Abstract
Microbial biotechnology plays a crucial role in improving industrial processes, particularly in the production of compounds with diverse applications. In this study, we used bioinformatic approaches to analyze the genomic architecture of Streptomyces albidoflavus MGMM6 and identify genes involved in various metabolic pathways that have significant biotechnological potential. Genome mining revealed that MGMM6 consists of a linear chromosome of 6,932,303 bp, with a high G+C content of 73.5%, lacking any plasmid contigs. Among the annotated genes, several are predicted to encode enzymes such as dye peroxidase, aromatic ring-opening dioxygenase, multicopper oxidase, cytochrome P450 monooxygenase, and aromatic ring hydroxylating dioxygenases which are responsible for the biodegradation of numerous endogenous and xenobiotic pollutants. In addition, we identified genes associated with heavy metal resistance, such as arsenic, cadmium, mercury, chromium, tellurium, antimony, and bismuth, suggesting the potential of MGMM6 for environmental remediation purposes. The analysis of secondary metabolites revealed the presence of multiple biosynthesis gene clusters responsible for producing compounds with potent antimicrobial and metal-chelating activities. Furthermore, laboratory tests conducted under controlled conditions demonstrated the effectiveness of MGMM6 in inhibiting phytopathogenic microbes, decolorizing and degrading aromatic triphenylmethane dyes, particularly Blue Brilliant G250, from wastewater by up to 98 ± 0.15%. Overall, the results of our study highlight the promising biotechnological potential of S. albidoflavus MGMM6.
1. Introduction
The negative impacts of industrial activities and urbanization, including the escalation of antibiotic resistance, soil degradation, industrial dye and heavy metal pollution, and contamination, are emerging as pressing global issues [1,2,3]. The pollution resulting from these factors is persistent, toxic, and poses a significant threat to the health of living organisms [4,5]. Antimicrobial resistance has emerged as one of the foremost concerns for human health, food safety, and sustainable development. For example, cadmium toxicity in crop plants delays the absorption and movement of nutrients and water, increases the occurrence of oxidative harm, disturbs plant metabolism, and hampers plant structure and function [6]. To address these challenges, researchers around the world are focusing on eco-friendly and economical solutions that can effectively solve this multifaceted problem. Heavy metals, soil remediation, and the discovery of novel sources of new antimicrobial compounds with high antibiotic activity can be managed with an eco-friendly approach using microorganisms [7,8]. Various microbial taxonomic genera, such as Actinomyces, Bacillus, Arthrobacter, and Photorhabdus, can survive harsh conditions [9]. They have developed resistance mechanisms such as adsorption of metals on the cell surface, increased activity of efflux pumps, production of extracellular chelating agents (siderophores), intracellular sequestration and biomineralization, and processes such as reduction, oxidation, or methylation that allow them to transform toxic metals into less harmful and more mobile forms [10,11,12,13]. Moreover, during their life cycle, they produce a variety of potentially bioactive compounds with agricultural, pharmaceutical, and biotechnological properties [14,15].
Streptomyces, as the largest bacterial genus, is considered a renowned genus for its ability to produce active metabolites (almost 80% of the world’s antibiotics), which makes it a subject of great interest in the development of novel therapeutic, agricultural, pharmaceutical, and biotechnological agents [16]. Several species of this genera isolated from diverse environmental samples are expected to act as plant growth-promoting and control agents against pathogens, as well as agents for biodegradation and bioremediation of insoluble polymers such as lignin and synthetic insecticides [16,17,18]. Streptomyces albidoflavus is a Gram-positive filamentous versatile soil bacterium with the ability to form a symbiotic relationship with both plants and animals. Strains of these species are considered remarkable producers of attractive metabolites with antimicrobial and plant growth-promoting activities [19,20,21,22]. Genomic analysis appears to be a promising approach for enhancing the selection of effective bioremediation agents for heavy metal pollutant elimination from environments and the identification of new active compounds with potent antimicrobial activity. In this study, we performed whole-genome sequencing of S. albidoflavus MGMM6 and employed bioinformatics approaches to identify genes, gene clusters, and metabolic pathways associated with S. albidoflavus MGMM6 which has potential applications in various fields of industrial and environmental biotechnology such as plant protection, biological remediation of dyes.
2. Materials and Methods
All microbial strains used in this study were provided by the Laboratory of Molecular Genetics and Microbiology Methods (FRC Kazan Scientific Center, Kazan, Russia). Bacterial strain MGMM6 was isolated from the rhizosphere soil of spring wheat (Triticum aestivum L.) Yoldyz variety (Republic of Tatarstan, Kazan, Russia). For this purpose, a 100 µL 5-fold-dilution of rhizospheric soil (100 mg) was performed with phosphate-buffered saline [(PBS): 140 mM NaCl, 5 mM KH2PO4, 1 mM NaHCO3, pH 7.4], plated on Gause #2 medium [(g/L): 2.5 g tryptone, 5 g peptone, 5 g NaCl, 10 g glucose, 20 g agar, pH 7.4], and incubated at 30 °C for 72 h. After incubation, single-growth colonies were replated on Gause #2 medium for molecular identification using 16S rRNA. Among the isolated bacterial strains, MGMM6 was identified as Streptomyces albidoflavus (GenBank: OR822207) and was further used in this study for its various biotechnology applications.
2.1. Phenotypic Characterization
Phenotypic characterization of MGMM6 was evaluated based on colony morphology (shape, color, and other characteristics such as form and transparency) after growth on KB (20 g Proteose peptone; 1.5 g K2HPO4; 0.75 g MgSO4 × 7H2O; 10 mL glycerol; 20 g agar, pH 7.2 ± 0.2 at 25 °C) and (International Streptomyces Project medium (ISP)-4 [(g/L): 10 g soluble starch; 1 g MgSO4 7H2O; 1 g NaCl; 2 g (NH4)2SO4; 2 g CaCO3. 1 mL trace salt solution (0.1 g FeSO4 × 7H2O; 0.1 g MnCl2 × 4H2O; 0.1 g ZnSO4 × 7H2O; 100 mL dH2O; 20 g agar)] medium. For this purpose, an overnight culture of MGMM6 grown on Gause #1 [(g/L): 20 g soluble starch; 1 g KNO3; 0.5 g NaCl; 0.5 g K2HPO4; 0.5 g MgSO4; 0.01 g FeSO4; 20 g agar] was streaked on KB and ISP-4 agar medium and incubated for up to 10 days at 30 ± 1 °C. After incubation, colony morphology was analyzed. All experiments were performed in triplicate.
2.2. Library Construction, Genome Sequencing, and Analysis
The genomic DNA of S. albidoflavus MGMM6 was isolated using a phenol-chloroform extraction method [23] and cleaned using a cleanup kit (Evrogen, Moscow, Russian Federation), according to the manufacturer’s instructions. The whole genome was sequenced using an Illumina HiSeq 2500 System with 2 × 125 bp paired-end reads. Low-quality reads, including adapters and N > 5% reads, were removed using Trimmomatic v. 0.36 and FastP v. 0.23.4-2 [24,25]. The genome was assembled using RAPT (Read Assembly and Annotation Pipeline Tool), Unicycler v.0.3.0, and SPAdes v. 3.15.4 [26,27,28]. Tetra correlation search using the web server tool JSpeciesWS (https://jspecies.ribohost.com/jspeciesws/#home, accessed on 20 March 2023) based on ANIb (average nucleotide identity based on BLAST) was used to select the close-related reference strain. A web server for contig scaffolding using algebraic rearrangements [29] was used to reorder contigs based on a comparison with the reference genome. The gaps in the scaffolds were filled and closed using GapBlaster and ntedit_sealer [30]. The presence of plasmids in the sequenced genome of MGMM6 was analyzed using Plasmid Spades [31], whereby Mob-recon [32,33] was applied to identify the presence of plasmids in the pre-assembled genomic contigs generated by Unicycler and SPAdes v. 3.15.4. Automated annotation of S. albidoflavus MGMM6 and the open reading frame was performed using Bakta v.1.8.1 and the NCBI Prokaryotic Genome Annotation Pipeline (PGAP) [34,35]. Quality assessment of S. albidoflavus MGMM6 was performed using CheckM v1.2.2 [36]. The detection of antimicrobial resistance genes in S. albidoflavus MGMM6 was predicted using the Comprehensive Antibiotic Resistance Database (CARD) [37]. The functional and organization of gene clusters involved in the synthesis of secondary metabolites (BGC) were analyzed using AntiSMASH v. 7.0 [38].
The xenobiotic biodegradation and biosynthesis pathways of prospective compounds with plant-beneficial functions involved in MGMM6 were analyzed using RAST and the KEGG Orthology-based Annotation System. MobileOG-db (beatrix-1.6) with default parameters [39] was used to analyze the genome of S. albidoflavus MGMM6 for the presence of mobile orthologous genes mediating integration/deletion, replication/recombination/repair, stability/protection, or transfer of bacterial mobile genetic elements and phages, as well as transcription regulators related to these processes.
2.3. In Silico Identification of Heavy Metal Resistance Genes
The identification of heavy metal and biocide resistance genes in MGMM6 was conducted using AMRFinderPlus v. 3.10 and BacMet [40,41]. Since heavy metal operons and AMR genes can often be found within bacterial chromosomes or plasmids, the annotated assembly of MGMM6 was used. The operons arsRBC were used as a reference to predict the presence of arsenic resistance in MGMM6. The operon PcoABCDSRE (multicopper oxidase PcoA, copper binding protein PcoB, copper resistance system metallochaperone PcoC and E, copper response regulator transcription factor PcoD, and copper resistance membrane protein PcoS) was used for copper. Operon cmtR (metal-responsive transcriptional repressor for the cmt operon) and cnrA (Fragment of nccA-like protein) were used as a reference to predict the ability of MGMM6 to be resistant to Co and Ni. The presence of metal genes resistant to antimony (Sb)-arsAB, bismuth-arsR, and cadC; cadmium (Cd)-actSR; chromium-chrA; cobalt-cmeAB; copper-actARP and baeR; gallium (Ga)-fbpABC; gold (Au)-gesA; lead (Pb)-cadC and nmtR; mercury (Hg)-dsbAB and merA; molybdenum (Mo)-modAB; selenium (Se)-recG, ruvB, and sodA; silver (Ag)-copAB, cueA, or others; tellurium (Te)-actP, tungsten (W)-baeRS, and modAB; vanadium (V)-mexI, perO, and yieF were analyzed.
2.4. Antibiosis Activity
The antibiosis assay of S. albidoflavus MGMM6 against the pathogens Fusarium oxysporum f. sp. radicis-lycopersic (Forl) ZUM2407, and F. proliferatum was performed using an agar diffusion assay on King’s B medium according to Bonev et al. [42]. Pseudomonas putida PCL1760 and Bacillus velezensis KS04AU were used as negative and positive controls, respectively.
2.5. Ability of S. albidoflavus MGMM6 to Decolorize and Degrade Dyes
The dye removal ability of MGMM6 was quantitatively (based on visual dye-discoloration) and qualitatively (by measuring the percentage of degraded dye) evaluated according to Aravind et al. [43]. For this purpose, an overnight culture of MGMM6 (with a cell density of 0.5 at 595 nm) was inoculated with a ratio of 1/100 in a two-fold diluted nutrient broth (NB) [(g/L) meat extract 1.0; peptone 5.0 g; NaCl 5.0 g; yeast extract 2.0 g; pH 7.4)] and wastewater medium (pH 8.0) amended with 1% (v/v) of Coomassie Brilliant Blue G-250 solution (prepared by dissolving 1 g of Coomassie Brilliant Blue G-250 in water and sterilized by filtration (passed through a 0.22 μm membrane filter (Type Millex-HA, Millipore, Burlington, MA, USA). Wastewater medium was prepared from wastewater sample (Figure 1) left at room temperature for one day for soil precipitation. After precipitation, the excess water was collected and used as a medium. Non-inoculated media were used as controls. All experiments were performed in triplicate.

Figure 1.
Wastewater used in this study to mimic the natural environment to evaluate the dye decoloration and degradation ability of S. albidoflavus MGMM6.
The obtained solution was incubated at 28 ± 1 °C for 7–14 days. After incubation, the cultures were centrifuged at 10,000 rpm for 10 min at room temperature. The obtained supernatants were used to determine the dye degradation percentage. The degradation of dye (dye removal) was measured using a UV/VIS spectrophotometer nanodrop with wavelength intervals from 534 to 716 nm. Non-inoculated media were used as blanks. The dye removal percentage (%) was expressed using the following formula:
2.6. Statistical Analysis
Statistical analysis of these obtained data was performed using the statistical program OriginLab pro-SR1 b9.5.1.195 (OriginLab Corp., Northampton, MA, USA). To determine the significant difference between groups, a one-way ANOVA and post hoc Tukey’s honestly significant difference test (p < 0.05) were conducted.
3. Results
3.1. Phenotypic Characterization and Genome Sequencing Analysis
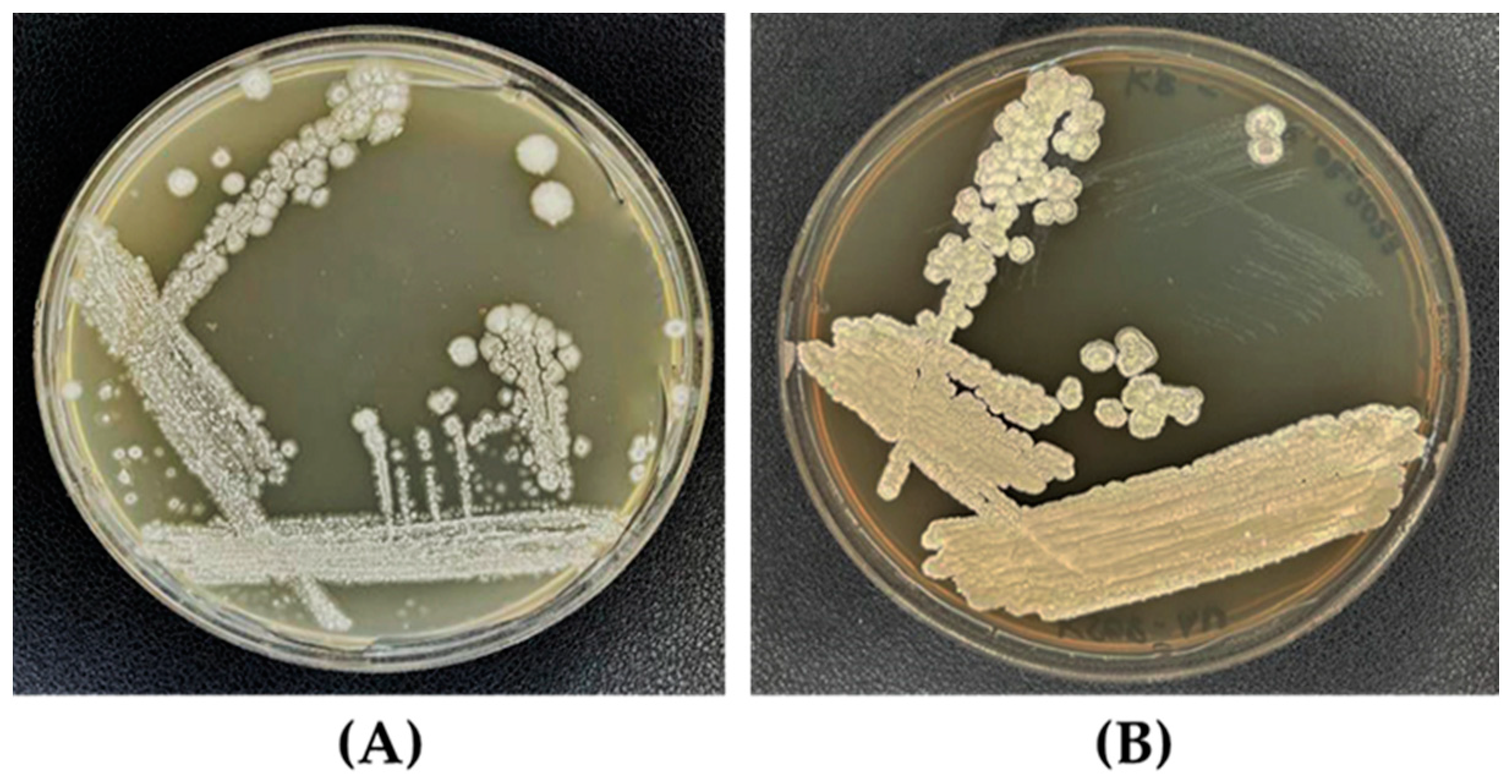
After incubation for 10 days at 30 ± 1 °C, the colony of S. albidoflavus MGMM6 developed buried colonies with hard consistency and dry appearance (Figure 2) and white (on ISP-4 agar medium, Figure 2A) and slightly whitish–yellow color (on KB agar medium, Figure 2B).
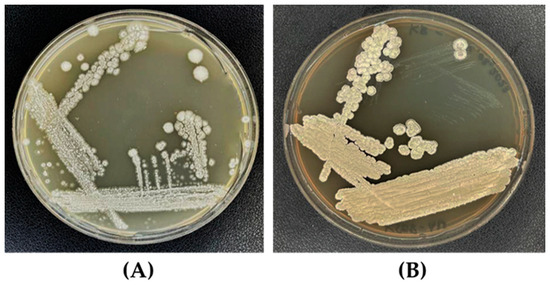
Figure 2.
Colony morphology of S. albidoflavus MGMM6 on KB (A) and ISP-4 (B) agar medium. Plates were incubated for up to 10 days at 30 ± 1 °C.
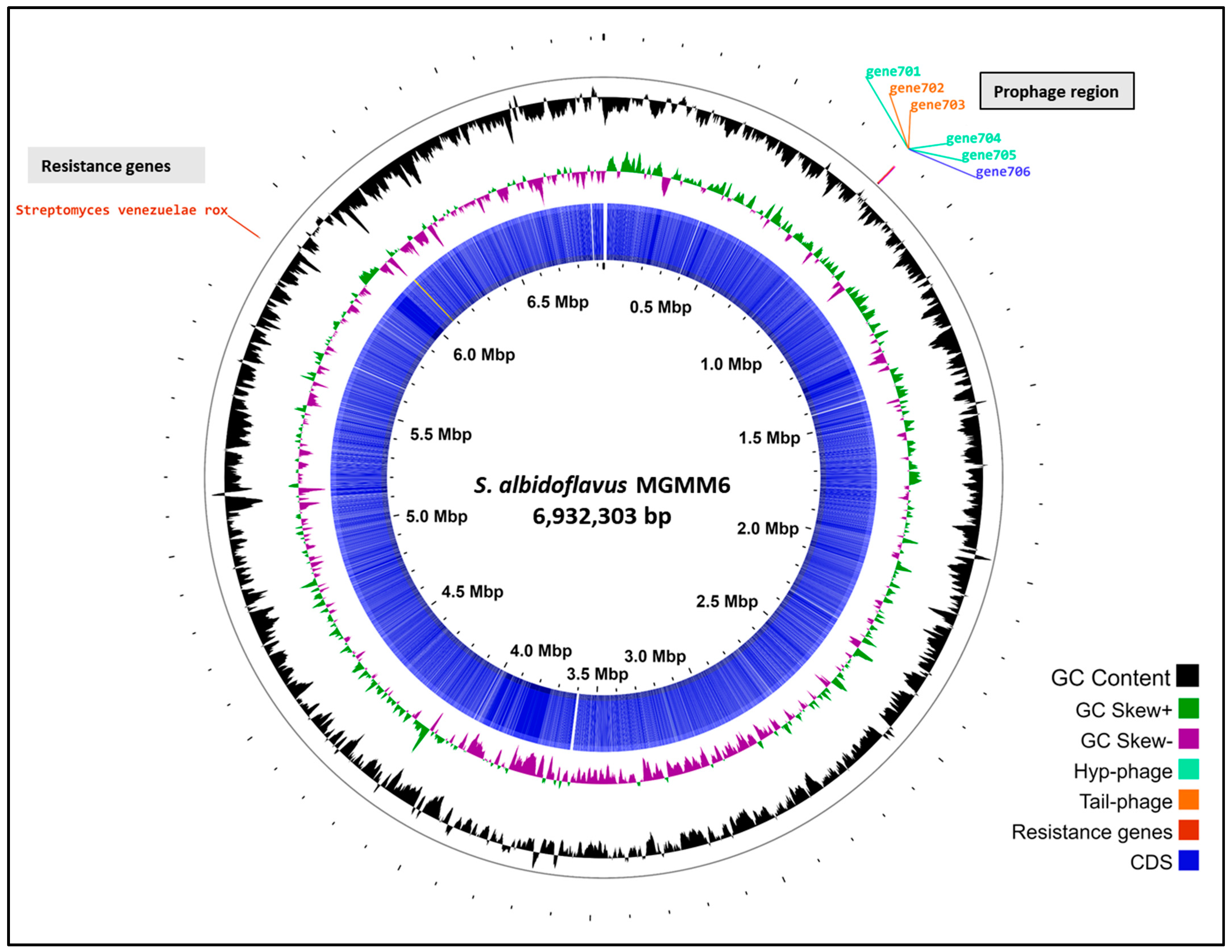
The genomic assembly of S. albidoflavus MGMM6 comprised a linear chromosome of 6,932,303 bp, with a high G + C content of 73.5% without plasmid contigs. Tetra correlation search using the Web server tool JSpeciesWS showed that S. albidoflavus MGMM6 was closely related to S. albidoflavus DSM 40233 (NCBI RefSeq assembly: GCF_004195775.1; BioSample ID: SAMN08225702) with 98.96% similarity based on ANIb.
Automated annotation using PGAP revealed that the genome of MGMM6 carries a total of 6024 genes, with 5851 coding genes. Streptomyces albidoflavus MGMM6 contains 69 genes (RNA), 63 of which are transfer ribonucleic acid (tRNA) and three are non-coding RNA (ncRNA)]) (Figure 3). The presence of 104 pseudogenes in S. albidoflavus MGMM6 was predicted using PGAP. Automated annotation using Bakta predicted the presence of 5949 coding genes, 64 genes (RNA), 11 pseudogenes, and 2 CRISPR arrays. The origin of replication (oriC) was predicted to be in the genome region from 5,331,504 to 5,332,639. Qualitative analysis using CheckM revealed that the completeness of the genome was 99.34% (Table 1).
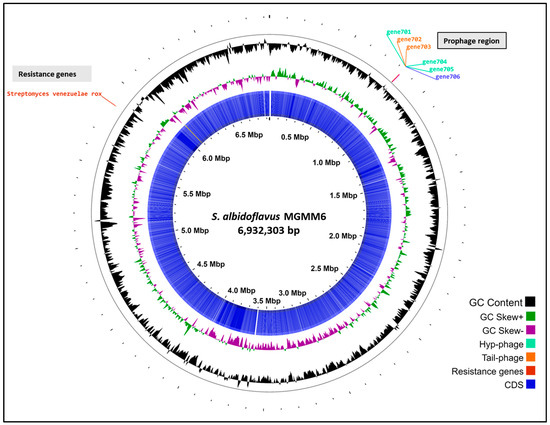
Figure 3.
The in silico circularization of the linear chromosome of S. albidoflavus MGMM6.

Table 1.
Summary of genome assembly and annotation of S. albidoflavus MGMM6.
Protein-encoding genes involved in chitin degradation, oxidation cleavage of glycosidic bonds, and reversible phosphorolysis of α-1,4-linked polysaccharides, namely chitinase, lytic polysaccharide monooxygenase, cellulase family glycosyl-hydrolase, and alpha-glucan family phosphorylase, were annotated in MGMM6. Moreover, a high proportion of genes involved in the degradation of aromatic compounds, such as multicopper oxidase, cytochrome P450 monooxygenase, DyP-type peroxidase, aromatic ring hydroxylating dioxygenases, aromatic ring-opening dioxygenase LigA, and nitric oxide dioxygenase, which play a key role in the biodegradation of numerous environmental pollutants, were predicted in S. albidoflavus MGMM6. The gene encoding 1-aminocyclopropane-1-carboxylate deaminase enzyme, which plays an important role in reducing biotic and abiotic stress in plants, was present in the annotated genome of S. albidoflavus MGMM6. The nitronate monooxygenase gene, a flavin-dependent enzyme that catalyzes the denitrification of propionate 3-nitronate (P3N) and other alkyl nitronates, was identified in the genome of MGMM6.
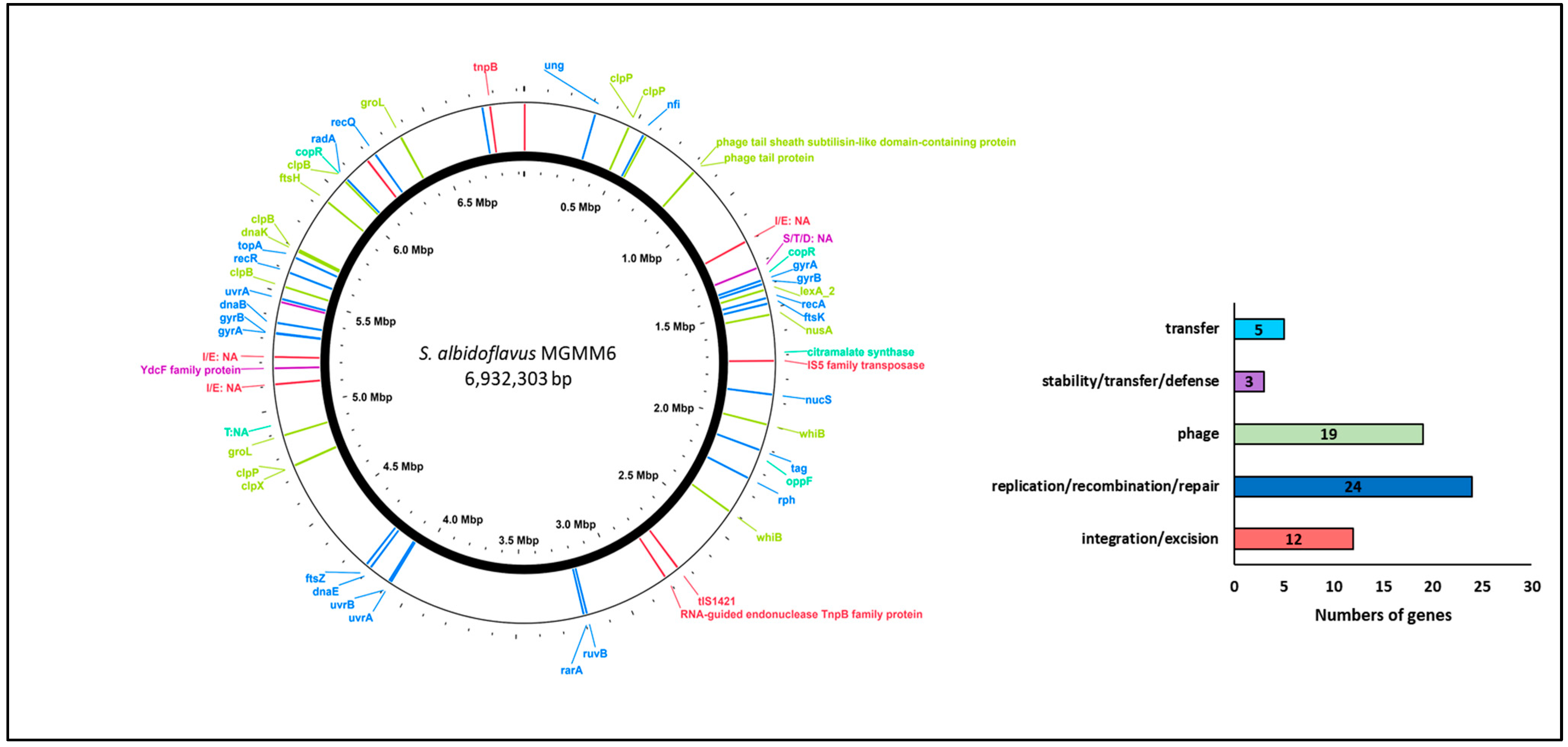
The analysis of gene families associated with integration and excision, replication and recombination, DNA repair, stability, defense, and the transfer of bacterial genetic elements and phages is presented in Figure 4. The results revealed that S. albidoflavus MGMM6 harbors 24 genes involved in the replication, recombination, or repair of mobile genetic elements, such as plasmid replication. Five genes associated with the mediation of inter-organism transfer of bacterial genetic elements were found in the genome of MGMM6. The presence of 12 genes (associated with integration and deletion) that control, mediate, or assist in site-specific recombination of genetic elements was predicted in S. albidoflavus MGMM6. The presence of 19 genes associated with the biological processes of bacteriophages, such as viral genome packaging lysis and lysogeny-associated machinery, was detected in MGMM6.
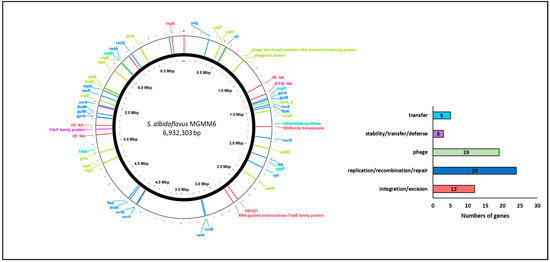
Figure 4.
Identification of genes associated with integration/excision, replication/recombination/repair, stability/defense, or transfer of bacterial genetic elements and phages.
The xenobiotic biodegradation pathway involved in MGMM6 is presented in Table 2. The obtained result revealed that S. albidoflavus MGMM6 harbors genes involved in the degradation of 1,3,4,6-Tetrachloro-1,4-cyclohexadiene to 2,5-Dichloro-2,5-cyclohexadiene-1,4-diol via alpha/beta hydrolase (LinB). In addition, among the distinct enzyme commission (EC) numbers involved in the metabolism of xenobiotics by cytochrome P450, 42.9% of the degradation pathway was predicted in the genome of MGMM6. Two (28.6%) ECs were associated with trinitrotoluene degradation. In addition, we found that MGMM6 can degrade (1R,2S)-Naphthalene-1.2-oxide to (91R)-OH-(2R)-Glutathionyl-1,2-dihydronaphthalene and (1R,2S)-Naphthalene-1.2-oxide to 1,2-Dhydroxy-1,2-dihydronaphthalene via epoxide hydrolase (EC 3.3.2.9), glutathione S-transferase (EC 2.5.1.18), alcohol dehydrogenase (EC 1.1.1.1), and acetaldehyde dehydrogenase (EC 1.2.1.10). The degradation ability of 1,2 dibromoethane and 2-bromo-acetaldehyde to S-[2-(N7-Guanyl) ethyl] N-acetyl-L-cysteine and S-(Formylmethyl)-glutathione via glutathione S-transferase (EC 2.5.1.18) was also found in the xenobiotic biodegradation pathway of S. albidoflavus MGMM6. The xenobiotic biodegradation of 4-nitrophenyl-phosphate to 4-nitrophenol via 4-nitrophenylphosphatase was predicted (Table 2).

Table 2.
Analysis of xenobiotic biodegradation predicted in the genome of S. albidoflavus MGMM6.
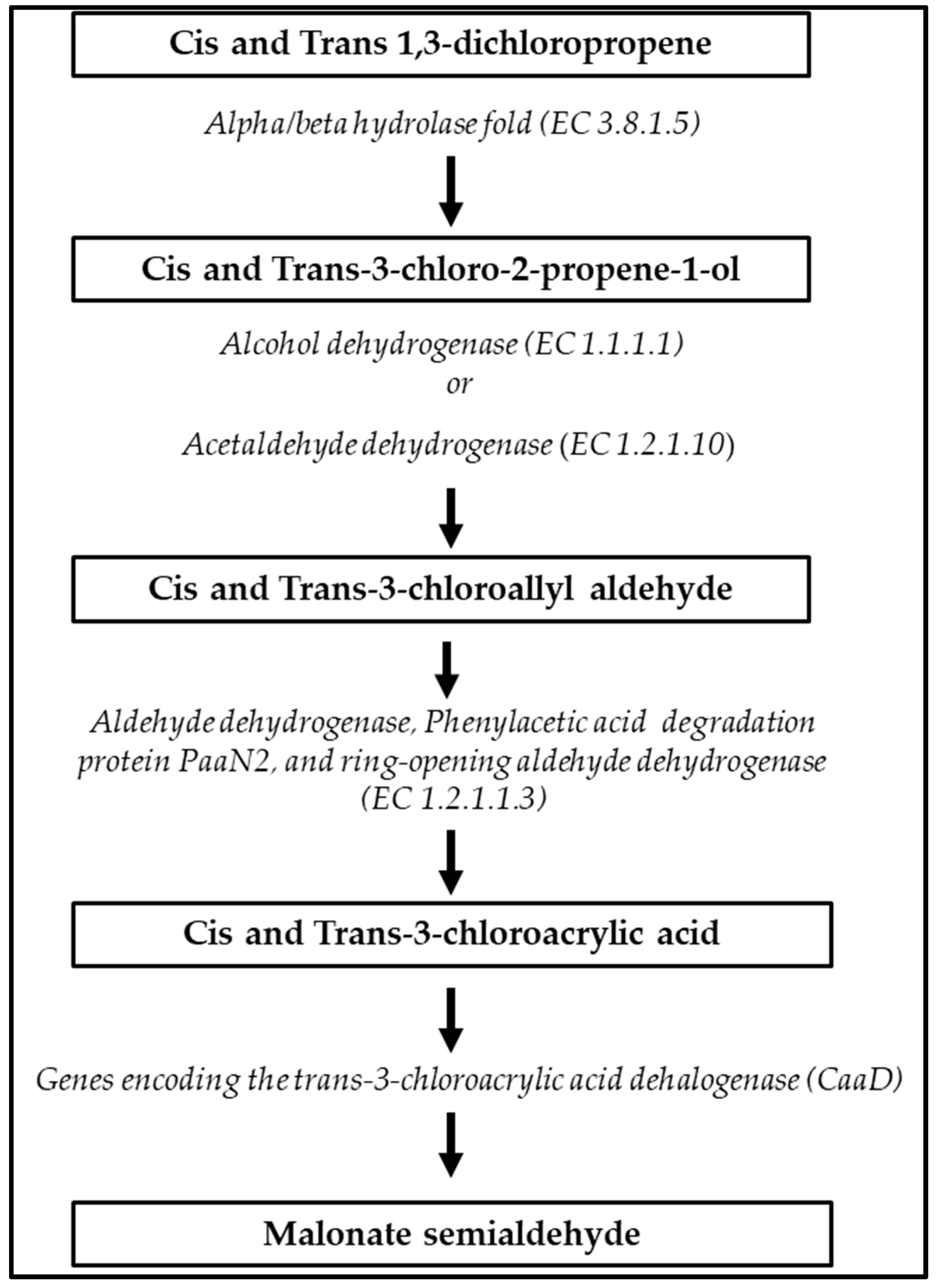
A full-degradation pathway of xenobiotic cis and trans-1,3-dichloropene, in which five distinct ECSs are involved, was predicted in S. albidoflavus MGMM6 (Figure 5). First, cis and trans-1,3-dichloropene are hydrolyzed into cis and trans-3-chloro-2-propene-1-ol; second, cis and trans-3-chloro-2-propene-1-ol are dehydrogenated to cis and trans-3-chlorophyll aldehyde, and then dehydrogenated to cis and trans-3-chloroacetic acid. Finally, cis and trans-3-chloroacetic acid undergoes dehydration to malonate semialdehyde.
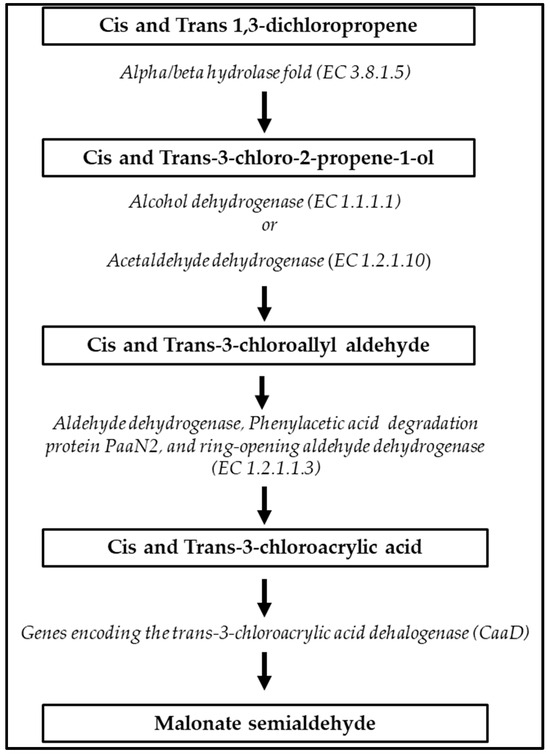
Figure 5.
Xenobiotic biodegradation pathway of cis and trans-1,3-dichloropropene by S. albidoflavus MGMM6.
Biosynthesis pathways of various metabolites with potentially beneficial activity for plants, including plant hormones, were predicted in the genome of S. albidoflavus MGMM6 (Table 3). The results showed that among various ECSs, MGMM6 produces 74 (56.5%) ECSs involved in the biosynthesis of plant hormones and 2 (66.7%) in the biosynthesis of brassinosteroids. The presence of pathways involved in the synthesis of auxin, abscisic acid, cytokinins, salicylic acid, gibberellin, strigolactone, jasmonic acid, and ethylene was predicted in MGMM6.

Table 3.
Biosynthesis pathways predicted in the genome of S. albidoflavus MGMM6.
Analysis of clusters involved in the biosynthesis of secondary metabolites using antiSMASH predicted the presence of 21 gene clusters in S. albidoflavus MGMM6 (Table 4).

Table 4.
Secondary metabolic gene clusters predicted in the genome of S. albidoflavus MGMM6.
Fifteen regions with similarity scores > 75% matched to the most similar known cluster were predicted in S. albidoflavus MGMM6. Among them, biosynthetic gene clusters (BGCs) encoding NRPS, terpene, ectoine, lanthipeptide-class-II and III, and NI-siderophore were found, which are responsible for the synthesis of antimycin, candicidin, minimycin, isorenieratene, ectoine, surugamide A/D, fredericamycin A, ectoine, hopene, AmfS, geosmin, and cyclofaulknamycin. Five BGCs encoding fNRPS, terpene/NRPS/NRPS-like, RiPP-like, and thiopeptide/LAP/RRE-containing peptides with less similarity (<70%) to most known BGCs were predicted to produce dudomycin A, fluostatins M-Q, hexacosalactone A, synechobactin, and julichrome.
3.2. Identification of Antimicrobial and Heavy Metal Resistance Genes in S. albidoflavus MGMM6
We analyzed the presence of heavy metal resistance and antimicrobial resistance genes harboring S. albidoflavus MGMM6 using bioinformatic approaches. Resistome analysis of S. albidoflavus MGMM6 using the CARD system revealed the presence of rifampin monooxygenase AMR gene family, which provides resistance to rifamycin antibiotic groups. This resistance occurs through the mechanism of antibiotic inactivation.
Several genes related to heavy metals were identified in the MGMM6 genome (Table 4). Genes providing resistance to copper, including multicopper oxidase, heme-copper oxidase subunit II, copper homeostasis cutC, copper ion binding), and copper chaperone PCu(A)C, were predicted in MGMM6. The annotated genome of MGMM6 harbors three arsenate reductase (arsC) genes that reduce arsenate As(V) to arsenite As(III). The operons recG and ruvB, which provide resistance to tellurium and selenium and are involved in repairing DNA damage caused by chromate or its derivatives, were identified in MGMM6. The gene modA involved in the transport of molybdenum and tungsten into cells was predicted in MGMM6. Two magnesium and cobalt transport proteins, CorA, which mediate the influx and efflux of magnesium, cobalt, and nickel ions, were identified in MGMM6. The F-box motif-containing protein (FBP) domain-containing protein, DsbA family protein, and 4 DsbA family oxidoreductases genes conferring resistance to cadmium, zinc, and mercury were predicted in S. albidoflavus MGMM6. Two magnesium and cobalt transport CorA operons that also confer resistance to magnesium, cobalt, nickel, and manganese were identified in MGMM6. In contrast, genes associated with resistance to vanadium and gold were not identified in the MGMM6 genome.
3.3. Antibiosis Activity
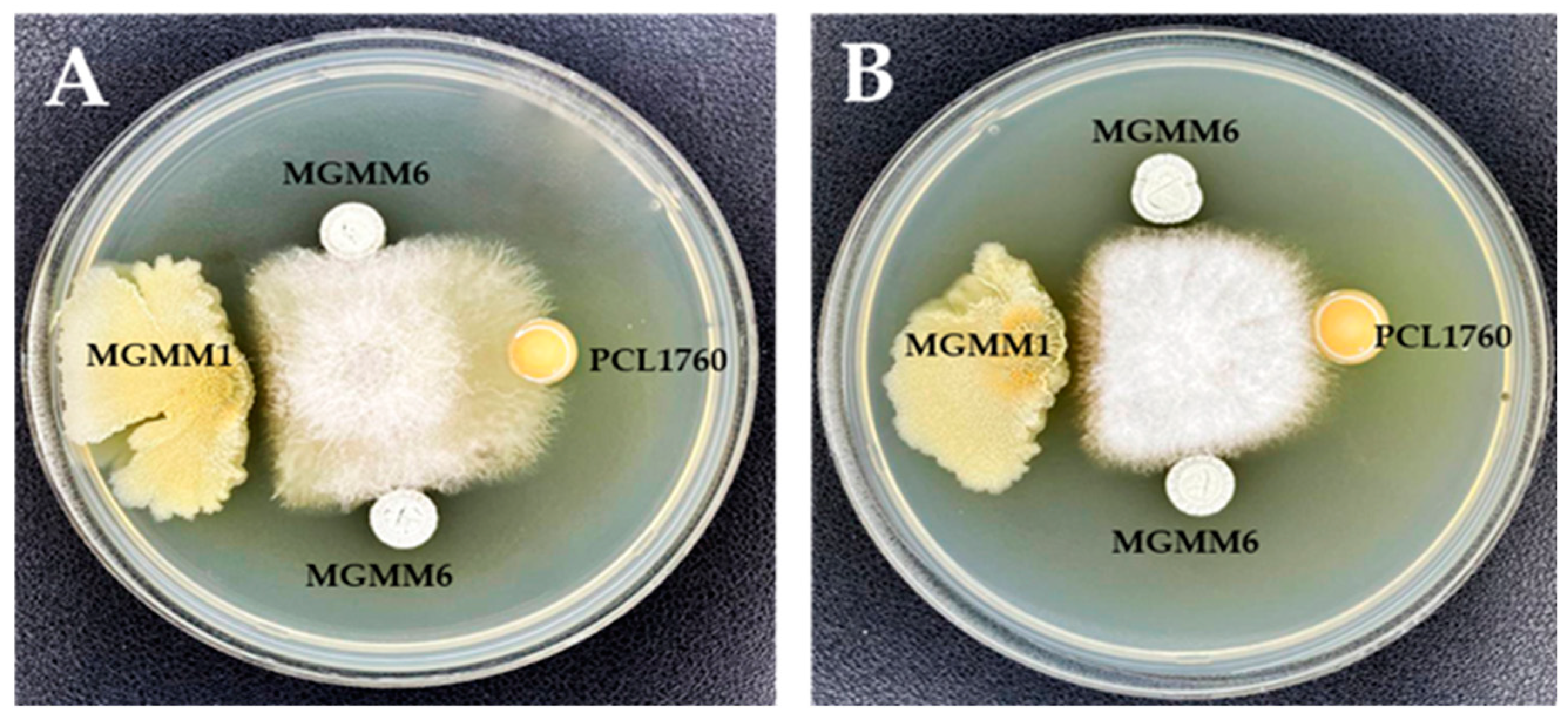
The antimicrobial activity of MGMM6 is shown in Figure 6. The obtained results indicate that MGMM6 inhibits the growth of the tested pathogens. A significant antagonistic effect was observed against the phytopathogen Fusarium oxysporium (Figure 6A).
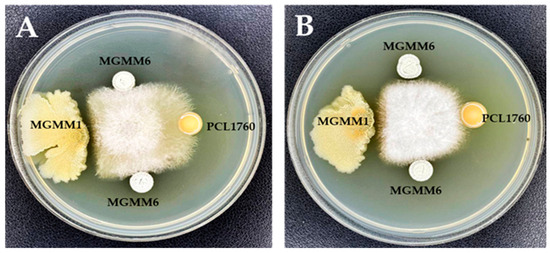
Figure 6.
Antimicrobial activity of S. albidoflavus MGMM6 against (A) F. oxysporum f. sp. radicis-lycopersici ZUM2407, and (B) F. proliferatum.
3.4. Dye Decolorization and Degradation Ability
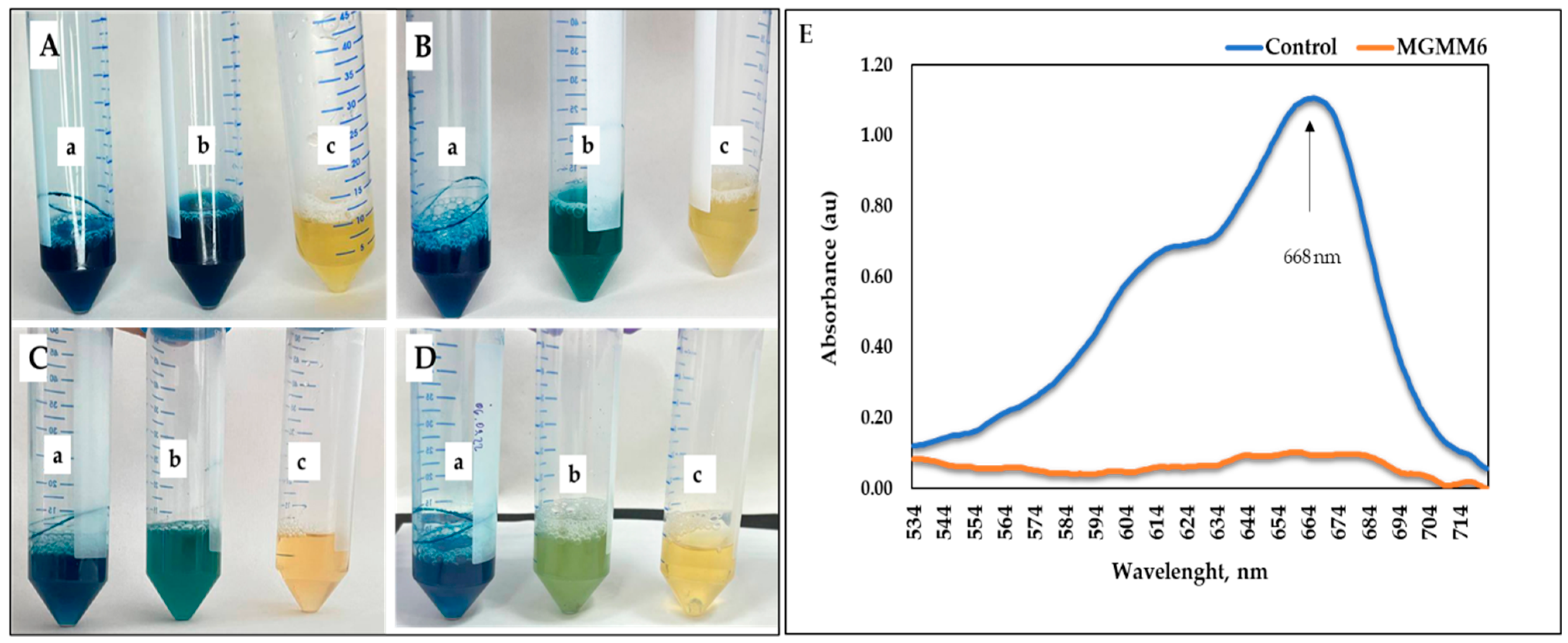
The ability of MGMM6 to decolorize and degrade dye is shown in Figure 7 and Figure 8. The results show that MGMM6 statistically decolorizes and degrades dyes. After incubation, decolorization of the solution was observed without changing the hydrogen index (pH) (Figure 7A–D). In addition, compared with the control, the UV–vis spectra of Coomassie brilliant blue G-250 of groups pre-treated with MGMM6 were not registered. Their absorption values were assayed as 1.51 ± 0.15 and 0.05 ± 0.03 au, respectively (Figure 7E). The dye removal ability of MGMM6 was measured as 96.67% ± 0.46.
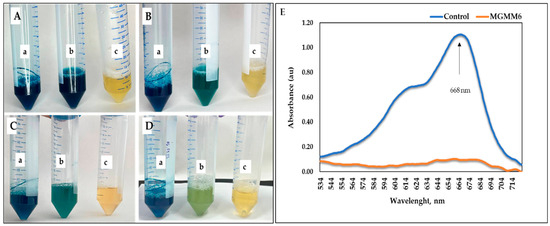
Figure 7.
S. albidoflavus MGMM6 was inoculated in a medium amended with 1% (v/v) Coomassie brilliant blue G-250 solution. (a) NB + 1% (v/v) of Coomassie brilliant blue G-250 solution; (b) NB + 1% Coomassie brilliant blue G-250 solution + MGMM6; (c) NB medium. Dye decolorization after inoculation (A) and 2 days (B), 4 days (C), and 7 days (D) after incubation at 28 ± 1 °C. The UV–vis spectra registered on the spectrophotometer 7 days after incubation (E)—where control and MGMG6 represent the UV–vis spectra of NB + 1% (v/v) of Coomassie brilliant blue G-250 solution and NB + 1% Coomassie brilliant blue G-250 solution + MGMM6.
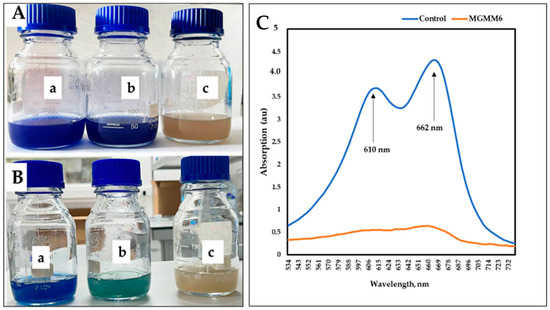
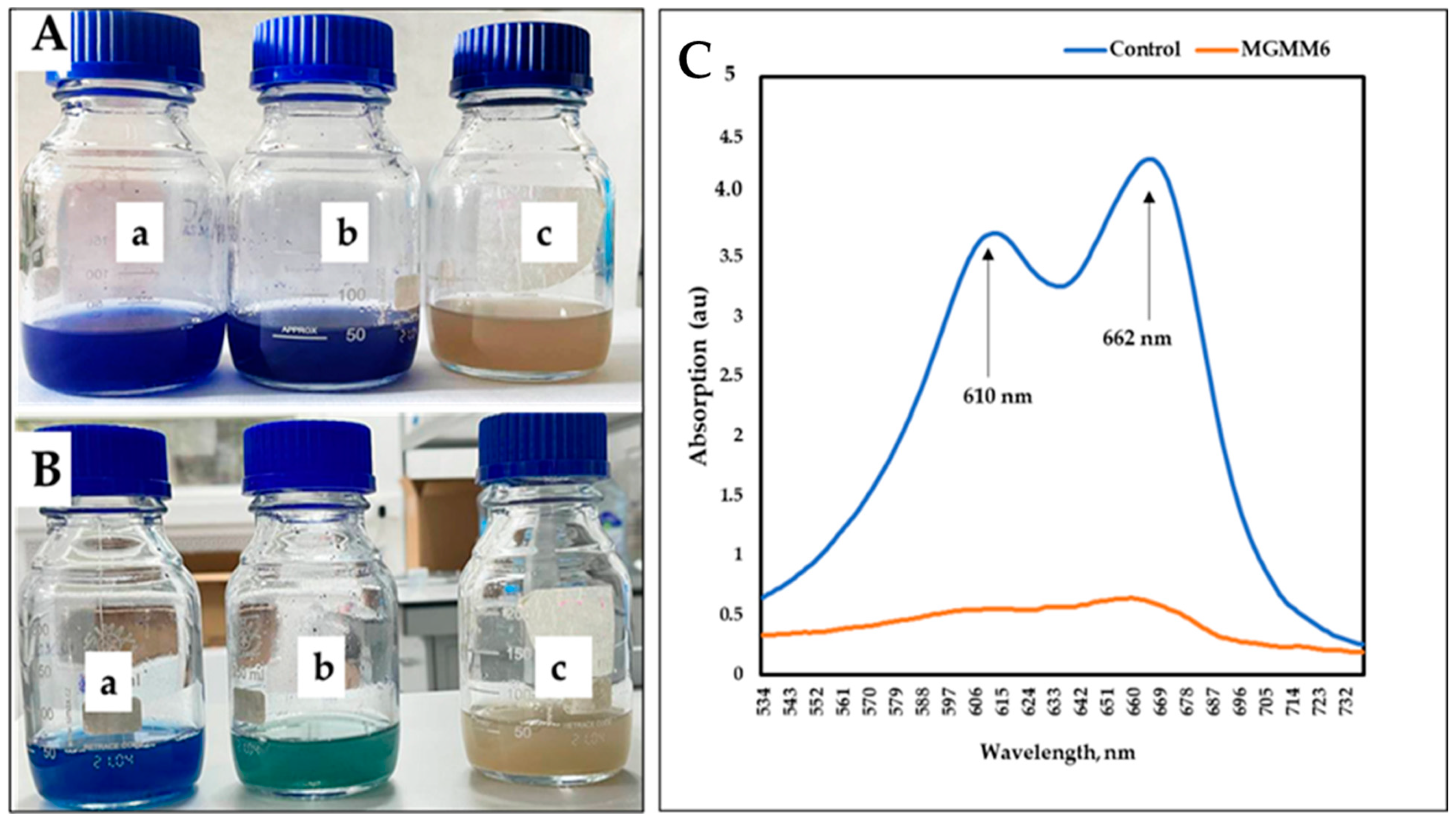
Figure 8.
The dye decolorization and degradation ability of S. albidoflavus MGMM6. Dye decolorization after one (A) and 14 (B) days of incubation at 28 ± 1 °C. The absorption spectrum of dye degradation after 14 days of incubation (C)—where control and MGMG6 represent the UV–vis spectra of wastewater + 1% (v/v) of Coomassie brilliant blue G-250 solution and wastewater + 1% Coomassie brilliant blue G-250 solution + MGMM6. Streptomyces albidoflavus MGMM6 was inoculated in wastewater amended with 1% (v/v) Coomassie brilliant blue G-250 solution. Wastewater + 1% (v/v) Coomassie brilliant blue G-250 solution (a); Wastewater + 1% (v/v) Coomassie brilliant blue G-250 solution + MGMM6 (b); wastewater (c).
Similar results were obtained using wastewater amended with 1% Coomassie Brilliant Blue G-250 (Figure 8A,B). After 14 days of incubation, two distinct peaks were detected in the control group at 610 and 662 nm, with absorption values of 4.28 ± 0.25 and 0.68 ± 0.05 a.u (absorbance units), respectively (Figure 8C). In comparison, the group pretreated with MGMM6 showed a slight UV-Vis spectrum of Coomassie Brilliant Blue G-250 at 662 nm, with a value of 0.63 a.u. The effectiveness of S. albidoflavus MGMM6 in degrading and decolorizing wastewater amended with 1% (v/v) Coomassie Brilliant Blue G-250 was up to 85.98 ± 0.25%. The effectiveness of S. albidoflavus MGMM6 in degrading and decolorizing wastewater amended with 1% Coomassie Brilliant Blue G-250 solution was up to 85.98 ± 0.25%.
4. Discussion
Microbial biotechnology is an emerging science field with significant potential applications, including food security, human health and nutrition, waste management, and plant protection [44,45,46]. The application of microbial strains in these related fields mainly depends on the specific arrangement of their genetic material, including the presence of certain genes and regulatory elements. In the context of potential sources of compounds with antimicrobial activity, according to the results of this extensive study, these abilities were identified in the genome of MGMM6 (Figure 6 and Table 3). The ability of S. albidoflavus MGMM6 to inhibit the growth of pathogens is mainly related to the presence of these putative gene clusters responsible for the synthesis of cyclic peptides such as valinomycin/montanastati, cyclofaulknamycin, hexacosalactone, and antimycin. These compounds possess strong antimicrobial activity against bacterial and fungal pathogens [47,48,49,50,51,52,53]. For example, surugamides and their derivatives, such as acyl-surugamide A, possess anticancer and antifungal activities [53,54]. Desferrioxamine B, which is a cation metal chelator, has been reported to be used in several medicinal and analytical applications, such as aluminum chelation therapy in people on dialysis [55,56]. Hence, S. albidoflavus MGMM6 can be applied in agriculture as a biocontrol agent and chelating agent to eliminate the deficiency of metals, such as iron, in plants. Polycyclic tetramine macrolactams (SGR PTM) and antimycin are well-known compounds secreted by streptomyces with fungicidal, insecticidal, and acaricidal activities [57,58]. Fusarium is a well-known genus that causes significant agricultural yield losses, reaching up to 14% annually [59,60,61]. Species of this genus produce mycotoxins in food and agricultural products and are suspected to be associated with various diseases in mammals and other organisms [62,63]. In this study, through dual-plate assay, S. albidoflavus MGMM6 inhibited the growth of Fusarium species. The obtained result consistent with these previously reported by [20], whereby S. albidoflavus strain CARA17 isolated from root diseases of grapevine plants (Vitis vinifera) showed the ability to inhibit the growth of fungal soil-borne pathogens such as Athelia rolfsii, F. oxysporum, Plectosphaerella ramiseptata, Sclerotinia sclerotiorum, and Verticillium dahlia. The role of microbial biotechnology in sustainable agriculture and environmental health is well documented. Microbial agents assimilate and acquire plant-essential nutrients, remediate and improve soil physicochemical properties, modulate the synthesis of plant hormones, and produce various signal compounds that inhibit the growth of several pathogens, as well as improve plant activities under stress conditions [64,65,66]. Moreover, through bioinformatics approaches, several beneficial plant genes responsible for degrading xenobiotic compounds, remediating heavy metals, and metabolic pathways involved in plant hormones and lytic enzyme synthesis were predicted in S. albidoflavus MGMM6. These genes were found to be involved in several biodegradation processes. Castillo et al. [67] reported the efficient ability of S. albidoflavus strain A7-9 to degrade up to 100% 4 mg/L of N′-3,4-Dichlorophenyl-N, N-dimethylurea (an herbicide of the aryl-urea class) for 15 days at 25 °C. These abilities to use aromatics as the sole carbon and energy sources are related to the presence of genes such as cytochrome P450 monooxygenase (P450s), oxidase, lignin peroxidase, manganese peroxidase, DyP-type peroxidase, versatile peroxidase, humic acid peroxidases, dioxygenase, and laccase [68,69,70,71,72]. The above genes were also found in the genome of S. albidoflavus MGMM6, which may refer to its ability to decompose the above aromatic compounds, as in the strain A7-9 of S. albidoflavus [67].
Considering the presence of these genes in the genome of MGMM6, we were interested in testing its ability to degrade polycyclic aromatic compounds such as triphenylmethane dyes. Different microorganisms isolated from different taxonomic groups have demonstrated several catabolic activities in the degradation of hazardous materials such as 2,4-dinitrophenol, 2,4,6-trichlorophenol, and pyridine [73,74]. In this study, S. albidoflavus MGMM6 demonstrated the ability to decolorize and degrade Coomassie Brilliant blue R250 from wastewater at 28 ± 1 °C at a constant pH (8.0). The obtained results correlated with previously reported data. For example, Musengi et al. [75] isolated an extracellular DyP-type peroxidase class from S. albidoflavus BSII#1, which showed the ability to degrade and decolonize reactive blue 4, reactive black 5, and Azure, as well as, to oxidize 2,4-dichlorophenol, 2,4-dichlorophenol, 2,6-dimethoxyphenol, 4-tert-butylcatechol, ABTS [2,2′-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid)], caffeic acid, catechol, guaiacol, L-DOPA, o-aminophenol, phenol, and pyrogallol. Similar results were obtained by Alaidaroos et al. [76], whereby Streptomyces flavus BA4 isolated from wastewater showed the ability to remove chromium up to 300 mg/L.
The survival and behavior of bacteria in soil mainly depend on their genomic architecture, including their resistome, mobilome, and metabolic features. Numerous studies have shown that antibiotics harm soil microbial communities [39,77,78,79,80]. For example, Liu et al. [81] found that sulfamethazine negatively affects the phosphatase activity and respiration of soil microflora. Shan et al. [82] demonstrated that the tetracycline antibiotic group can affect the denitrification process of soil microbial communities. Moreover, analysis of various genomes has shown that species with a lower number of transposons tend to exhibit greater genomic stability [83,84].
Microbes can be successfully used to sequester heavy metals from environments [85,86]. In this study, through bioinformatic methods, we found that MGMM6 harbors several heavy metal resistance genes, such as cadmium, mercury, chromium, tellurium, arsenic (As), and antimony (Sb). These findings are consistent with those reported by Kaliyaraj et al. [87], whereby S. albidoflavus T N10 isolated from insect nests demonstrated the ability to recover various heavy metals, including Ca, Cu, Cd, Ni, Ag, and Pb, under laboratory conditions. Thus, these results suggest that S. albidoflavus is a promising candidate for its application as a heavy metal remediator. Understanding the relationship between microbial metabolites and their host plants is crucial for developing effective biological agents [88,89]. Streptomyces albidoflavus MGMM6 uses multiple metabolite pathways, such as siderophore secretion, nitrogen fixation, and phytohormones, to establish itself in the rhizosphere, compete with other microorganisms in the soil, and protect the plant from pathogens. Furthermore, the limited number of transposable elements within the genome of MGMM6, along with the presence of genes involved in replication, recombination, and metabolic pathways for the degradation of xenobiotic pollutants such as cis and trans-1,3-chloroprene, provide advantages in the genetic manipulation of MGMM6 for various biotechnology applications to plant protection, bioremediation, and biodegradation of persistent organic pollutants. The versatility of S. albidoflavus MGMM6 makes it an excellent model organism for novel biotechnological applications.
5. Conclusions
Microbiological biotechnology has numerous applications across various scientific fields, each of which can benefit from the unique capabilities of microorganisms. In agriculture, microbiological biotechnology can lead to the development of more resilient and productive crops through the manipulation of plant-microbe interactions, nutrient cycling, and pest management. In this study, S. albidoflavus MGMM6 was demonstrated to be an effective bioagent for degrading xenobiotic compounds, remediating heavy metals, and controlling pathogen growth. In addition, the genome of MGMM6 harbors contains several genes involved in replication, recombination, and metabolic pathways for the degradation of xenobiotic pollutants. These features provide distinct advantages for genetic manipulation, making S. albidoflavus an attractive candidate for various biotechnology applications such as plant protection, bioremediation, and engineering manipulation of secondary metabolite biosynthesis.
Author Contributions
Conceptualization, R.G.C.D.; methodology, R.G.C.D.; data acquisition, R.G.C.D.; software, R.G.C.D.; investigation, R.G.C.D., M.F. and S.K.; analysis, R.G.C.D.; writing—original draft preparation, R.G.C.D.; writing—review and editing, D.M.A. and S.Z.V.; supervision, D.M.A. and S.Z.V. All authors have read and agreed to the published version of the manuscript.
Funding
This study was conducted with financial support provided by the Ministry of Education and Science of the Russian Federation, Grant # RF-1930.61321X0001/15.IP.21.0020, and from the government assignment for the FRC Kazan Scientific Center of RAS.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
This genome sequencing project of S. albidoflavus MGMM6 has been deposited in the NCBI genome database under NCBI Reference Sequence: CP128384.1. The BioProject and BioSample accessions are PRJNA224116 and SAMN34378109, respectively.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Dincer, I. Renewable Energy and Sustainable Development: A Crucial Review. Renew. Sustain. Energy Rev. 2000, 4, 157–175. [Google Scholar] [CrossRef]
- Bondarczuk, K.; Markowicz, A.; Piotrowska-Seget, Z. The Urgent Need for Risk Assessment on the Antibiotic Resistance Spread via Sewage Sludge Land Application. Environ. Int. 2016, 87, 49–55. [Google Scholar] [CrossRef]
- Cesare, A.D.; Eckert, E.; Corno, G. Co-Selection of Antibiotic and Heavy Metal Resistance in Freshwater Bacteria. J. Limnol. 2016, 75, 9–66. [Google Scholar] [CrossRef]
- Yang, X.; Wang, J.; Zhao, X.; Wang, Q.; Xue, R. Increasing Manganese Peroxidase Production and Biodecolorization of Triphenylmethane Dyes by Novel Fungal Consortium. Bioresour. Technol. 2011, 102, 10535–10541. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Hu, H.; Wang, G.; Li, Z.; Wang, B.; Jia, X.; Zhao, Y. Removal of Malachite Green from Aqueous Solution by Immobilized Pseudomonas Sp. DY1 with Aspergillus Oryzae. Int. Biodeterior. Biodegrad. 2011, 65, 429–434. [Google Scholar] [CrossRef]
- Valls, M.; de Lorenzo, V. Exploiting the Genetic and Biochemical Capacities of Bacteria for the Remediation of Heavy Metal Pollution. FEMS Microbiol. Rev. 2002, 26, 327–338. [Google Scholar] [CrossRef] [PubMed]
- Álvarez, A.; Catalano, S.A.; Amoroso, M.J. Heavy Metal Resistant Strains Are Widespread along Streptomyces Phylogeny. Mol. Phylogenet. Evol. 2013, 66, 1083–1088. [Google Scholar] [CrossRef] [PubMed]
- Ojuederie, O.B.; Babalola, O.O. Microbial and Plant-Assisted Bioremediation of Heavy Metal Polluted Environments: A Review. Int. J. Environ. Res. Public Health 2017, 14, 1504. [Google Scholar] [CrossRef]
- Gadd, G.M. Metals, minerals and microbes: Geomicrobiology and bioremediation. Microbiology 2010, 156, 609–643. [Google Scholar] [CrossRef]
- Lei, Q.; Zhong, J.; Chen, S.-F.; Wu, S.; Huang, Y.; Guo, P.; Mishra, S.; Bhatt, K.; Chen, S. Microbial Degradation as a Powerful Weapon in the Removal of Sulfonylurea Herbicides. Environ. Res. 2023, 235, 116570. [Google Scholar] [CrossRef]
- Mishra, S.; Huang, Y.; Li, J.; Wu, X.; Zhou, Z.; Lei, Q.; Bhatt, P.; Chen, S. Biofilm-Mediated Bioremediation Is a Powerful Tool for the Removal of Environmental Pollutants. Chemosphere 2022, 294, 133609. [Google Scholar] [CrossRef] [PubMed]
- Rastogi, R.P.; Sinha, R.P. Biotechnological and Industrial Significance of Cyanobacterial Secondary Metabolites. Biotechnol. Adv. 2009, 27, 521–539. [Google Scholar] [CrossRef] [PubMed]
- Yadav, A.N. Microbial biotechnology for bioprospecting of microbial bioactive compounds and secondary metabolites. J. Appl. Biol. Biotechnol. 2021, 9, 1–6. [Google Scholar] [CrossRef]
- Bugg, T.D.; Ahmad, M.; Hardiman, E.M.; Singh, R. The Emerging Role for Bacteria in Lignin Degradation and Bio-Product Formation. Curr. Opin. Biotechnol. 2011, 22, 394–400. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Geng, P.; Xiao, Y.; Hu, M. Bioremediation of β-Cypermethrin and 3-Phenoxybenzaldehyde Contaminated Soils Using Streptomyces Aureus HP-S-01. Appl. Microbiol. Biotechnol. 2012, 94, 505–515. [Google Scholar] [CrossRef] [PubMed]
- Bérdy, J. Thoughts and Facts about Antibiotics: Where We Are Now and Where We Are Heading. J. Antibiot. 2012, 65, 385–395. [Google Scholar] [CrossRef]
- Rong, X.; Guo, Y.; Huang, Y. Proposal to Reclassify the Streptomyces albidoflavus Clade on the Basis of Multilocus Sequence Analysis and DNA–DNA Hybridization, and Taxonomic Elucidation of Streptomyces griseus Subsp. Solvifaciens. Syst. Appl. Microbiol. 2009, 32, 314–322. [Google Scholar] [CrossRef]
- Seipke, R.F.; Hutchings, M.I. The Regulation and Biosynthesis of Antimycins. Beilstein J. Org. Chem. 2013, 9, 2556–2563. [Google Scholar] [CrossRef]
- Du, Y.; Wang, T.; Jiang, J.; Wang, Y.; Lv, C.; Sun, K.; Sun, J.; Yan, B.; Kang, C.; Guo, L. Biological Control and Plant Growth Promotion Properties of Streptomyces Albidoflavus St-220 Isolated from Salvia Miltiorrhiza Rhizosphere. Front. Plant Sci. 2022, 13, 976813. [Google Scholar] [CrossRef]
- Carlucci, A.; Raimondo, M.L.; Colucci, D.; Lops, F. Streptomyces albidoflavus Strain CARA17 as a Biocontrol Agent against Fungal Soil-Borne Pathogens of Fennel Plants. Plants 2022, 11, 1420. [Google Scholar] [CrossRef]
- Green, M.R.; Sambrook, J. Isolation of High-Molecular-Weight DNA Using Organic Solvents. Cold Spring Harb. Protoc. 2017, 2017, pdb.prot093450. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.; Pirrung, M.; McCue, L.A. FQC Dashboard: Integrates FastQC Results into a Web-Based, Interactive, and Extensible FASTQ Quality Control Tool. Bioinformatics 2017, 33, 3137–3139. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An Ultra-Fast All-in-One FASTQ Preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Souvorov, A.; Agarwala, R.; Lipman, D.J. SKESA: Strategic k-Mer Extension for Scrupulous Assemblies. Genome Biol. 2018, 19, 153. [Google Scholar] [CrossRef] [PubMed]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving Bacterial Genome Assemblies from Short and Long Sequencing Reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Chen, K.-T.; Lu, C.L. CSAR-Web: A Web Server of Contig Scaffolding Using Algebraic Rearrangements. Nucleic Acids Res. 2018, 46, W55–W59. [Google Scholar] [CrossRef]
- De Sa, P.H.; Miranda, F.; Veras, A.; de Melo, D.M.; Soares, S.; Pinheiro, K.; Ramos, R.T. GapBlaster—A graphical gap filler for prokaryote genomes. PLoS ONE 2016, 11, e0155327. [Google Scholar] [CrossRef]
- Antipov, D.; Hartwick, N.; Shen, M.; Raiko, M.; Lapidus, A.; Pevzner, P.A. plasmidSPAdes: Assembling Plasmids from Whole Genome Sequencing Data. Bioinformatics 2016, 32, 3380–3387. [Google Scholar] [CrossRef]
- Robertson, J.; Nash, J.H.E. MOB-Suite: Software Tools for Clustering, Reconstruction and Typing of Plasmids from Draft Assemblies. Microb. Genom. 2018, 4, e000206. [Google Scholar] [CrossRef]
- Robertson, J.; Bessonov, K.; Schonfeld, J.; Nash, J.H.E. Universal Whole-Sequence-Based Plasmid Typing and Its Utility to Prediction of Host Range and Epidemiological Surveillance. Microb. Genom. 2020, 6, mgen000435. [Google Scholar] [CrossRef] [PubMed]
- Tatusova, T.; DiCuccio, M.; Badretdin, A.; Chetvernin, V.; Nawrocki, E.P.; Zaslavsky, L.; Lomsadze, A.; Pruitt, K.D.; Borodovsky, M.; Ostell, J. NCBI Prokaryotic Genome Annotation Pipeline. Nucleic Acids Res. 2016, 44, 6614–6624. [Google Scholar] [CrossRef]
- Schwengers, O.; Jelonek, L.; Dieckmann, M.A.; Beyvers, S.; Blom, J.; Goesmann, A. Bakta: Rapid and Standardized Annotation of Bacterial Genomes via Alignment-Free Sequence Identification. Microb. Genom. 2021, 7, 000685. [Google Scholar] [CrossRef] [PubMed]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the Quality of Microbial Genomes Recovered from Isolates, Single Cells, and Metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef] [PubMed]
- McArthur, A.G.; Waglechner, N.; Nizam, F.; Yan, A.; Azad, M.A.; Baylay, A.J.; Bhullar, K.; Canova, M.J.; De Pascale, G.; Ejim, L.; et al. The Comprehensive Antibiotic Resistance Database. Antimicrob. Agents Chemother. 2013, 57, 3348–3357. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Shaw, S.; Augustijn, H.E.; Reitz, Z.L.; Biermann, F.; Alanjary, M.; Fetter, A.; Terlouw, B.R.; Metcalf, W.W.; Helfrich, E.J.N.; et al. antiSMASH 7.0: New and Improved Predictions for Detection, Regulation, Chemical Structures and Visualisation. Nucleic Acids Res. 2023, 51, W46–W50. [Google Scholar] [CrossRef]
- Brown, C.L.; Mullet, J.; Hindi, F.; Stoll, J.E.; Gupta, S.; Choi, M.; Keenum, I.; Vikesland, P.; Pruden, A.; Zhang, L. mobileOG-Db: A Manually Curated Database of Protein Families Mediating the Life Cycle of Bacterial Mobile Genetic Elements. Appl. Environ. Microbiol. 2022, 88, e00991-22. [Google Scholar] [CrossRef]
- Pal, C.; Bengtsson-Palme, J.; Rensing, C.; Kristiansson, E.; Larsson, D.G.J. BacMet: Antibacterial Biocide and Metal Resistance Genes Database. Nucleic Acids Res. 2014, 42, D737–D743. [Google Scholar] [CrossRef]
- Feldgarden, M.; Brover, V.; Gonzalez-Escalona, N.; Frye, J.G.; Haendiges, J.; Haft, D.H.; Hoffmann, M.; Pettengill, J.B.; Prasad, A.B.; Tillman, G.E.; et al. AMRFinderPlus and the Reference Gene Catalog Facilitate Examination of the Genomic Links among Antimicrobial Resistance, Stress Response, and Virulence. Sci. Rep. 2021, 11, 12728. [Google Scholar] [CrossRef]
- Bonev, B.; Hooper, J.; Parisot, J. Principles of Assessing Bacterial Susceptibility to Antibiotics Using the Agar Diffusion Method. J. Antimicrob. Chemother. 2008, 61, 1295–1301. [Google Scholar] [CrossRef]
- Aravind, P.; Selvaraj, H.; Ferro, S.; Sundaram, M. An Integrated (Electro- and Bio-Oxidation) Approach for Remediation of Industrial Wastewater Containing Azo-Dyes: Understanding the Degradation Mechanism and Toxicity Assessment. J. Hazard. Mater. 2016, 318, 203–215. [Google Scholar] [CrossRef] [PubMed]
- Diabankana, R.G.C.; Afordoanyi, D.M.; Safin, R.I.; Nizamov, R.M.; Karimova, L.Z.; Validov, S.Z. Antifungal Properties, Abiotic Stress Resistance, and Biocontrol Ability of Bacillus mojavensis PS17. Curr. Microbiol. 2021, 78, 3124–3132. [Google Scholar] [CrossRef] [PubMed]
- Czaja, W.; Krystynowicz, A.; Bielecki, S.; Brown, R.M. Microbial Cellulose—The Natural Power to Heal Wounds. Biomaterials 2006, 27, 145–151. [Google Scholar] [CrossRef] [PubMed]
- Sen, R. Biotechnology in Petroleum Recovery: The Microbial EOR. Prog. Energy Combust. Sci. 2008, 34, 714–724. [Google Scholar] [CrossRef]
- Yadav, A.N.; Singh, S.; Mishra, S.; Gupta, A. Recent Advancement in White Biotechnology through Fungi; Springer International Publishing: Cham, Switzerland, 2019; p. 528. [Google Scholar]
- Haeder, S.; Wirth, R.; Herz, H.; Spiteller, D. Candicidin-Producing Streptomyces Support Leaf-Cutting Ants to Protect Their Fungus Garden against the Pathogenic Fungus Escovopsis. Proc. Natl. Acad Sci. USA 2009, 106, 4742–4746. [Google Scholar] [CrossRef] [PubMed]
- Dias, D.A.; Urban, S.; Roessner, U. A Historical Overview of Natural Products in Drug Discovery. Metabolites 2012, 2, 303–336. [Google Scholar] [CrossRef] [PubMed]
- Marcone, G.L.; Binda, E.; Berini, F.; Marinelli, F. Old and New Glycopeptide Antibiotics: From Product to Gene and Back in the Post-Genomic Era. Biotechnol. Adv. 2018, 36, 534–554. [Google Scholar] [CrossRef]
- Pishchany, G.; Mevers, E.; Ndousse-Fetter, S.; Horvath, D.J.; Paludo, C.R.; Silva-Junior, E.A.; Koren, S.; Skaar, E.P.; Clardy, J.; Kolter, R. Amycomicin Is a Potent and Specific Antibiotic Discovered with a Targeted Interaction Screen. Proc. Natl. Acad. Sci. USA 2018, 115, 10124–10129. [Google Scholar] [CrossRef]
- Zhang, D.; Lu, Y.; Chen, H.; Wu, C.; Zhang, H.; Chen, L.; Chen, X. Antifungal Peptides Produced by Actinomycetes and Their Biological Activities against Plant Diseases. J. Antibiot. 2020, 73, 265–282. [Google Scholar] [CrossRef]
- Shi, P.; Li, Y.; Zhu, J.; Shen, Y.; Wang, H. Targeted Discovery of the Polyene Macrolide Hexacosalactone A from Streptomyces by Reporter-Guided Selection of Fermentation Media. J. Nat. Prod. 2021, 84, 1924–1929. [Google Scholar] [CrossRef]
- Takada, K.; Ninomiya, A.; Naruse, M.; Sun, Y.; Miyazaki, M.; Nogi, Y.; Okada, S.; Matsunaga, S. Surugamides A-E, Cyclic Octapeptides with Four D-Amino Acid Residues, from a Marine Streptomyces sp.: LC-MS-Aided Inspection of Partial Hydrolysates for the Distinction of D- and L-Amino Acid Residues in the Sequence. J. Org. Chem. 2013, 78, 6746–6750. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Nazari, B.; Moon, K.; Bushin, L.B.; Seyedsayamdost, M.R. Discovery of a Cryptic Antifungal Compound from Streptomyces Albus J1074 Using High-Throughput Elicitor Screens. J. Am. Chem. Soc. 2017, 139, 9203–9212. [Google Scholar] [CrossRef]
- Codd, R.; Richardson-Sanchez, T.; Telfer, T.J.; Gotsbacher, M.P. Advances in the Chemical Biology of Desferrioxamine B. ACS Chem. Biol. 2018, 13, 11–25. [Google Scholar] [CrossRef] [PubMed]
- Bellotti, D.; Remelli, M. Deferoxamine B: A Natural, Excellent and Versatile Metal Chelator. Molecules 2021, 26, 3255. [Google Scholar] [CrossRef] [PubMed]
- Kido, G.S.; Spyhalski, E. Antimycin A, an Antibiotic with Insecticidal and Miticidal Properties. Science 1950, 112, 172–173. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Huang, H.; Liang, J.; Wang, M.; Lu, L.; Shao, Z.; Cobb, R.E.; Zhao, H. Activation and Characterization of a Cryptic Polycyclic Tetramate Macrolactam Biosynthetic Gene Cluster. Nat. Commun. 2013, 4, 2894. [Google Scholar] [CrossRef] [PubMed]
- Trabelsi, D.; Mhamdi, R. Microbial Inoculants and Their Impact on Soil Microbial Communities: A Review. Biomed. Res. Int. 2013, 2013, 863240. [Google Scholar] [CrossRef] [PubMed]
- Bai, G.; Shaner, G. Management and resistance in wheat and barley to Fusarium head blight. Annu. Rev. Phytopathol. 2004, 42, 135–161. [Google Scholar] [CrossRef]
- Oldenburg, E.; Höppner, F.; Ellner, F.; Weinert, J. Fusarium diseases of maize associated with mycotoxin contamination of agricultural products intended to be used for food and feed. Mycotoxin. Res. 2017, 33, 167–182. [Google Scholar] [CrossRef]
- Anand, G.; Rajeshkumar, K.C. Challenges and Threats Posed by Plant Pathogenic Fungi on Agricultural Productivity and Economy. In Fungal Diversity, Ecology and Control Management; Springer Nature Singapore: Singapore, 2022; pp. 483–493. [Google Scholar] [CrossRef]
- Tekauz, A.; McCallum, B.; Gilbert, J. Fusarium head blight of barley in western Canada. Can. J. Plant Pathol. 2000, 22, 9–16. [Google Scholar] [CrossRef]
- Mudili, V.; Siddaih, C.N.; Nagesh, M.; Garapati, P.; Naveen Kumar, K.; Murali, H.S.; Yli Mattila, T.; Batra, H.V. Mould incidence and mycotoxin contamination in freshly harvested maize kernels originated from India. J. Sci. Food Agric. 2014, 94, 2674–2683. [Google Scholar] [CrossRef] [PubMed]
- Nelson, M.S.; Sadowsky, M.J. Secretion Systems and Signal Exchange between Nitrogen-Fixing Rhizobia and Legumes. Front. Plant Sci. 2015, 6, 491. [Google Scholar] [CrossRef] [PubMed]
- Massalha, H.; Korenblum, E.; Tholl, D.; Aharoni, A. Small Molecules Below-Ground: The Role of Specialized Metabolites in the Rhizosphere. Plant J. 2017, 90, 788–807. [Google Scholar] [CrossRef] [PubMed]
- Castillo, M.A.; Felis, N.; Aragón, P.; Cuesta, G.; Sabater, C. Biodegradation of the Herbicide Diuron by Streptomycetes Isolated from Soil. Int. Biodeterior. Biodegrad. 2006, 58, 196–202. [Google Scholar] [CrossRef]
- Seo, J.-S.; Keum, Y.-S.; Li, Q.X. Bacterial Degradation of Aromatic Compounds. Int. J. Environ. Res. Public Health 2009, 6, 278–309. [Google Scholar] [CrossRef] [PubMed]
- Jaouadi, B.; Rekik, H.; Badis, A.; Jaouadi, N.Z.; Belhoul, M.; Hmidi, M.; Kourdali, S.; Fodil, D.; Bejar, S. Production, Purification, and Characterization of a Highly Thermostable and Humic Acid Biodegrading Peroxidase from a Decolorizing Streptomyces albidoflavus strain TN644 Isolated from a Tunisian off-Shore Oil Field. Int. Biodeterior. Biodegrad. 2014, 90, 36–44. [Google Scholar] [CrossRef]
- Tian, M.; Du, D.; Zhou, W.; Zeng, X.; Cheng, G. Phenol Degradation and Genotypic Analysis of Dioxygenase Genes in Bacteria Isolated from Sediments. Braz. J. Microbiol. 2016, 48, 305–313. [Google Scholar] [CrossRef]
- Wang, S.-S.; Ning, Y.-J.; Wang, S.-N.; Zhang, J.; Zhang, G.-Q.; Chen, Q.-J. Purification, Characterization, and Cloning of an Extracellular Laccase with Potent Dye Decolorizing Ability from White Rot Fungus Cerrena Unicolor GSM-01. Int. J. Biol. Macromol. 2017, 95, 920–927. [Google Scholar] [CrossRef]
- Murugesan, K.; Nam, I.-H.; Kim, Y.-M.; Chang, Y.-S. Decolorization of Reactive Dyes by a Thermostable Laccase Produced by Ganoderma lucidum in Solid State Culture. Enzyme Microb. 2007, 40, 1662–1672. [Google Scholar] [CrossRef]
- Zhou, N.; Wang, J.; Wang, W.; Wu, X. Purification, Characterization, and Catalytic Mechanism of N-Isopropylammelide Isopropylaminohydrolase (AtzC) Involved in the Degradation of s-Triazine Herbicides. Environ. Pollut. 2021, 268, 115803. [Google Scholar] [CrossRef]
- Musengi, A.; Durrell, K.; Prins, A.; Khan, N.; Agunbiade, M.; Kudanga, T.; Kirby-McCullough, B.; Pletschke, B.I.; Burton, S.G.; Le Roes-Hill, M. Production and Characterisation of a Novel Actinobacterial DyP-Type Peroxidase and Its Application in Coupling of Phenolic Monomers. Enzyme Microb. 2020, 141, 109654. [Google Scholar] [CrossRef] [PubMed]
- Alaidaroos, B.A. Improvement of Phenol and Heavy Metal Removal by Streptomyces flavabus BA4 Used for Wastewater Treatments. Eur. Online J. Nat. Soc. 2022, 11, 837–851. [Google Scholar]
- Costanzo, S.D.; Murby, J.; Bates, J. Ecosystem Response to Antibiotics Entering the Aquatic Environment. Mar. Pollut. Bull. 2005, 51, 218–223. [Google Scholar] [CrossRef] [PubMed]
- Vittoria Pinna, M.; Castaldi, P.; Deiana, P.; Pusino, A.; Garau, G. Sorption Behavior of Sulfamethazine on Unamended and Manure-Amended Soils and Short-Term Impact on Soil Microbial Community. Ecotoxicol. Environ. Saf. 2012, 84, 234–242. [Google Scholar] [CrossRef] [PubMed]
- Proia, L.; Lupini, G.; Osorio, V.; Pérez, S.; Barceló, D.; Schwartz, T.; Amalfitano, S.; Fazi, S.; Romaní, A.M.; Sabater, S. Response of Biofilm Bacterial Communities to Antibiotic Pollutants in a Mediterranean River. Chemosphere 2013, 92, 1126–1135. [Google Scholar] [CrossRef]
- Cycoń, M.; Mrozik, A.; Piotrowska-Seget, Z. Antibiotics in the soil environment-degradation and their impact on microbial activity and diversity. Front. Microbiol. 2019, 10, 338. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Ying, G.-G.; Tao, R.; Zhao, J.-L.; Yang, J.-F.; Zhao, L.-F. Effects of Six Selected Antibiotics on Plant Growth and Soil Microbial and Enzymatic Activities. Environ. Pollut. 2009, 157, 1636–1642. [Google Scholar] [CrossRef] [PubMed]
- Shan, J.; Yang, P.; Rahman, M.M.; Shang, X.; Yan, X. Tetracycline and Sulfamethazine Alter Dissimilatory Nitrate Reduction Processes and Increase N2O Release in Rice Fields. Environ. Pollut. 2018, 242, 788–796. [Google Scholar] [CrossRef]
- Oliver, K.R.; Greene, W.K. Transposable Elements: Powerful Facilitators of Evolution. Bioessays 2009, 31, 703–714. [Google Scholar] [CrossRef]
- Schubert, I.; Vu, G.T.H. Genome Stability and Evolution: Attempting a Holistic View. Trends Plant Sci. 2016, 21, 749–757. [Google Scholar] [CrossRef]
- Mitra, S.; Chakraborty, A.J.; Tareq, A.M.; Emran, T.B.; Nainu, F.; Khusro, A.; Idris, A.M.; Khandaker, M.U.; Osman, H.; Alhumaydhi, F.A.; et al. Impact of Heavy Metals on the Environment and Human Health: Novel Therapeutic Insights to Counter the Toxicity. J. King Saud Univ. Sci. 2022, 34, 101865. [Google Scholar] [CrossRef]
- Wu, X.; Cobbina, S.J.; Mao, G.; Xu, H.; Zhang, Z.; Yang, L. A Review of Toxicity and Mechanisms of Individual and Mixtures of Heavy Metals in the Environment. Environ. Sci. Pollut. Res. Int. 2016, 23, 8244–8259. [Google Scholar] [CrossRef] [PubMed]
- Haider, F.U.; Liqun, C.; Coulter, J.A.; Cheema, S.A.; Wu, J.; Zhang, R.; Wenjun, M.; Farooq, M. Cadmium Toxicity in Plants: Impacts and Remediation Strategies. Ecotoxicol. Environ. Saf. 2021, 211, 111887. [Google Scholar] [CrossRef] [PubMed]
- de Mora, A.P.; Ortega-Calvo, J.J.; Cabrera, F.; Madejón, E. Changes in Enzyme Activities and Microbial Biomass after “in Situ” Remediation of a Heavy Metal-Contaminated Soil. Appl. Soil. Ecol. 2005, 28, 125–137. [Google Scholar] [CrossRef]
- Rana, A.; Sindhu, M.; Kumar, A.; Dhaka, R.K.; Chahar, M.; Singh, S.; Nain, L. Restoration of Heavy Metal-Contaminated Soil and Water through Biosorbents: A Review of Current Understanding and Future Challenges. Physiol. Plant 2021, 173, 394–417. [Google Scholar] [CrossRef]
- Kaliyaraj, D.; Rajendran, M.; Angamuthu, V.; Antony, A.R.; Kaari, M.; Thangavel, S.; Venugopal, G.; Joseph, J.; Manikkam, R. Bioleaching of Heavy Metals from Printed Circuit Board (PCB) by Streptomyces albidoflavus TN10 Isolated from Insect Nest. Bioresour. Bioprocess. 2019, 6, 47. [Google Scholar] [CrossRef]
- Hussain, T.; Akthar, N.; Aminedi, R.; Danish, M.; Nishat, Y.; Patel, S. Role of the Potent Microbial Based Bioagents and Their Emerging Strategies for the Ecofriendly Management of Agricultural Phytopathogens. In Natural Bioactive Products in Sustainable Agriculture; Singh, J., Yadav, A.N., Eds.; Springer: Singapore, 2020; pp. 45–66. ISBN 9789811530241. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).