||||
| 8.5 | 69.65 ± 1.10 | ||||||||
| 9.0 | 56.66 ± 1.68 | ||||||||
| 36 | 35 | 8.5 | Glucose | - | Succinic acid | 0.2 | - | - | 75.69 ± 1.28 |
| Sucrose | 72.38 ± 1.56 | ||||||||
| Fructose | 66.44 ± 2.78 | ||||||||
| Maltose | 69.58 ± 2.51 | ||||||||
| Xylose | 66.05 ± 2.16 | ||||||||
| 36 | 35 | 8.5 | Glucose | Peptone | Succinic acid | 0.2 | - | - | 70.35 ± 2.19 |
| Yeast extract | 71.32 ± 2.90 | ||||||||
| Sodium nitrate | 75.71 ± 2.44 | ||||||||
| Ammonium sulfate | 81.40 ± 1.26 | ||||||||
| Urea | 75.47 ± 3.12 | ||||||||
| 36 | 35 | 8.5 | Glucose | Ammonium sulfate | Succinic acid | 0.2 | - | - | 78.59 ± 2.87 |
| Oxalic acid | 65.66 ± 2.61 | ||||||||
| Citric acid | 87.25 ± 1.23 | ||||||||
| 36 | 35 | 8.5 | Glucose | Ammonium sulfate | Citric acid | 0.2 | - | - | 85.65 ± 1.15 |
| 0.4 | 91.17 ± 0.96 | ||||||||
| 0.6 | 80.70 ± 2.48 | ||||||||
| 0.8 | 71.43 ± 3.33 | ||||||||
| 1.0 | 68.26 ± 3.04 | ||||||||
| 36 | 35 | 8.5 | Glucose | Ammonium sulfate | Citric acid | 0.4 | 0.01 | - | 83.77 ± 1.19 |
| 0.10 | 73.10 ± 1.87 | ||||||||
| 1.00 | 52.80 ± 3.29 | ||||||||
| 10.0 | 38.61 ± 2.06 | ||||||||
| 36 | 35 | 8.5 | Glucose | Ammonium sulfate | Citric acid | 0.4 | - | Fe+3 | 38.57 ± 1.72 |
| Cu+2 | 62.01 ± 2.82 | ||||||||
| Mn+2 | 52.97 ± 3.70 | ||||||||
| Zn+2 | 68.26 ± 1.35 |
| Micro-Organism | Habitat | Incubation Time (h) | Temp (°C) | pH | Carbon Source | Nitrogen Source | Organic Acid | Yield | Ref. |
|---|---|---|---|---|---|---|---|---|---|
| P. aeruginosa FP6 | Soil sample | - | - | - | Sucrose Mannitol | Urea | - | 104.8 mM 92.9 mM | [43] |
| B. cereus P. weihenstephanensis | Marine | 100–150 | 25 | 8.5 | - | - | - | - | [44] |
| Brevibacillus brevis GZDF3 | Rhizosphere soil | 48 | 32 | 7 | Sucrose | Asparagine | - | - | [6] |
| P. aeruginosa RZS9 | - | 24 | 27.8 | 7.1 | - | - | Succinic acid | 69.03 %SU | [45] |
| Bacillus sp. PZ-1 | Soil sample | 48 | 30 | 6.2 | Glucose | Asparagine | 90.52 %SU | [46] | |
| P. fluorescens | - | 24 | 29 | 7 | Glucose | Urea | Succinic acid | 96% | [47] |
| Bacillus sp. (VITVK5) Enterobacter sp. (VITVK6) | Soil sample | - | 37 | 8 | Sucrose Glucose | Sodium Nitrate | Citric acid | ~60–80% | [48] |
| E. coli Bacillus spp. ST13 Streptomyces pilosus | - | - | 55 | 6 | Sucrose Glucose | - | - | 48 μg/mL 31 μg/mL 32 μg/mL | [49] |
| P. aeruginosa | - | - | 27.8 | 7.1 | - | - | - | 68.41% | [50] |
| P. aeruginosa azar 11 | Aquatic soil | 72 | 37 | 7 | Maltose | Ammonium nitrate | Citric acid | 59.18% | [51] |
| Marinobacter hydrocarbonoclausticus SVU_3 | Marine | 48 | 30 | 8.5 | Glucose | Sodium nitrate | Succinic acid | 82.75 %SU | [52] |
| B. teanensis SMI_1 | Marine | 48 | 30 | 8 | Sucrose | Sodium nitrate | Succinic acid | 96.48 %SU | Present study |
| Enterobacter sp. AABM_9 | Marine | 36 | 30 | 8 | Sucrose | Ammonium sulfate | Succinic acid | 86.14 %SU | Present study |
| P. mendocina AMPPS_5 | Marine | 36 | 35 | 8.5 | Glucose | Ammonium sulfate | Citric acid | 91.17 %SU | Present study |
| Metal Salt(s) (1 mM) | Metal Ion(s) | Marine Bacterial Isolate(s) | ||
|---|---|---|---|---|
| SMI_1 | AABM_9 | AMPPS_5 | ||
| FeCl3.7H2O | Fe+3 | +++ | ++ | ++ |
| AgNO3 | Ag+2 | ++ | - | - |
| Al2(SO4)3 | Al+3 | ++ | +++ | ++ |
| CdCl2 | Cd+2 | - | ++ | ++ |
| CoCl2.6H2O | Co+2 | +++ | ++ | ++ |
| K2Cr2O7 | Cr+6 | +++ | ++ | ++ |
| HgCl2 | Hg+2 | +++ | ++ | ++ |
| La2O3 | La+3 | ++ | +++ | ++ |
| Na2MoO4.2H2O | Mo+6 | ++ | - | - |
| NiCl2.6H2O | Ni+2 | + | - | - |
| C4H6O4Pb.3H2O | Pb+2 | - | ++ | ++ |
| PdCl2 | Pd+2 | ++ | +++ | + |
| Y2O3 | Y+3 | ++ | +++ | + |
| Species | Cell-Free Supernatant | Length (cm) at Incubation Time (h) | % GP | ||
|---|---|---|---|---|---|
| 12 | 24 | 36 | |||
| Brown chickpea (Cicer arietinum L.) | Control (Tap Water) | 0 | 0.16 ± 0.16 | 0.38 ± 0.08 | 36.2 |
| SMI_1 | 0.32 ± 0.13 | 0.82 ± 0.22 | 1.76 ± 0.35 | 92.4 | |
| AABM_9 | 0.23 ± 0.04 | 0.6 ± 0.12 | 1.28 ± 0.14 | 89.1 | |
| AMPPS_5 | 0.14 ± 0.08 | 0.36 ± 0.05 | 0.84 ± 0.13 | 67.3 | |
| Peanut (Arachis hypogaea) | Control (Tap Water) | 0 | 0.18 ± 0.11 | 0.42 ± 0.08 | 28.1 |
| SMI_1 | 0.22 ± 0.04 | 0.52 ± 0.08 | 1.12 ± 0.13 | 84.9 | |
| AABM_9 | 0.24 ± 0.05 | 0.46 ± 0.09 | 0.74 ± 0.20 | 71.4 | |
| AMPPS_5 | 0.14 ± 0.05 | 0.66 ± 0.16 | 0.9 ± 0.12 | 63.2 | |
| Green gram (Vigna radiata) | Control (Tap Water) | 0.26 ± 0.08 | 0.94 ± 0.36 | 1.8 ± 0.33 | 67.4 |
| SMI_1 | 0.34 ± 0.13 | 0.74 ± 0.11 | 2.22 ± 0.4 | 96.2 | |
| AABM_9 | 0.32 ± 0.08 | 0.86 ± 0.86 | 1.58 ± 0.19 | 93.7 | |
| AMPPS_5 | 0.28 ± 0.08 | 0.58 ± 0.08 | 1.54 ± 0.33 | 81.3 | |
| Kabuli chana (Cicer arietinum) | Control (Tap Water) | 0 | 0.98 ± 0.39 | 1.96 ± 0.4 | 52.6 |
| SMI_1 | 0.28 ± 0.13 | 0.92 ± 0.27 | 1.86 ± 0.39 | 89.2 | |
| AABM_9 | 0.22 ± 0.08 | 1.4 ± 0.29 | 2.04 ± 0.37 | 82.5 | |
| AMPPS_5 | 0.04 ± 0.05 | 0.6 ± 0.2 | 1.44 ± 0.4 | 78.3 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sarvepalli, M.; Velidandi, A.; Korrapati, N. Optimization of Siderophore Production in Three Marine Bacterial Isolates along with Their Heavy-Metal Chelation and Seed Germination Potential Determination. Microorganisms 2023, 11, 2873. https://doi.org/10.3390/microorganisms11122873
Sarvepalli M, Velidandi A, Korrapati N. Optimization of Siderophore Production in Three Marine Bacterial Isolates along with Their Heavy-Metal Chelation and Seed Germination Potential Determination. Microorganisms. 2023; 11(12):2873. https://doi.org/10.3390/microorganisms11122873
Chicago/Turabian StyleSarvepalli, Mounika, Aditya Velidandi, and Narasimhulu Korrapati. 2023. "Optimization of Siderophore Production in Three Marine Bacterial Isolates along with Their Heavy-Metal Chelation and Seed Germination Potential Determination" Microorganisms 11, no. 12: 2873. https://doi.org/10.3390/microorganisms11122873
APA StyleSarvepalli, M., Velidandi, A., & Korrapati, N. (2023). Optimization of Siderophore Production in Three Marine Bacterial Isolates along with Their Heavy-Metal Chelation and Seed Germination Potential Determination. Microorganisms, 11(12), 2873. https://doi.org/10.3390/microorganisms11122873







