Conversion of Escherichia coli into Mixotrophic CO2 Assimilation with Malate and Hydrogen Based on Recombinant Expression of 2-Oxoglutarate:Ferredoxin Oxidoreductase Using Adaptive Laboratory Evolution
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains and Genomic Modification
2.2. Enzymatic Activity Assays of 2-Oxoglutarate:Ferredoxin Oxidoreductase
2.3. Growth Condition and Data Processing
2.4. Evolution in the Bioreactor
2.5. Next-Generation Sequencing and Transcriptome Analysis
2.5.1. Genomic DNA Isolation and Sequencing
2.5.2. Genomic DNA Variant Analysis
2.5.3. RNA Isolation, Library Preparation, and Sequencing
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- The Intergovernmental Panel on Climate Change. Synthesis Report. Contribution of Working Groups I. II and III to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change; The Intergovernmental Panel on Climate Change: Geneva, Switzerland, 2014.
- Zhuang, Z.Y.; Li, S.Y. RuBisCO-based engineered Escherichia coli for in situ carbon dioxide recycling. Bioresour. Technol. 2013, 150, 79–88. [Google Scholar] [CrossRef] [PubMed]
- Gong, F.; Liu, G.; Zhai, X.; Zhou, J.; Cai, Z.; Li, Y. Quantitative analysis of an engineered CO2-fixing Escherichia coli reve eveals great potential of heterotrophic CO2 fixation. Biotechnol. Biofuels 2015, 8, 86. [Google Scholar] [CrossRef] [Green Version]
- Antonovsky, N.; Gleizer, S.; Noor, E.; Zohar, Y.; Herz, E.; Barenholz, U.; Zelcbuch, L.; Amram, S.; Wides, A.; Tepper, N.; et al. Sugar synthesis from CO2 in Escherichia coli. Cell 2016, 166, 115–125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gleizer, S.; Ben-Nissan, R.; Bar-On, Y.M.; Antonovsky, N.; Noor, E.; Zohar, Y.; Jona, G.; Krieger, E.; Shamshoum, M.; Bar-Even, A.; et al. Conversion of Escherichia coli to generate all biomass carbon from CO2. Cell 2019, 179, 1255–1263. [Google Scholar] [CrossRef] [Green Version]
- Gassler, T.; Steiger, M.G. The industrial yeast Pichia pastoris is converted from a heterotroph into an autotroph capable of growth on CO2. Nat. Biotechnol. 2020, 38, 210–216. [Google Scholar] [CrossRef]
- Chen, F.Y.; Jung, H.W.; Tsuei, C.Y.; Liao, J.C. Converting Escherichia coli to a synthetic methylotroph growing solely on methanol. Cell 2020, 182, 933–946. [Google Scholar] [CrossRef]
- Folsom, J.P.; Parker, A.E.; Carlson, R.P. Physiological and proteomic analysis of Escherichia coli iron-limited chemostat growth. J. Bacteriol. 2014, 196, 2748–2761. [Google Scholar] [CrossRef] [Green Version]
- Mei, H.; Arbeithuber, B.; Cremona, M.A.; DeGiorgio, M.; Nekrutenko, A. A high-resolution view of adaptive event dynamics in a plasmid. Genome Biol. Evol. 2019, 11, 3022–3034. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Toprak, E.; Veres, A.; Michel, J.B.; Chait, R.; Hartl, D.L.; Kishony, R. Evolutionary paths to antibiotic resistance under dynamically sustained drug selection. Nat. Genet. 2011, 44, 101–105. [Google Scholar] [CrossRef]
- Gresham, D.; Hong, J. The functional basis of adaptive evolution in chemostats. FEMS Microbiol. Rev. 2015, 39, 2–16. [Google Scholar] [CrossRef]
- Bar-Even, A.; Milo, R. A survey of carbon fixation pathways through a quantitative lens. J. Exp. Bot. 2012, 63, 2325–2342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berg, I.; Fuchs, G. Autotrophic carbon fixation in archaea. Nat. Rev. Microbiol. 2010, 8, 447–460. [Google Scholar] [CrossRef] [PubMed]
- Berg, I. Ecological aspects of the distribution of different autotrophic CO2 fixation pathways. Appl. Environ. Microbiol. 2011, 77, 1925–1936. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buchanan, B.B.; Evans, M.C. The synthesis of alpha-ketoglutarate from succinate and carbon dioxide by a subcellular preparation of a photosynthetic bacterium. Proc. Natl. Acad. Sci. USA 1965, 54, 1212–1218. [Google Scholar] [CrossRef] [Green Version]
- Evans, M.C.; Arnon, D. A new ferredoxin-dependent carbon reduction cycle in a photosynthetic bacterium. Proc. Natl. Acad. Sci. USA 1966, 55, 928–934. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamamoto, M.; Igarashi, Y. Characterization of two different 2-oxoglutarate:ferredoxin oxidoreductases from Hydrogenobacter thermophilus TK-6. Biochem. Biophys. Res. Commun. 2003, 312, 1297–1302. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, M.; Igarashi, Y. Role of two 2-oxoglutarate:ferredoxin oxidoreductases in Hydrogenobacter thermophilus under aerobic and anaerobic conditions. FEMS Microbiol. Lett. 2006, 263, 189–193. [Google Scholar] [CrossRef] [Green Version]
- Mall, A.; Berg, I. Reversibility of citrate synthase allows autotrophic growth of a thermophilic bacterium. Science 2018, 359, 563–567. [Google Scholar] [CrossRef] [Green Version]
- Nunoura, T.; Takai, K. A primordial and reversible TCA cycle in a facultatively chemolithoautotrophic thermophile. Science 2018, 359, 559–563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, P.Y.T.; Li, B.; Drennan, C.L.; Elliot, S.J. A reverse TCA cycle 2-oxoacid;ferredoxin oxidoreuctase that maks C-C bonds from CO2. Joule 2019, 3, 595–611. [Google Scholar] [CrossRef]
- Cheng, H.-T.-Y.; Lo, S.-C.; Huang, C.-C.; Yang, Y.-T. Detailed profiling of carbon fixation of in silico synthetic autotrophy with reductive tricarboxylic acid cycle and Calvin-Benson-Bassham cycle in Esherichia coli using hydrogen as an energy source. Synth. Sys. Biotech. 2019, 4, 165–172. [Google Scholar] [CrossRef] [PubMed]
- Lo, S.-C.; Chiang, E.-P.I.; Yang, Y.-T.; Li, S.-Y.; Peng, J.-H.; Tsai, S.-Y.; Wu, D.-Y.; Yu, C.-H.; Huang, C.-H.; Su, T.-T.; et al. Growth Enhancement Facilitated by Gaseous CO2 through Heterologous Expression of Reductive Tricarboxylic Acid Cycle Genes in Escherichia coli. Fermentation 2021, 7, 98. [Google Scholar] [CrossRef]
- Macy, J.; Kulla, H.; Gottshohalk, G. H2 dependent anaerobic growth of Escherichia coli on L-Malate: Succinate formation. J. Bacteriol. 1976, 125, 423–428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsuge, K.; Matsui, K.; Itaya, M. One step assembly of multiple DNA fragments with a designed order and orientation in Bacillus subtilis plasmid. Nucleic Acids Res. 2003, 31, e133. [Google Scholar] [CrossRef] [Green Version]
- Kaneko, S.; Akioka, M.; Tsuge, K.; Itaya, M. DNA shuttling between plasmid vectors and a genome vector: Systematic conversion and preservation of DNA Libraries using the Bacillus subtilis Genome (BGM) Vector. J. Mol. Biol. 2005, 349, 1036–1044. [Google Scholar] [CrossRef] [PubMed]
- Dragosits, M.; Mattanovich, D. Adaptive laboratory evolution—Principles and applications for biotechnology. Microb. Cell Factories 2013, 12, 64. [Google Scholar] [CrossRef] [Green Version]
- Lin, D.S.; Yang, Y.T. Wireless bioreactor for anaerobic cultivation of bacteria. Biotech. Prog. 2020, 36, e3009. [Google Scholar] [CrossRef]
- Swain, P.S.; Pilizota, T. Inferring time-derivatives, including cell growth rates, using Gaussian processes. Nat. Commun. 2016, 7, 13766. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef] [Green Version]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef]
- Sawers, G. The anaerobic degradation of l-serine and l-threonine in enterobacteria: Networks of pathways and regulatory signals. Arch. Microbiol. 1998, 171, 1–5. [Google Scholar] [CrossRef]
- Zinser, E.R.; Kolter, R. Mutations enhancing amino acid catabolism confer a growth advantage in stationary phase. J. Bacteriol. 1999, 181, 5800–5807. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Debs-Louka, E.; Louka, N.; Abraham, G.; Chabot, V.; Allaf, K. Effect of compressed carbon dioxide on microbial cell viability. Appl. Environ. Microbiol. 1999, 65, 626–631. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flamholtz, A.; Noor, E.; Bar-Even, A.; Milo, R. eQullibrator—The biochemical thermodynamic calculator. Nucleic Acid Res. Syst. 2012, 40, D770–D775. [Google Scholar] [CrossRef] [Green Version]
- Shinar, G.; Rabinowitz, J.D.; Alon, U. Robustness in Glyoxylate bypass Regulation. PLoS Comput. Biol. 2009, 5, e1000297. [Google Scholar] [CrossRef] [Green Version]
- LaPorte, D.C.; Thorsness, P.E.; Koshland, D.E., Jr. Compensatory phosphorylation of isocitrate dehydrogenase. A mechanism for adaptation to the intracellular environment. J. Biol. Chem. 1985, 260, 10563–10568. [Google Scholar] [CrossRef] [PubMed]
- Lebeuf-Taylor, E.; McCloskey, N.; Bailey, S.F.; Hinz, A.; Kassen, R. The distribution of fitness effects among synonymous mutations in a gene under directional selection. Elife 2019, 8, e45952. [Google Scholar] [CrossRef]
- Bailey, S.F.; Hinz, A.; Kassen, R. Adaptive synonymous mutations in an experimentally evolved Pseudomonas fluorescens population. Nat. Commun. 2014, 5, 4076. [Google Scholar] [CrossRef] [Green Version]
- Plotkin, J.B.; Kudla, G. Synonymous but not the same: The causes and consequences of codon bias. Nat. Rev. Genet. 2011, 12, 32–42. [Google Scholar] [CrossRef] [Green Version]
- Agashe, D.; Sane, M.; Phalnikar, K.; Diwan, G.D.; Habibullah, A.; Martinez-Gomez, N.C.; Sahasrabuddhe, V.; Polachek, W.; Wang, J.; Chubiz, L.M.; et al. Large-Effect beneficial synonymous mutations mediate rapid and parallel adaptation in a bacterium. Mol. Biol. Evol. 2016, 33, 1542–1553. [Google Scholar] [CrossRef]
- Phaneuf, P.V.; Gosting, D.; Palsson, B.O.; Feist, A.M. ALEdb 1.0: A database of mutations from adaptive laboratory evolution experimentation. Nucleic Acids Res. 2019, 47, D1164–D1171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Utrilla, J.; O’Brien, E.J.; Chen, K.; McCloskey, D.; Cheung, J.; Wang, H.; Armenta-Medina, D.; Feist, A.M.; Palsson, B.O. Global Rebalancing of Cellular Resources by Pleiotropic Point Mutations Illustrates a Multi-scale Mechanism of Adaptive Evolution. Cell Syst. 2016, 2, 260–271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sievert, C.; Nieves, L.M.; Panyon, L.A.; Loeffler, T.; Morris, C.; Cartwright, R.A.; Wang, X. Experimental evolution reveals an effective avenue to release catabolite repression via mutations in XylR. Proc. Natl. Acad. Sci. USA 2017, 114, 7349–7354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Savageau, M.A. Design of molecular control mechanism and the demand for gene expression. Proc. Natl. Acad. Sci. USA 1977, 74, 5647–5651. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neidhardt, F.C.; Ingraham, J.L.; Schaechter, M. Physiology of the Bacterial Cell: A Molecular Approach; Sinauer Associates: Sunderland, MA, USA, 1990. [Google Scholar]
- Macomber, L.; Elsey, S.P.; Haisinger, R. P Fructose-1,6-bisphosphate aldolase (class II) is the primary site of nickel toxicity in Escherichia coli. Mol. Microbiol. 2011, 82, 1291–1300. [Google Scholar] [CrossRef] [Green Version]
- Blount, Z.D.; Borland, C.Z.; Lenski, R.E. Historical contingency and the evolution of a key innovation in an experimental population of Escherichia coli. Proc. Natl. Acad. Sci. USA 2008, 105, 7899–7906. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quandt, E.M.; Gollihar, J.; Blount, Z.D.; Ellington, A.D.; Georgiou, G.; Barrick, J.E. Fine-tuning citrate synthase flux potentiates and refines metabolic innovation in the Lenski evolution experiment. eLife 2015, 4, e09696. [Google Scholar] [CrossRef]
- Blaschkowski, H.P.; Neuer, G.; Ludwig-Festl, M.; Knappe, J. Routes of flavodoxin and ferredoxin reduction in Escherichia coli. CoA-acylating pyruvate: Flavodoxin and NADPH: Fla-vodoxin oxidoreductases participating in the activation of pyruvate formate-lyase. Eur. J. Biochem. 1982, 123, 563–569. [Google Scholar] [CrossRef]
- Nilekani, S.; SivaRaman, C. Purification and properties of citrate lyase from Escherichia coli. Biochemistry 1983, 22, 4657–4663. [Google Scholar] [CrossRef]
- Bar-Even, A.; Noor, E.; Lewis, N.E.; Milo, R. Design and analysis of synthetic carbon fixation pathways. Proc. Natl. Acad. Sci. USA 2010, 107, 8889–8894. [Google Scholar] [CrossRef]
- Gokam, R.R.; Eiteman, M.A.; Altman, E. Metabolic Analysis of Escherichia coli in the presence and absence of the carboxylating enzymes phosphoenolpyruvate carboxylase and pyruvate carboxylase. Appl. Environ. Microbiol. 2000, 66, 1844–1850. [Google Scholar]
- Volpers, M.; Claassens, N.J.; Noor, E.; van der Oost, J.; de Vos, W.M.; Kengen, S.W.; Martins Dos Santos, V.A. Integrated In Silico Analysis of Pathway Designs for Synthetic Photo-Electro-Autotrophy. PLoS ONE 2016, 11, e0157851. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rabinowitz, J.D.; Shihavy, T.J. Metabolite turns mater regulator. Nature 2013, 500, 283–284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Towbin, B.D.; Korem, Y.; Bren, A.; Doron, S.; Sorek, R.; Alon, U. Optimality and sub-optimality in a bacterial growth law. Nat. Commun. 2017, 8, 14123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sawers, R.G.; Ballantine, S.P.; Boxer, D.H. Differential expression of hydrogenase isoenzymes in Escherichia coli K-12: Evidence for a third isoenzyme. J. Bacteriol. 1985, 164, 1324–1331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, L.-F.; Mandrand, M.A. Microbial hydrogenases: Primary structure, classification, signatures and phylogeny. FEMS Microbiol. Rev. 1993, 104, 243–270. [Google Scholar] [CrossRef] [PubMed]
- Sargent, F. The Model [NiFe]-Hydrogenases of Escherichia coli. Microb. Physiol. 2016, 68, 433–507. [Google Scholar]
- Claassens, N.J.; Sa´nchez-Andrea, I.; Sousa, D.Z.; Bar-Even, A. Towards sustainable feedstocks: A guide to electron donors for microbial carbon fixation. Curr. Opin. Biotechnol. 2018, 50, 195–205. [Google Scholar] [CrossRef]
- Torella, J.P.; Gagliardi, C.J.; Chen, J.S.; Bediako, D.K.; Colón, B.; Way, J.C.; Silver, P.A.; Nocera, D.G. Efficient solar-to-fuels production from a hybrid microbial–water-splitting catalyst system. Proc. Natl Acad. Sci. USA 2015, 112, 2337–2342. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.; Colón, B.C.; Ziesack, M.; Silver, P.A.; Nocera, D.G. Water splitting-biosynthetic system with CO2 reduction efficiencies exceeding photosynthesis. Science 2016, 352, 1210–1213. [Google Scholar] [CrossRef]
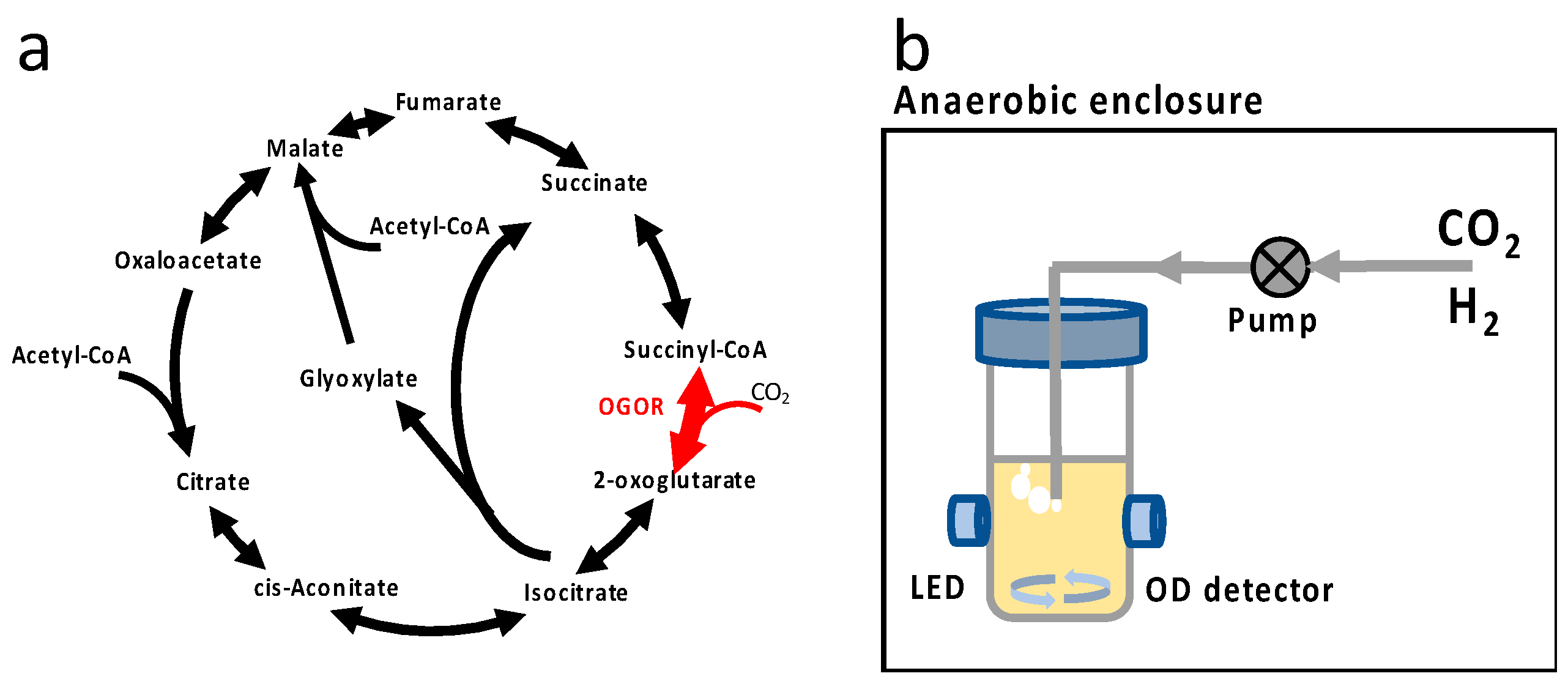
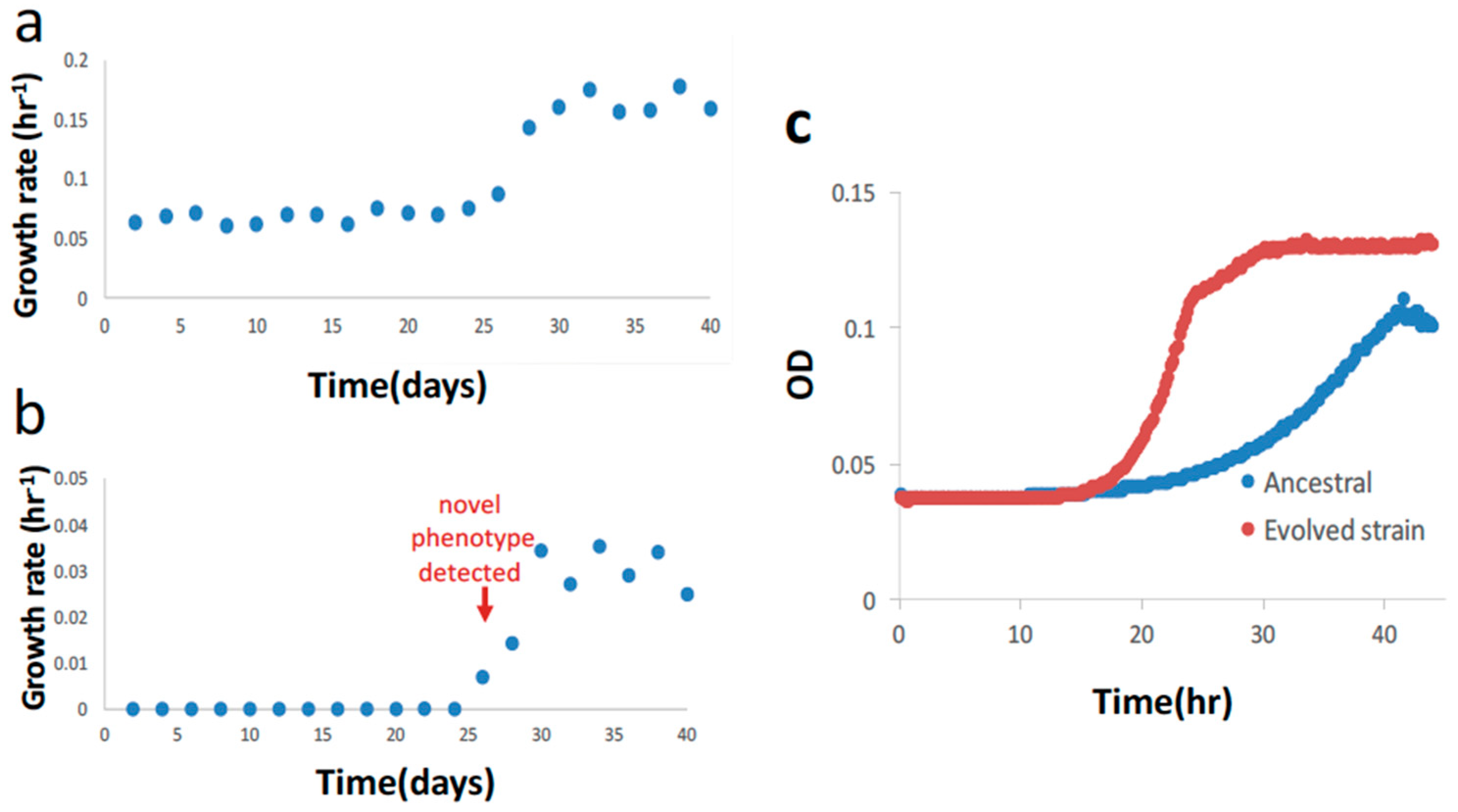
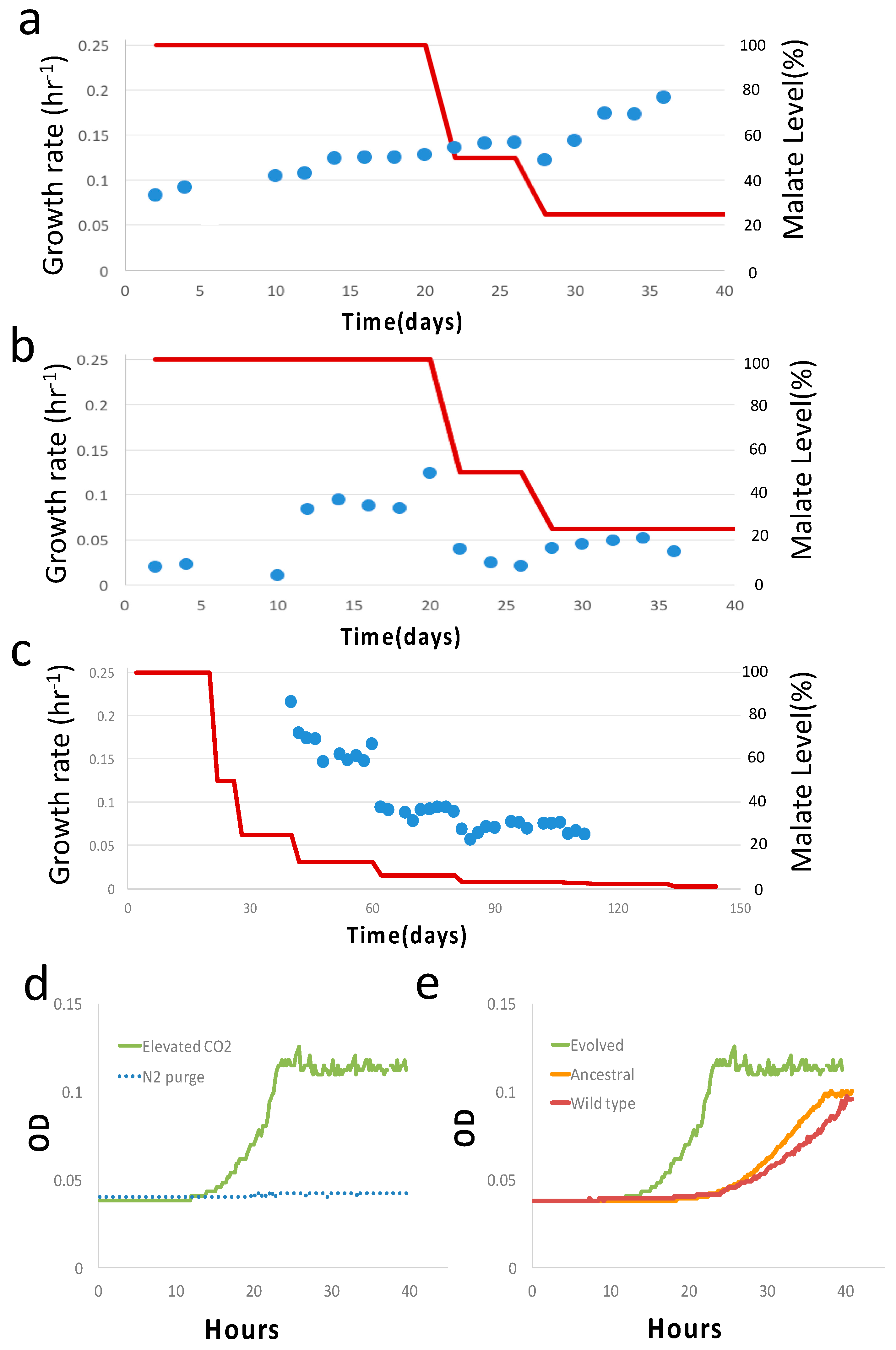
| Primer a | Primer Sequence (5′ to 3′) b |
|---|---|
| KOR-DraIII-f | ATGCACGTTGTGGAAAAAAGGAAGAGGGGATACCCATGAGTGAC |
| KOR-PflMI-r | ATGCCAAAGATTGGTCAGTTGATCGTCCAGGTGCTGTTGC |
| Enzyme Activity (nmol min−1 mg−1 Protein) | |
|---|---|
| Strains | 2-oxoglutarate:ferredoxin oxidoreductase |
| wild-type a | <0.01 |
| ancestral b | 49.6 ± 12.2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheng, Y.-C.; Huang, W.-H.; Lo, S.-C.; Huang, E.; Chiang, E.-P.I.; Huang, C.-C.; Yang, Y.-T. Conversion of Escherichia coli into Mixotrophic CO2 Assimilation with Malate and Hydrogen Based on Recombinant Expression of 2-Oxoglutarate:Ferredoxin Oxidoreductase Using Adaptive Laboratory Evolution. Microorganisms 2023, 11, 253. https://doi.org/10.3390/microorganisms11020253
Cheng Y-C, Huang W-H, Lo S-C, Huang E, Chiang E-PI, Huang C-C, Yang Y-T. Conversion of Escherichia coli into Mixotrophic CO2 Assimilation with Malate and Hydrogen Based on Recombinant Expression of 2-Oxoglutarate:Ferredoxin Oxidoreductase Using Adaptive Laboratory Evolution. Microorganisms. 2023; 11(2):253. https://doi.org/10.3390/microorganisms11020253
Chicago/Turabian StyleCheng, Yu-Chen, Wei-Han Huang, Shou-Chen Lo, Eugene Huang, En-Pei Isabel Chiang, Chieh-Chen Huang, and Ya-Tang Yang. 2023. "Conversion of Escherichia coli into Mixotrophic CO2 Assimilation with Malate and Hydrogen Based on Recombinant Expression of 2-Oxoglutarate:Ferredoxin Oxidoreductase Using Adaptive Laboratory Evolution" Microorganisms 11, no. 2: 253. https://doi.org/10.3390/microorganisms11020253
APA StyleCheng, Y.-C., Huang, W.-H., Lo, S.-C., Huang, E., Chiang, E.-P. I., Huang, C.-C., & Yang, Y.-T. (2023). Conversion of Escherichia coli into Mixotrophic CO2 Assimilation with Malate and Hydrogen Based on Recombinant Expression of 2-Oxoglutarate:Ferredoxin Oxidoreductase Using Adaptive Laboratory Evolution. Microorganisms, 11(2), 253. https://doi.org/10.3390/microorganisms11020253






