Beyond the Gut, Emerging Microbiome Areas of Research: A Focus on Early-Life Microbial Colonization
Abstract
:1. Introduction
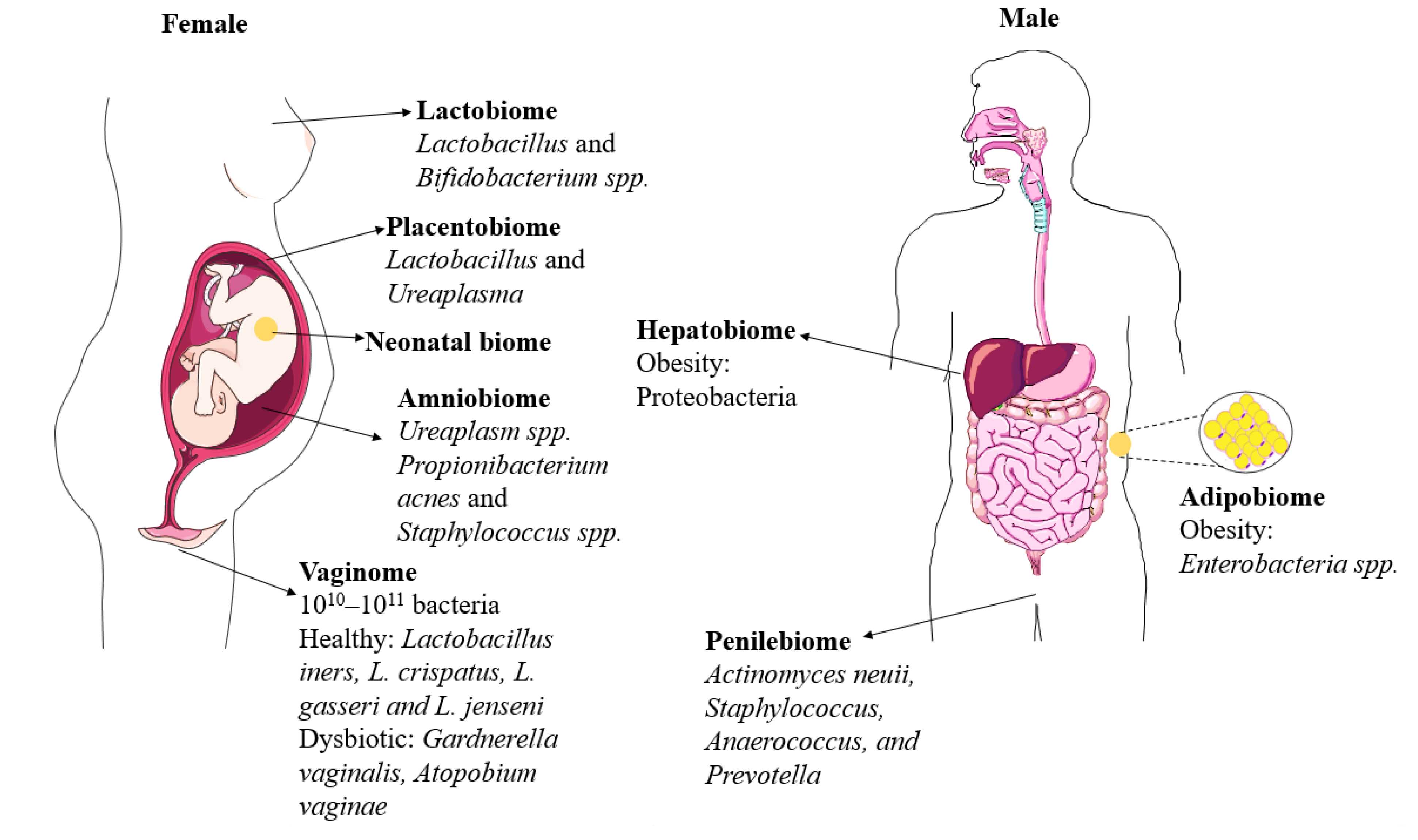
2. Genital Microbiomes
2.1. Vaginobiome
2.2. Penilebiome
3. Microbiomes Associated with the Early Life Development
3.1. Amniobiome
3.2. Placentalbiome
3.3. Meconiobiome
3.4. Lactobiome
4. Microbiomes Associated with Later-Life Metabolic Health: Emerging Research Areas
4.1. Adipobiome
4.2. Hepatobiome
5. Discussion
6. Conclusions and Future Directions
- (i)
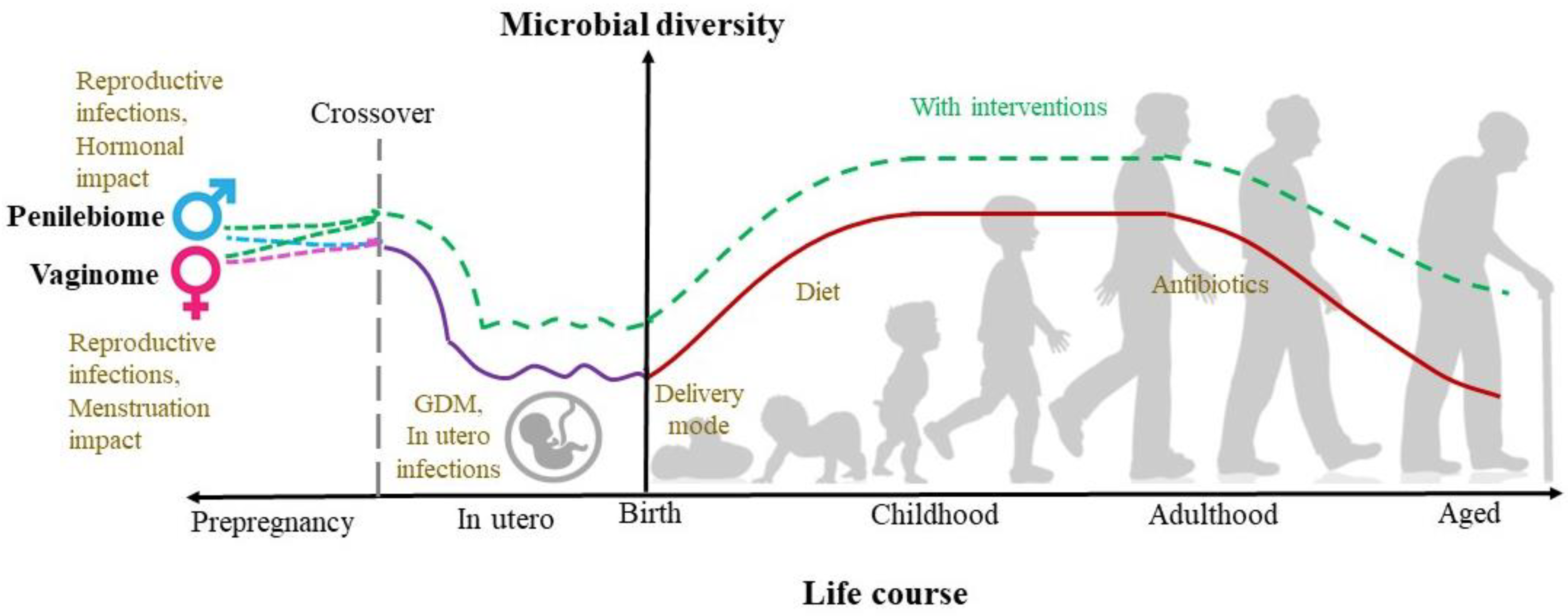
- Microbial progression phases during a human lifecycle, as mentioned in the earlier sections, may not be established post-birth. Emerging research challenges the sterile womb hypothesis with evidence of the microbial presence in various sites in the womb.
- (ii)
- The role of contamination is paramount in the identification of the true representation of the “microbiome” due to their low biomass. Low biomass data is impacted by analyzing a relatively small sample size. The small sample size or underpowered microbiome research can reduce the amplification of low biomass samples, in turn lowering the detection sensitivity and resolution. To gain more insight into bacteriomes, a more robust and optimized 16S gene sequencing pipeline with longer reads will be beneficial to catalog the bacterial DNA profiles of different tissues and provide a database to analyze host/bacterial interactions in relation to homeostasis and disease.
- (iii)
- The reasons for low biomass in the observed bacteriome in amniotic fluid passed via the maternal, as seen in cord blood, but may not be able to survive the womb due to host defenses but be transient in nature.
- (iv)
- Another interesting reason could be the presence of microbes beyond bacteriomes, such as viromes. Virome majorly includes phages, which may play a role in maintaining low biomass and the lack of proper colonization, such as the gut. NGS technologies such as WGS should be utilized, which is a more robust and high-resolution platform.
- (v)
- A more important aspect to consider is the role of female health, not only during pregnancy and post-pregnancy but also during pre-pregnancy and copulation. Identification of vaginome and penilebiome has revolutionized microbiome research and can provide information on possible in-utero infections, preterm labor, and microbial sources. These genital biomes should be considered important in understanding the microbiome establishment, which will provide scope to modulate these biomes, in turn, neonatal health.
- (vi)
- During the transition and stable phases, the lactobiome plays a key role in growth and metabolism. More research into the lactobiome-hepatobiome-adipobiome axis will be beneficial in understanding the future risk of metabolic diseases.
- (vii)
- As the “omics” technologies are getting updated as we speak, more research utilizing advanced sequencing methods (proteomics, transcriptomics, metabolomics, RNA-seq, and Immuno-seq) is necessary to understand the in-utero colonization, which perhaps will enable us to prevent the origins of many diseases.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Berg, G.; Rybakova, D.; Fischer, D.; Cernava, T.; Vergès, M.-C.C.; Charles, T.; Chen, X.; Cocolin, L.; Eversole, K.; Corral, G.H. Microbiome definition re-visited: Old concepts and new challenges. Microbiome 2020, 8, 103. [Google Scholar] [CrossRef] [PubMed]
- Marchesi, J.R.; Ravel, J. The Vocabulary of Microbiome Research: A Proposal; Springer: Berlin/Heidelberg, Germany, 2015; Volume 3, pp. 1–3. [Google Scholar]
- Vemuri, R.; Shankar, E.M.; Chieppa, M.; Eri, R.; Kavanagh, K. Beyond just bacteria: Functional biomes in the gut ecosystem including virome, mycobiome, archaeome and helminths. Microorganisms 2020, 8, 483. [Google Scholar] [CrossRef] [Green Version]
- Aagaard, K.; Ma, J.; Antony, K.M.; Ganu, R.; Petrosino, J.; Versalovic, J. The placenta harbors a unique microbiome. Sci. Transl. Med. 2014, 6, 237ra65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Collado, M.C.; Rautava, S.; Aakko, J.; Isolauri, E.; Salminen, S. Human gut colonisation may be initiated in utero by distinct microbial communities in the placenta and amniotic fluid. Sci. Rep. 2016, 6, 23129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, J.; Nomura, Y.; Bashir, A.; Fernandez-Hernandez, H.; Itzkowitz, S.; Pei, Z.; Stone, J.; Loudon, H.; Peter, I. Diversified microbiota of meconium is affected by maternal diabetes status. PLoS ONE 2013, 8, e78257. [Google Scholar] [CrossRef] [Green Version]
- Ferretti, P.; Pasolli, E.; Tett, A.; Asnicar, F.; Gorfer, V.; Fedi, S.; Armanini, F.; Truong, D.T.; Manara, S.; Zolfo, M. Mother-to-infant microbial transmission from different body sites shapes the developing infant gut microbiome. Cell Host Microbe 2018, 24, 133–145. [Google Scholar] [CrossRef] [PubMed]
- Roswall, J.; Olsson, L.M.; Kovatcheva-Datchary, P.; Nilsson, S.; Tremaroli, V.; Simon, M.-C.; Kiilerich, P.; Akrami, R.; Krämer, M.; Uhlén, M. Developmental trajectory of the healthy human gut microbiota during the first 5 years of life. Cell Host Microbe 2021, 29, 765–776. [Google Scholar] [CrossRef]
- Shukla, G.; Arya, S.; Goyal, P.; Channa, U. Comparative microbial analysis and assessment of paired umbilical cord blood culture and peripheral venous blood culture in neonates with high risk factors to detect early onset neonatal sepsis: A study from tertiary care hospital of central India. Asian J. Med. Sci. 2022, 13, 134–139. [Google Scholar] [CrossRef]
- Vemuri, R.; Sylvia, K.E.; Klein, S.L.; Forster, S.C.; Plebanski, M.; Eri, R.; Flanagan, K.L. The microgenderome revealed: Sex differences in bidirectional interactions between the microbiota, hormones, immunity and disease susceptibility. Semin. Immunopathol. 2019, 41, 265–275. [Google Scholar] [CrossRef]
- Serrano, M.G.; Parikh, H.I.; Brooks, J.P.; Edwards, D.J.; Arodz, T.J.; Edupuganti, L.; Huang, B.; Girerd, P.H.; Bokhari, Y.A.; Bradley, S.P. Racioethnic diversity in the dynamics of the vaginal microbiome during pregnancy. Nat. Med. 2019, 25, 1001–1011. [Google Scholar] [CrossRef]
- Fettweis, J.M.; Brooks, J.P.; Serrano, M.G.; Sheth, N.U.; Girerd, P.H.; Edwards, D.J.; Strauss Iii, J.F.; Jefferson, K.K.; Buck, G.A.; Vaginal Microbiome, C. Differences in vaginal microbiome in African American women versus women of European ancestry. Microbiology 2014, 160, 2272. [Google Scholar] [CrossRef] [Green Version]
- MacIntyre, D.A.; Chandiramani, M.; Lee, Y.S.; Kindinger, L.; Smith, A.; Angelopoulos, N.; Lehne, B.; Arulkumaran, S.; Brown, R.; Teoh, T.G. The vaginal microbiome during pregnancy and the postpartum period in a European population. Sci. Rep. 2015, 5, 8988. [Google Scholar] [CrossRef] [Green Version]
- Mehta, S.D.; Zhao, D.; Green, S.J.; Agingu, W.; Otieno, F.; Bhaumik, R.; Bhaumik, D.; Bailey, R.C. The microbiome composition of a man’s penis predicts incident bacterial vaginosis in his female sex partner with high accuracy. Front. Cell. Infect. Microbiol. 2020, 433. [Google Scholar] [CrossRef]
- DiGiulio, D.B.; Callahan, B.J.; McMurdie, P.J.; Costello, E.K.; Lyell, D.J.; Robaczewska, A.; Sun, C.L.; Goltsman, D.S.A.; Wong, R.J.; Shaw, G. Temporal and spatial variation of the human microbiota during pregnancy. Proc. Natl. Acad. Sci. USA 2015, 112, 11060–11065. [Google Scholar] [CrossRef] [Green Version]
- Mehta, S.D.; Nandi, D.; Agingu, W.; Green, S.J.; Bhaumik, D.K.; Bailey, R.C.; Otieno, F. Vaginal and penile microbiome associations with herpes simplex virus type 2 in women and their male sex partners. J. Infect. Dis. 2022, 226, 644–654. [Google Scholar] [CrossRef]
- Kayem, G.; Doloy, A.; Schmitz, T.; Chitrit, Y.; Bouhanna, P.; Carbonne, B.; Jouannic, J.M.; Mandelbrot, L.; Benachi, A.; Azria, E.; et al. Antibiotics for amniotic-fluid colonization by Ureaplasma and/or Mycoplasma spp. to prevent preterm birth: A randomized trial. PLoS ONE 2018, 13, e0206290. [Google Scholar] [CrossRef] [Green Version]
- Jiménez, E.; Marín, M.L.; Martín, R.; Odriozola, J.M.; Olivares, M.; Xaus, J.; Fernández, L.; Rodríguez, J.M. Is meconium from healthy newborns actually sterile? Res. Microbiol. 2008, 159, 187–193. [Google Scholar] [CrossRef]
- Williams, N.; Vella, R.; Zhou, Y.; Gao, H.; Mass, K.; Townsel, C.; Campbell, W.; Luo, G. Investigating the origin of the fetal gut and placenta microbiome in twins. J. Matern. Fetal Neonatal Med. 2021, 35, 7025–7035. [Google Scholar] [CrossRef]
- Pace, R.M.; Williams, J.E.; Robertson, B.; Lackey, K.A.; Meehan, C.L.; Price, W.J.; Foster, J.A.; Sellen, D.W.; Kamau-Mbuthia, E.W.; Kamundia, E.W. Variation in human milk composition is related to differences in milk and infant fecal microbial communities. Microorganisms 2021, 9, 1153. [Google Scholar] [CrossRef]
- Hunt, K.M.; Foster, J.A.; Forney, L.J.; Schütte, U.M.E.; Beck, D.L.; Abdo, Z.; Fox, L.K.; Williams, J.E.; McGuire, M.K.; McGuire, M.A. Characterization of the diversity and temporal stability of bacterial communities in human milk. PLoS ONE 2011, 6, e21313. [Google Scholar] [CrossRef]
- Massier, L.; Chakaroun, R.; Tabei, S.; Crane, A.; Didt, K.D.; Fallmann, J.; von Bergen, M.; Haange, S.-B.; Heyne, H.; Stumvoll, M. Adipose tissue derived bacteria are associated with inflammation in obesity and type 2 diabetes. Gut 2020, 69, 1796–1806. [Google Scholar] [CrossRef] [PubMed]
- Anhê, F.F.; Jensen, B.A.H.; Varin, T.V.; Servant, F.; Van Blerk, S.; Richard, D.; Marceau, S.; Surette, M.; Biertho, L.; Lelouvier, B. Type 2 diabetes influences bacterial tissue compartmentalisation in human obesity. Nat. Metab. 2020, 2, 233–242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suppli, M.P.; Bagger, J.I.; Lelouvier, B.; Broha, A.; Demant, M.; Kønig, M.J.; Strandberg, C.; Lund, A.; Vilsbøll, T.; Knop, F.K. Hepatic microbiome in healthy lean and obese humans. JHEP Rep. 2021, 3, 100299. [Google Scholar] [CrossRef] [PubMed]
- Leinwand, J.C.; Paul, B.; Chen, R.; Xu, F.; Sierra, M.A.; Paluru, M.M.; Nanduri, S.; Alcantara, C.G.; Shadaloey, S.A.A.; Yang, F. Intrahepatic microbes govern liver immunity by programming NKT cells. J. Clin. Investig. 2022, 132, e151725. [Google Scholar] [CrossRef]
- Chu, D.M.; Seferovic, M.; Pace, R.M.; Aagaard, K.M. The microbiome in preterm birth. Best Pract. Res. Clin. Obstet. Gynaecol. 2018, 52, 103–113. [Google Scholar] [CrossRef]
- Parris, K.M.; Amabebe, E.; Cohen, M.C.; Anumba, D.O. Placental microbial–metabolite profiles and inflammatory mechanisms associated with preterm birth. J. Clin. Pathol. 2021, 74, 10–18. [Google Scholar] [CrossRef]
- Olaniyi, K.S.; Moodley, J.; Mahabeer, Y.; Mackraj, I. Placental microbial colonization and its association with pre-eclampsia. Front. Cell. Infect. Microbiol. 2020, 10, 413. [Google Scholar] [CrossRef]
- Singh, A.; Mittal, M. Neonatal microbiome–a brief review. J. Matern. Fetal Neonatal Med. 2020, 33, 3841–3848. [Google Scholar] [CrossRef]
- Lundgren, P.; Thaiss, C.A. The microbiome-adipose tissue axis in systemic metabolism. Am. J. Physiol. Gastrointest. Liver Physiol. 2020, 318, G717–G724. [Google Scholar] [CrossRef]
- Tilg, H.; Burcelin, R.; Tremaroli, V. Liver tissue microbiome in NAFLD: Next step in understanding the gut–liver axis? Gut 2020, 69, 1373–1374. [Google Scholar] [CrossRef]
- Vemuri, R.; Gundamaraju, R.; Shastri, M.D.; Shukla, S.D.; Kalpurath, K.; Ball, M.; Tristram, S.; Shankar, E.M.; Ahuja, K.; Eri, R. Gut microbial changes, interactions, and their implications on human lifecycle: An ageing perspective. BioMed Res. Int. 2018, 2018, 4178607. [Google Scholar] [CrossRef] [PubMed]
- Seferovic, M.D.; Pace, R.M.; Carroll, M.; Belfort, B.; Major, A.M.; Chu, D.M.; Racusin, D.A.; Castro, E.C.C.; Muldrew, K.L.; Versalovic, J. Visualization of microbes by 16S in situ hybridization in term and preterm placentas without intraamniotic infection. Am. J. Obstet. Gynecol. 2019, 221, 146.e1–146.e23. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, S.; Friedberg, I.; Ivanov, I.V.; Davidson, L.A.; Goldsby, J.S.; Dahl, D.B.; Herman, D.; Wang, M.; Donovan, S.M.; Chapkin, R.S. A metagenomic study of diet-dependent interaction between gut microbiota and host in infants reveals differences in immune response. Genome Biol. 2012, 13, r32. [Google Scholar] [CrossRef] [Green Version]
- Koenig, J.E.; Spor, A.; Scalfone, N.; Fricker, A.D.; Stombaugh, J.; Knight, R.; Angenent, L.T.; Ley, R.E. Succession of microbial consortia in the developing infant gut microbiome. Proc. Natl. Acad. Sci. USA 2011, 108, 4578–4585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monira, S.; Nakamura, S.; Gotoh, K.; Izutsu, K.; Watanabe, H.; Alam, N.H.; Endtz, H.P.; Cravioto, A.; Ali, S.I.; Nakaya, T. Gut microbiota of healthy and malnourished children in Bangladesh. Front. Microbiol. 2011, 2, 228. [Google Scholar] [CrossRef] [Green Version]
- Biagi, E.; Nylund, L.; Candela, M.; Ostan, R.; Bucci, L.; Pini, E.; Nikkïla, J.; Monti, D.; Satokari, R.; Franceschi, C. Through ageing, and beyond: Gut microbiota and inflammatory status in seniors and centenarians. PLoS ONE 2010, 5, e10667. [Google Scholar] [CrossRef]
- Ghosh, T.S.; Shanahan, F.; O’Toole, P.W. The gut microbiome as a modulator of healthy ageing. Nat. Rev. Gastroenterol. Hepatol. 2022, 19, 565–584. [Google Scholar] [CrossRef]
- Aleman, F.D.D.; Valenzano, D.R. Microbiome evolution during host aging. PLoS Pathog. 2019, 15, e1007727. [Google Scholar] [CrossRef]
- Perez-Muñoz, M.E.; Arrieta, M.-C.; Ramer-Tait, A.E.; Walter, J. A critical assessment of the “sterile womb” and “in utero colonization” hypotheses: Implications for research on the pioneer infant microbiome. Microbiome 2017, 5, 48. [Google Scholar] [CrossRef] [Green Version]
- Balasubramanian, H.; Patole, S. Early probiotics to prevent childhood metabolic syndrome: A systematic review. World J. Methodol. 2015, 5, 157–163. [Google Scholar] [CrossRef]
- Pärtty, A.; Kalliomäki, M.; Wacklin, P.; Salminen, S.; Isolauri, E. A possible link between early probiotic intervention and the risk of neuropsychiatric disorders later in childhood: A randomized trial. Pediatr. Res. 2015, 77, 823–828. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lim, E.S.; Rodriguez, C.; Holtz, L.R. Amniotic fluid from healthy term pregnancies does not harbor a detectable microbial community. Microbiome 2018, 6, 87. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Yang, G.X.; Hu, Y.; Lam, P.; Sangha, K.; Siciliano, D.; Swenerton, A.; Miller, R.; Tilley, P.; Von Dadelszen, P. Comprehensive human amniotic fluid metagenomics supports the sterile womb hypothesis. Sci. Rep. 2022, 12, 6875. [Google Scholar] [CrossRef] [PubMed]
- Lluch, J.; Servant, F.; Païssé, S.; Valle, C.; Valiere, S.; Kuchly, C.; Vilchez, G.; Donnadieu, C.; Courtney, M.; Burcelin, R. The characterization of novel tissue microbiota using an optimized 16S metagenomic sequencing pipeline. PLoS ONE 2015, 10, e0142334. [Google Scholar] [CrossRef]
- Liu, Y.; Li, X.; Zhu, B.; Zhao, H.; Ai, Q.; Tong, Y.; Qin, S.; Feng, Y.; Wang, Y.; Wang, S. Midtrimester amniotic fluid from healthy pregnancies has no microorganisms using multiple methods of microbiologic inquiry. Am. J. Obstet. Gynecol. 2020, 223, 248.e1–248.e21. [Google Scholar] [CrossRef] [Green Version]
- Mendz, G.L.; Kaakoush, N.O.; Quinlivan, J.A. Bacterial aetiological agents of intra-amniotic infections and preterm birth in pregnant women. Front. Cell. Infect. Microbiol. 2013, 3, 58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Romero, R.; Gomez-Lopez, N.; Winters, A.D.; Jung, E.; Shaman, M.; Bieda, J.; Panaitescu, B.; Pacora, P.; Erez, O.; Greenberg, J.M. Evidence that intra-amniotic infections are often the result of an ascending invasion–a molecular microbiological study. J. Perinat. Med. 2019, 47, 915–931. [Google Scholar] [CrossRef] [PubMed]
- Kurath-Koller, S.; Neumann, C.; Moissl-Eichinger, C.; Kraschl, R.; Kanduth, C.; Hopfer, B.; Pausan, M.-R.; Urlesberger, B.; Resch, B. Hospital regimens including probiotics guide the individual development of the gut microbiome of very low birth weight infants in the first two weeks of life. Nutrients 2020, 12, 1256. [Google Scholar] [CrossRef]
- Xu, J.; Bian, G.; Zheng, M.; Lu, G.; Chan, W.Y.; Li, W.; Yang, K.; Chen, Z.J.; Du, Y. Fertility factors affect the vaginal microbiome in women of reproductive age. Am. J. Reprod. Immunol. 2020, 83, e13220. [Google Scholar] [CrossRef] [Green Version]
- Perrotta, A.R.; Borrelli, G.M.; Martins, C.O.; Kallas, E.G.; Sanabani, S.S.; Griffith, L.G.; Alm, E.J.; Abrao, M.S. The vaginal microbiome as a tool to predict rASRM stage of disease in endometriosis: A pilot study. Reprod. Sci. 2020, 27, 1064–1073. [Google Scholar] [CrossRef]
- Chaban, B.; Links, M.G.; Jayaprakash, T.P.; Wagner, E.C.; Bourque, D.K.; Lohn, Z.; Albert, A.Y.K.; van Schalkwyk, J.; Reid, G.; Hemmingsen, S.M. Characterization of the vaginal microbiota of healthy Canadian women through the menstrual cycle. Microbiome 2014, 2, 23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miller, E.A.; Beasley, D.E.; Dunn, R.R.; Archie, E.A. Lactobacilli dominance and vaginal pH: Why is the human vaginal microbiome unique? Front. Microbiol. 2016, 7, 1936. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krog, M.C.; Hugerth, L.W.; Fransson, E.; Bashir, Z.; Nyboe Andersen, A.; Edfeldt, G.; Engstrand, L.; Schuppe-Koistinen, I.; Nielsen, H.S. The healthy female microbiome across body sites: Effect of hormonal contraceptives and the menstrual cycle. Hum. Reprod. 2022, 37, 1525–1543. [Google Scholar] [CrossRef] [PubMed]
- Mohanty, T.; Doke, P.P.; Khuroo, S.R. Effect of bacterial vaginosis on preterm birth: A meta-analysis. Arch. Gynecol. Obstet. 2022, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Lu, Y.; Chen, T.; Li, R. The female vaginal microbiome in health and bacterial vaginosis. Front. Cell. Infect. Microbiol. 2021, 11, 631972. [Google Scholar] [CrossRef] [PubMed]
- Song, S.D.; Acharya, K.D.; Zhu, J.E.; Deveney, C.M.; Walther-Antonio, M.R.S.; Tetel, M.J.; Chia, N. Daily Vaginal Microbiota Fluctuations Associated with Natural Hormonal Cycle, Contraceptives, Diet, and Exercise. mSphere 2020, 5, e00593-20. [Google Scholar] [CrossRef]
- Zozaya, M.; Ferris, M.J.; Siren, J.D.; Lillis, R.; Myers, L.; Nsuami, M.J.; Eren, A.M.; Brown, J.; Taylor, C.M.; Martin, D.H. Bacterial communities in penile skin, male urethra, and vaginas of heterosexual couples with and without bacterial vaginosis. Microbiome 2016, 4, 16. [Google Scholar] [CrossRef] [Green Version]
- Byrd, A.L.; Belkaid, Y.; Segre, J.A. The human skin microbiome. Nat. Rev. Microbiol. 2018, 16, 143–155. [Google Scholar] [CrossRef]
- Capone, K.A.; Dowd, S.E.; Stamatas, G.N.; Nikolovski, J. Diversity of the human skin microbiome early in life. J. Investig. Dermatol. 2011, 131, 2026–2032. [Google Scholar] [CrossRef] [Green Version]
- Sánchez, C.; Franco, L.; Regal, P.; Lamas, A.; Cepeda, A.; Fente, C. Breast milk: A source of functional compounds with potential application in nutrition and therapy. Nutrients 2021, 13, 1026. [Google Scholar] [CrossRef]
- Sepúlveda-Valbuena, N.; Nieto-Ruiz, A.; Diéguez, E.; Herrmann, F.; Escudero-Marín, M.; De-Castellar, R.; Rodríguez-Palmero, M.; Miranda, M.T.; García-Santos, J.A.; Bermúdez, M.G. Growth patterns and breast milk/infant formula energetic efficiency in healthy infants up to 18 months of life: The COGNIS study. Br. J. Nutr. 2021, 126, 1809–1822. [Google Scholar] [CrossRef]
- Eshriqui, I.; Viljakainen, H.T.; Ferreira, S.R.G.; Raju, S.C.; Weiderpass, E.; Figueiredo, R.A.O. Breastfeeding may have a long-term effect on oral microbiota: Results from the Fin-HIT cohort. Int. Breastfeed. J. 2020, 15, 42. [Google Scholar] [CrossRef]
- Burcham, Z.M.; Garneau, N.L.; Comstock, S.S.; Tucker, R.M.; Knight, R.; Metcalf, J.L. Patterns of oral microbiota diversity in adults and children: A crowdsourced population study. Sci. Rep. 2020, 10, 2133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gonzalez, E.; Brereton, N.J.B.; Li, C.; Lopez Leyva, L.; Solomons, N.W.; Agellon, L.B.; Scott, M.E.; Koski, K.G. Distinct changes occur in the human breast milk microbiome between early and established lactation in breastfeeding Guatemalan mothers. Front. Microbiol. 2021, 12, 557180. [Google Scholar] [CrossRef] [PubMed]
- Haslam, D.W.; James, W.P. Obesity. Lancet 2005, 366, 1197–1209. [Google Scholar] [CrossRef] [PubMed]
- Sultan, S.E. Developmental plasticity: Re-conceiving the genotype. Interface Focus 2017, 7, 20170009. [Google Scholar] [CrossRef] [PubMed]
- Barker, D.J.P. Sir Richard Doll Lecture. Developmental origins of chronic disease. Public Health 2012, 126, 185–189. [Google Scholar] [CrossRef] [PubMed]
- Wells, J.C.K. Adaptive variability in the duration of critical windows of plasticity: Implications for the programming of obesity. Evol. Med. Public Health 2014, 2014, 109–121. [Google Scholar] [CrossRef] [Green Version]
- Sahoo, K.; Sahoo, B.; Choudhury, A.K.; Sofi, N.Y.; Kumar, R.; Bhadoria, A.S. Childhood obesity: Causes and consequences. J Fam. Med. Prim. Care 2015, 4, 187–192. [Google Scholar] [CrossRef]
- Bennett, A.E.; Kearney, J.M. Maternal sociodemographic and health behaviours associated with adiposity in infants as measured by air displacement plethysmography. Early Hum. Dev. 2020, 140, 104887. [Google Scholar] [CrossRef]
- Herath, M.P.; Ahuja, K.D.K.; Beckett, J.M.; Jayasinghe, S.; Byrne, N.M.; Hills, A.P. Determinants of Infant Adiposity across the First 6 Months of Life: Evidence from the Baby-bod study. J. Clin. Med. 2021, 10, 1770. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zheng, J.; Shi, W.; Du, N.; Xu, X.; Zhang, Y.; Ji, P.; Zhang, F.; Jia, Z.; Wang, Y. Dysbiosis of maternal and neonatal microbiota associated with gestational diabetes mellitus. Gut 2018, 67, 1614–1625. [Google Scholar] [CrossRef] [PubMed]
- Koleva, P.T.; Bridgman, S.L.; Kozyrskyj, A.L. The infant gut microbiome: Evidence for obesity risk and dietary intervention. Nutrients 2015, 7, 2237–2260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Branton, W.G.; Ellestad, K.K.; Maingat, F.; Wheatley, B.M.; Rud, E.; Warren, R.L.; Holt, R.A.; Surette, M.G.; Power, C. Brain microbial populations in HIV/AIDS: α-proteobacteria predominate independent of host immune status. PLoS ONE 2013, 8, e54673. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zuppi, M.; Hendrickson, H.L.; O’Sullivan, J.M.; Vatanen, T. Phages in the Gut Ecosystem. Front. Cell. Infect. Microbiol. 2021, 11, 822562. [Google Scholar] [CrossRef]
- O’Hara, A.M.; Shanahan, F. The gut flora as a forgotten organ. EMBO Rep. 2006, 7, 688–693. [Google Scholar] [CrossRef]
| Biome | Study Type | Dominant Bacteria | Method Used | Reference |
|---|---|---|---|---|
| 1. Vaginome | Human (pregnant and non-pregnant) | Pregnant: Lactobacillus vagitypes (L. crispatus, L. iners, L. gasseri and L. jensenii) Non-pregnant: Lactobacillus | V1–V3 16S rRNA | [11] |
| Vaginal microbial profiles in European (E) | Pre-pregnancy: L. crispatus Pregnancy: L. jensenii, L. crispatus PP: BV- associated taxa Prevotella spp., Clostridium spp., Atopobium spp. and Megasphaera spp. | V1-V2 16S rRNA | [12] | |
| Vaginal microbial profiles in African American (AA) versus European (E) ancestry women | BV: Gardnerella vaginalis (AA) AA: L. iners E: L. crispatus, L. iners, G. vaginalis AA/E differences: Mycoplasma, Gardnerella, Prevotella and Sneathia | V1–V3 16S rRNA | [13] | |
| Vaginome during pregnancy, preterm and PP | Pregnancy: L. crispatus, L. gasseri, L. iners, L. jensenii Preterm: Gardnerella and Ureaplasma PP: Peptoniphilus, Prevotella, and Anaerococcus | V3–V5 16S rRNA | [15] | |
| 2. Penilebiome | Penile (both meatal and glans/coronal sulcus/circumcised) and vaginal microbial profiles related to BV | Penile: Corynebacterium (circumcised), Streptococcus, Anaerococcus, Finegoldia. BV: Parvimonas, L. iners, L. crispatus, Fastidiosipila, and Prevotella | V3–V4 16S rRNA | [14] |
| Vaginal and penile microbiomes related to herpes simplex virus type 2 (HSV-2) | BV: G. vaginalis and L. iners Penile: Ureaplasma and Aerococcus (HSV-2) | V3–V4 16S rRNA | [16] | |
| 3. Amniobiome | Preterm in 2nd trimester, asymptomatic | Ureaplasma and/or Mycoplasma spp. | 16S rRNA | [17] |
| In utero to first 4 days of birth | Enterobacter, Escherichia/Shigella and Propionibacterium | V1-V3 16S rRNA | [5,18] | |
| 4. Placentalbiome | In utero to first 4 days of birth | Propionibacterium, Enterobacter and Escherichia/Shigella | V1-V3 16S rRNA | [5] |
| Meconium in twins | Salinibacter and Enterobacteriaceae_unclassified | V3-V4 16S rRNA | [19] | |
| 5. Meconiobiome | In utero to first 4 days of birth | Propionibacterium, Escherichia/Shigella, and Lactobacillus | V1-V3 16S rRNA | [5] |
| Meconium in twins | Enterobacteriaceae_unclassified | V3-V4 16S rRNA | [19] | |
| Temporal and spatial variation in early-life microbiome | Lactobacillus, Bifidobacterium, Staphylococcus, and Enterococcus spp. | V3–V5 16S | [15] | |
| 6. Lactobiome | Milk microbiome | Staphylococcus and Streptococcus | V1-V3 16S rRNA | [20] |
| Milk microbiome | Streptococcus, Staphylococcus, Serratia and Corynebacteria | V1-V2 16S rRNA | [21] | |
| 7. Adipobiome | Adipose tissue microbiome | Proteobacteria and Firmicutes | V4-V5 16S rRNA | [22] |
| Adipose tissue microbiome related to type 2 diabetes (T2D) and obesity humans | Pseudomonas, Faecalibacterium, Bacteroides and Enterobacter | V3-V4 16S rRNA | [23] | |
| 8. Hepatobiome | Liver microbiome in obese and non-obese humans | Obese: Proteobacteria, Massilia spp. | V3-V4 16S rRNA | [24] |
| Liver tissue microbiome related to diabetes and obesity humans | Obese: Pseudomonas, Arthrobacter and Ruminococcus | V3-V4 16S rRNA | [23] | |
| Liver microbiome in Humans and Mice | Mice: Pseudomonas, Delftia and Coprococcus Humans: Proteobacteria | 16S rRNA | [25] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vemuri, R.; Herath, M.P. Beyond the Gut, Emerging Microbiome Areas of Research: A Focus on Early-Life Microbial Colonization. Microorganisms 2023, 11, 239. https://doi.org/10.3390/microorganisms11020239
Vemuri R, Herath MP. Beyond the Gut, Emerging Microbiome Areas of Research: A Focus on Early-Life Microbial Colonization. Microorganisms. 2023; 11(2):239. https://doi.org/10.3390/microorganisms11020239
Chicago/Turabian StyleVemuri, Ravichandra, and Manoja P. Herath. 2023. "Beyond the Gut, Emerging Microbiome Areas of Research: A Focus on Early-Life Microbial Colonization" Microorganisms 11, no. 2: 239. https://doi.org/10.3390/microorganisms11020239
APA StyleVemuri, R., & Herath, M. P. (2023). Beyond the Gut, Emerging Microbiome Areas of Research: A Focus on Early-Life Microbial Colonization. Microorganisms, 11(2), 239. https://doi.org/10.3390/microorganisms11020239






