Genetic Diversity and Primary Drug Resistance of Mycobacterium tuberculosis Beijing Genotype Strains in Northwestern Russia
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Setting and Bacterial Strains
2.2. Drug Susceptibility Testing
2.3. Genotyping
3. Results
3.1. Main Genotype Clusters
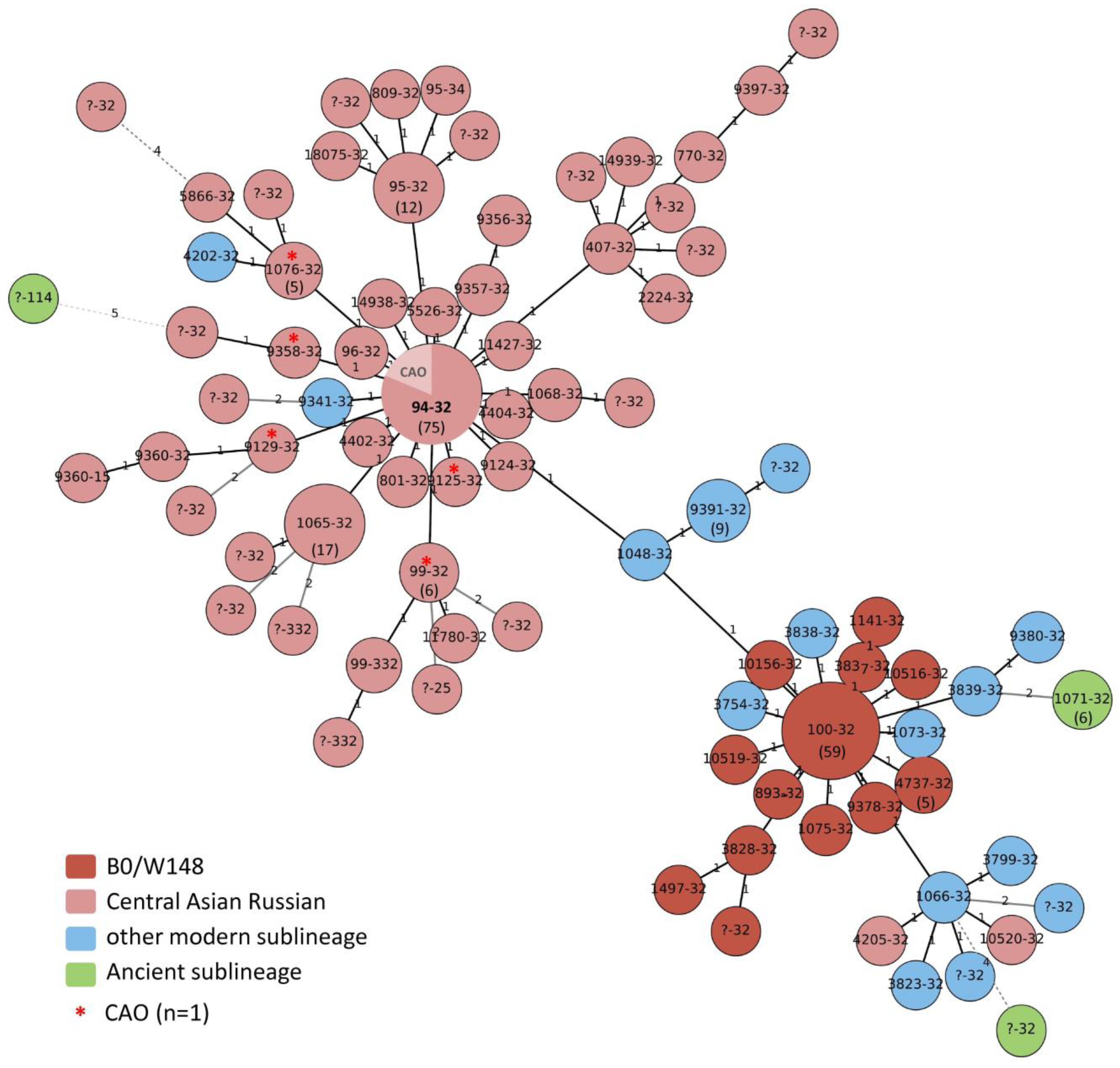
3.2. High-Resolution Population Structure
3.3. Drug Resistance versus Genotypes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- van Soolingen, D.; Qian, L.; de Haas, P.E.W.; Douglas, J.T.; Traore, H.; Portaels, F.; Qing, H.Z.; Enkhsaikan, D.; Nymadawa, P.; van Embden, J.D. Predominance of a single genotype of Mycobacterium tuberculosis in countries of East Asia. J. Clin. Microbiol. 1995, 33, 3234–3238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Merker, M.; Blin, C.; Mona, S.; Duforet-Frebourg, N.; Lecher, S.; Willery, E.; Blum, M.G.; Rüsch-Gerdes, S.; Mokrousov, I.; Aleksic, E.; et al. Evolutionary history and global spread of the Mycobacterium tuberculosis Beijing lineage. Nat. Genet. 2015, 47, 242–249. [Google Scholar] [CrossRef] [PubMed]
- Glynn, J.R.; Whiteley, J.; Bifani, P.J.; Kremer, K.; van Soolingen, D. Worldwide occurrence of Beijing/W strains of Mycobacterium tuberculosis: A systematic review. Emerg. Infect. Dis. 2002, 8, 843–849. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ford, C.B.; Shah, R.R.; Maeda, M.K.; Gagneux, S.; Murray, M.B.; Cohen, T.; Johnston, J.C.; Gardy, J.; Lipsitch, M.; Fortune, S.M. Mycobacterium tuberculosis mutation rate estimates from different lineages predict substantial differences in the emergence of drug-resistant tuberculosis. Nat. Genet. 2013, 45, 784–790. [Google Scholar] [CrossRef] [Green Version]
- Hakamata, M.; Takihara, H.; Iwamoto, T.; Tamaru, A.; Hashimoto, A.; Tanaka, T.; Kaboso, S.A.; Gebretsadik, G.; Ilinov, A.; Yokoyama, A.; et al. Higher genome mutation rates of Beijing lineage of Mycobacterium tuberculosis during human infection. Sci. Rep. 2020, 10, 17997. [Google Scholar] [CrossRef]
- Werngren, J.; Hoffner, S.E. Drug-susceptible Mycobacterium tuberculosis Beijing genotype does not develop mutation-conferred resistance to rifampin at an elevated rate. J. Clin. Microbiol. 2003, 41, 1520–1524. [Google Scholar] [CrossRef] [Green Version]
- Mokrousov, I.; Narvskaya, O.; Otten, T.; Vyazovaya, A.; Limeschenko, E.; Steklova, L.; Vyshnevskyi, B. Phylogenetic reconstruction within Mycobacterium tuberculosis Beijing genotype in northwestern Russia. Res. Microbiol. 2002, 153, 629–637. [Google Scholar] [CrossRef]
- Ebrahimi-Rad, M.; Bifani, P.; Martin, C.; Kremer, K.; Samper, S.; Rauzier, J.; Kreiswirth, B.; Blazquez, J.; Jouan, M.; van Soolingen, D.; et al. Mutations in putative mutator genes of Mycobacterium tuberculosis strains of the W-Beijing family. Emerg. Infect. Dis. 2003, 9, 838–845. [Google Scholar] [CrossRef] [Green Version]
- Tsolaki, A.G.; Hirsh, A.E.; De Riemer, K.; Enciso, J.A.; Wong, M.Z.; Hannan, M.; de la Salmoniere, Y.O.L.G.; Aman, K.; Kato-Maeda, M.; Small, P.M. Functional and evolutionary genomics of Mycobacterium tuberculosis: Insights from genomic deletions in 100 strains. Proc. Natl. Acad. Sci. USA 2004, 101, 4865–4870. [Google Scholar] [CrossRef] [Green Version]
- Mokrousov, I.; Ly, H.M.; Otten, T.; Lan, N.N.; Vyshnevskyi, B.; Hoffner, S.; Narvskaya, O. Origin and primary dispersal of the Mycobacterium tuberculosis Beijing genotype: Clues from human phylogeography. Genome Res. 2005, 15, 1357–1364. [Google Scholar] [CrossRef]
- Tsolaki, A.G.; Gagneux, S.; Pym, A.S.; Goguet de la Salmoniere, Y.O.; Kreiswirth, B.N.; van Soolingen, D.; Small, P.M. Genomic deletions classify the Beijing/W strains as a distinct genetic lineage of Mycobacterium tuberculosis. J. Clin. Microbiol. 2005, 43, 3185–3191. [Google Scholar] [CrossRef] [Green Version]
- Yin, Q.Q.; Liu, H.C.; Jiao, W.W.; Li, Q.J.; Han, R.; Tian, J.L.; Liu, Z.G.; Zhao, X.Q.; Li, Y.J.; Wan, K.L.; et al. Evolutionary History and Ongoing Transmission of Phylogenetic Sublineages of Mycobacterium tuberculosis Beijing Genotype in China. Sci. Rep. 2016, 6, 34353. [Google Scholar] [CrossRef]
- Mourik, B.C.; de Steenwinkel, J.E.M.; de Knegt, G.J.; Huizinga, R.; Verbon, A.; Ottenhoff, T.H.M.; van Soolingen, D.; Leenen, P.J.M. Mycobacterium tuberculosis clinical isolates of the Beijing and East-African Indian lineage induce fundamentally different host responses in mice compared to H37Rv. Sci. Rep. 2019, 9, 19922. [Google Scholar] [CrossRef] [Green Version]
- Maeda, S.; Hang, N.T.; Lien, L.T.; Thuong, P.H.; Hung, N.V.; Hoang, N.P.; Cuong, V.C.; Hijikata, M.; Sakurada, S.; Keicho, N. Mycobacterium tuberculosis strains spreading in Hanoi, Vietnam: Beijing sublineages, genotypes, drug susceptibility patterns, and host factors. Tuberculosis 2014, 94, 649–656. [Google Scholar] [CrossRef] [Green Version]
- Mokrousov, I.; Vyazovaya, A.; Pasechnik, O.; Gerasimova, A.; Dymova, M.; Chernyaeva, E.; Tatarintseva, M.; Stasenko, V. Early ancient sublineages of Mycobacterium tuberculosis Beijing genotype: Unexpected clues from phylogenomics of the pathogen and human history. Clin. Microbiol. Infect. 2019, 25, 1039.e1–1039.e6. [Google Scholar] [CrossRef]
- Vinogradova, T.; Dogonadze, M.; Zabolotnykh, N.; Badleeva, M.; Yarusova, I.; Vyazovaya, A.; Gerasimova, A.; Zhdanova, S.; Vitovskaya, M.; Solovieva, N.; et al. Extremely lethal and hypervirulent Mycobacterium tuberculosis strain cluster emerging in Far East, Russia. Emerg. Microbes Infect. 2021, 10, 1691–1701. [Google Scholar] [CrossRef]
- Bespyatykh, J.A.; Vinogradova, T.I.; Manicheva, O.A.; Zabolotnykh, N.V.; Dogonadze, M.Z.; Vitovskaya, M.L.; Guliaev, A.S.; Zhuravlev, V.Y.; Shitikov, E.A.; Ilina, E.N. In vivo virulence of Beijing genotype Mycobacterium tuberculosis. Russ. J. Infect. Immun. 2019, 9, 173–182. (In Russian) [Google Scholar] [CrossRef] [Green Version]
- Shitikov, E.; Vyazovaya, A.; Malakhova, M.; Guliaev, A.; Bespyatykh, J.; Proshina, E.; Pasechnik, O.; Mokrousov, I. Simple Assay for detection of the Central Asia outbreak clade of the Mycobacterium tuberculosis Beijing genotype. J. Clin. Microbiol. 2019, 57, e00215-19. [Google Scholar] [CrossRef] [Green Version]
- van Embden, J.D.; Cave, M.D.; Crawford, J.T.; Dale, J.W.; Eisenach, K.D.; Gicquel, B.; Hermans, P.; Martin, C.; McAdam, R.; Shinnick, T.M.; et al. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: Recommendations for a standardized methodology. J. Clin. Microbiol. 1993, 31, 406–409. [Google Scholar] [CrossRef] [Green Version]
- Mokrousov, I.; Vyazovaya, A.; Zhuravlev, V.; Otten, T.; Millet, J.; Jiao, W.W.; Shen, A.D.; Rastogi, N.; Vishnevsky, B.; Narvskaya, O. Real-time PCR assay for rapid detection of epidemiologically and clinically significant Mycobacterium tuberculosis Beijing genotype isolates. J. Clin. Microbiol. 2014, 52, 1691–1693. [Google Scholar] [CrossRef]
- Mokrousov, I.; Narvskaya, O.; Vyazovaya, A.; Otten, T.; Jiao, W.W.; Gomes, L.L.; Suffys, P.N.; Shen, A.D.; Vishnevsky, B. Russian “successful” clone B0/W148 of Mycobacterium tuberculosis Beijing genotype: A multiplex PCR assay for rapid detection and global screening. J. Clin. Microbiol. 2012, 50, 3757–3759. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mokrousov, I.; Chernyaeva, E.; Vyazovaya, A.; Skiba, Y.; Solovieva, N.; Valcheva, V.; Levina, K.; Malakhova, N.; Jiao, W.W.; Gomes, L.L.; et al. Rapid assay for detection of the epidemiologically important Central Asian/Russian strain of the Mycobacterium tuberculosis Beijing genotype. J. Clin. Microbiol. 2018, 56, e01551-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shitikov, E.; Kolchenko, S.; Mokrousov, I.; Bespyatykh, J.; Ischenko, D.; Ilina, E.; Govorun, V. Evolutionary pathway analysis and unified classification of East Asian lineage of Mycobacterium tuberculosis. Sci. Rep. 2017, 7, 9227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iwamoto, T.; Grandjean, L.; Arikawa, K.; Nakanishi, N.; Caviedes, L.; Coronel, J.; Sheen, P.; Wada, T.; Taype, C.A.; Shaw, M.A.; et al. Genetic diversity and transmission characteristics of Beijing family strains of Mycobacterium tuberculosis in Peru. PLoS ONE 2012, 7, e49651. [Google Scholar] [CrossRef] [PubMed]
- Supply, P.; Allix, C.; Lesjean, S.; Cardoso-Oelemann, M.; Rüsch-Gerdes, S.; Willery, E.; Savine, E.; de Haas, P.; van Deutekom, H.; Roring, S.; et al. Proposal for standardization of optimized Mycobacterial interspersed repetitive Unit-Variable-Number Tandem Repeat Typing of Mycobacterium tuberculosis. J. Clin. Microbiol. 2006, 44, 4498–4510. [Google Scholar] [CrossRef] [Green Version]
- Mokrousov, I. Revisiting the Hunter Gaston discriminatory index: Note of caution and courses of change. Tuberculosis. 2017, 104, 20–23. [Google Scholar] [CrossRef]
- Supply, P.; Lesjean, S.; Savine, E.; Kremer, K.; van Soolingen, D.; Locht, C. Automated high-throughput genotyping for study of global epidemiology of Mycobacterium tuberculosis based on mycobacterial interspersed repetitive units. J. Clin. Microbiol. 2001, 39, 3563–3571. [Google Scholar] [CrossRef] [Green Version]
- Nechaeva, O.B.; Gordina, A.V.; Sterlikov, S.A.; Kucheryavaya, D.A.; Dergachev, A.V.; Ponomarev, S.B.; Burykhin, V.S. Resources and Activities of TB Organizations in the Russian Federation in 2018–2019 (Statistical Materials); RIO TsNIIOIZ: Moscow, Russia, 2020; 99p. [Google Scholar]
- Galkin, V.B.; Sterlikov, S.A.; Yablonsky, P.K.; Beltyukov, M.V.; Grishko, A.N.; Baglina, S.S.; Vasilyeva, T.V.; Danilova, T.I.; Kononenko, Y.S.; Kulizhskaya, A.I.; et al. Dynamics of the Prevalence of Multidrug-Resistant Tuberculosis and HIV Infection in the North-West Region of Russia. Meditsinskiy Alians 2019, 6–23. (In Russian) [Google Scholar]
- Mokrousov, I.; Valcheva, V.; Sovhozova, N.; Aldashev, A.; Rastogi, N.; Isakova, J. Penitentiary population of Mycobacterium tuberculosis in Kyrgyzstan: Exceptionally high prevalence of the Beijing genotype and its Russia-specific subtype. Infect. Genet. Evol. 2009, 9, 1400–1405. [Google Scholar] [CrossRef]
- Vyazovaya, A.A.; Pasechnik, O.A.; Gerasimova, A.A.; Mokrousov, I.V. Population structure of the Beijing Mycobacterium tuberculosis genetic family in Western Siberia. Tuberc. Lung Dis. 2020, 98, 32–36. (In Russian) [Google Scholar] [CrossRef]
- Dymova, M.A.; Kinsht, V.N.; Cherednichenko, A.G.; Khrapov, E.A.; Svistelnik, A.V.; Filipenko, M.L. Highest prevalence of the Mycobacterium tuberculosis Beijing genotype isolates in patients newly diagnosed with tuberculosis in the Novosibirsk oblast, Russian Federation. J. Med. Microbiol. 2011, 60 Pt 7, 1003–1009. [Google Scholar] [CrossRef]
- Afanas’ev, M.V.; Ikryannikova, L.N.; Il’ina, E.N.; Kuz’min, A.V.; Larionova, E.E.; Smirnova, T.G.; Chernousova, L.N.; Govorun, V.M. Molecular typing of Mycobacterium tuberculosis circulated in Moscow, Russian Federation. Eur. J. Clin. Microbiol. Infect. Dis. 2011, 30, 181–191. [Google Scholar] [CrossRef]
- Zhdanova, S.; Mokrousov, I.; Orlova, E.; Sinkov, V.; Ogarkov, O. Transborder molecular analysis of drug-resistant tuberculosis in Mongolia and Eastern Siberia, Russia. Transbound. Emerg. Dis. 2022, 69, e1800–e1814. [Google Scholar] [CrossRef]
- Zhdanova, S.N.; Ogarkov, O.B.; Alekseeva, G.I.; Vinokurova, M.K.; Sinkov, V.V.; Savilov, E.D.; Kravchenko, A.F. Genetic diversity of Mycobacterium tuberculosis isolates from Sakha (Yakutia), Russia. Mol. Genet. Microbiol. Virol. 2016, 31, 51–57. [Google Scholar] [CrossRef]
- Khromova, P.A.; Kornilov, M.S.; Zhdanova, S.N.; Yakovleva, A.A.; Ogarkov, O.B. Identification of epidemic subtypes of the Beijing Mycobacterium tuberculosis genotype circulating in Primorsky Krai. Acta Biomed. Sci. 2018, 3, 154–158. (In Russian) [Google Scholar]
- Mokrousov, I. Insights into the origin, emergence, and current spread of a successful Russian clone of Mycobacterium tuberculosis. Clin. Microbiol. Rev. 2013, 26, 342–360. [Google Scholar] [CrossRef] [Green Version]
- De Beer, J.L.; Kodmon, C.; van der Werf, M.J.; van Ingen, J.; van Soolingen, D. ECDC MDR-TB Molecular Surveillance Project Participants. Molecular surveillance of multi- and extensively drug-resistant tuberculosis transmission in the European Union from 2003 to 2011. Euro Surveill 2014, 19, 20742. [Google Scholar] [CrossRef] [Green Version]
- Pole, I.; Trofimova, J.; Norvaisa, I.; Supply, P.; Skenders, G.; Nodieva, A.; Ozere, I.; Riekstina, V.; Igumnova, V.; Storozenko, J.; et al. Analysis of Mycobacterium tuberculosis genetic lineages circulating in Riga and Riga region, Latvia, isolated between 2008 and 2012. Infect. Genet. Evol. 2020, 78, 104126. [Google Scholar] [CrossRef]
- Skiba, Y.; Mokrousov, I.; Ismagulova, G.; Maltseva, E.; Yurkevich, N.; Bismilda, V.; Chingissova, L.; Abildaev, T.; Aitkhozhina, N. Molecular snapshot of Mycobacterium tuberculosis population in Kazakhstan: A country-wide study. Tuberculosis 2015, 95, 538–546. [Google Scholar] [CrossRef]
- Mokrousov, I.; Vyazovaya, A.; Levina, K.; Gerasimova, A.; Zhuravlev, V.; Viiklepp, P.; Kütt, M. Spatiotemporal dynamics of drug-resistant Mycobacterium tuberculosis: Contrasting trends and implications for tuberculosis control in EU high-priority country. Transbound. Emerg. Dis. 2021, 68, 896–906. [Google Scholar] [CrossRef]
- Merker, M.; Rasigade, J.P.; Barbier, M.; Cox, H.; Feuerriegel, S.; Kohl, T.A.; Shitikov, E.; Klaos, K.; Gaudin, C.; Antoine, R.; et al. Transcontinental spread and evolution of Mycobacterium tuberculosis W148 European/Russian clade toward extensively drug resistant tuberculosis. Nat. Commun. 2022, 13, 5105. [Google Scholar] [CrossRef] [PubMed]
- Merker, M.; Barbier, M.; Cox, H.; Rasigade, J.P.; Feuerriegel, S.; Kohl, T.A.; Diel, R.; Borrell, S.; Gagneux, S.; Nikolayevskyy, V.; et al. Compensatory evolution drives multidrug-resistant tuberculosis in Central Asia. Elife 2018, 7, e38200. [Google Scholar] [CrossRef] [PubMed]
- Casali, N.; Nikolayevskyy, V.; Balabanova, Y.; Harris, S.R.; Ignatyeva, O.; Kontsevaya, I.; Corander, J.; Bryant, J.; Parkhill, J.; Nejentsev, S.; et al. Evolution and transmission of drug-resistant tuberculosis in a Russian population. Nat. Genet. 2014, 46, 279–286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pasechnik, O.; Vyazovaya, A.; Vitriv, S.; Tatarintseva, M.; Blokh, A.; Stasenko, V.; Mokrousov, I. Major genotype families and epidemic clones of Mycobacterium tuberculosis in Omsk region, Western Siberia, Russia, marked by a high burden of tuberculosis-HIV coinfection. Tuberculosis 2018, 108, 163–168. [Google Scholar] [CrossRef]
- Mokrousov, I. Mycobacterium tuberculosis phylogeography in the context of human migration and pathogen’s pathobiology: Insights from Beijing and Ural families. Tuberculosis 2015, 95 (Suppl. 1), S167–S176. [Google Scholar] [CrossRef]
- Mokrousov, I.; Vyazovaya, A.; Sinkov, V.; Gerasimova, A.; Ioannidis, P.; Jiao, W.; Khromova, P.; Papaventsis, D.; Pasechnik, O.; Perdigão, J.; et al. Practical approach to detection and surveillance of emerging highly resistant Mycobacterium tuberculosis Beijing 1071-32-cluster. Sci. Rep. 2021, 11, 21392. [Google Scholar] [CrossRef]
- European Centre for Disease Prevention and Control. Molecular Typing for Surveillance of Multidrug-Resistant Tuberculosis in the EU/EEA: Surveillance Report. Stockholm, 2017. Available online: https://www.ecdc.europa.eu/en/publications-data/molecular-typing-surveillance-multidrug-resistant-tuberculosis-eueea (accessed on 27 December 2022).
- Perdigão, J.; Silva, C.; Maltez, F.; Machado, D.; Miranda, A.; Couto, I.; Rabna, P.; de Sessions, P.F.; Phelan, J.; Pain, A.; et al. Emergence of multidrug-resistant Mycobacterium tuberculosis of the Beijing lineage in Portugal and Guinea-Bissau: A snapshot of moving clones by whole-genome sequencing. Emerg. Microbes Infect. 2020, 9, 1342–1353. [Google Scholar] [CrossRef]
- Vyazovaya, A.; Levina, K.; Zhuravlev, V.; Viiklepp, P.; Kütt, M.; Mokrousov, I. Emerging resistant clones of Mycobacterium tuberculosis in a spatiotemporal context. J. Antimicrob. Chemother. 2018, 73, 325–331. [Google Scholar] [CrossRef]
- In the 1990s, the Influx of Migrants to Russia from the Former Soviet Republics of the USSR Decreased. Available online: http://www.demoscope.ru/weekly/037/tema01.php (accessed on 11 January 2023). (In Russian).
- Migration in Post-Soviet Countries. Available online: https://russiancouncil.ru/analytics-and-comments/analytics/migratsiya-v-postsovetskikh-stranakh/ (accessed on 11 January 2023). (In Russian).
- In Central Asia, the Number of Ethnic Russians fell to 5.7 Million. Available online: https://www.asiaplustj.info/ru/news/tajikistan/20071120/v-tsentralnoi-azii-chislennost-etnicheskikh-russkikh-snizilas-do-57-mln (accessed on 11 January 2023). (In Russian).

| Region, Number of Isolates | Number of Isolates (% of All Beijing) | All Beijing | ||||
|---|---|---|---|---|---|---|
| B0/W148 | Central Asian/Russian (Other Than CAO) | Central Asian/Russian CAO | Other Modern Beijing | Ancient Beijing | ||
| Vologda, n = 82 | 6 (11.8) | 34 (66.7) | 7 (13.7) | 3 (5.9) | 1 (2.0) | 51 (62.2) |
| Kaliningrad, n = 73 | 17 (37.0) | 17 (37.0) | 4 (8.7) | 6 (13.0) | 2 (4.3) | 46 (63.0) |
| Karelia n = 67 | 13 (36.1) | 15 (41.7) | 3 (8.3) | 2 (5.6) | 3 (8.3) | 36 (53.7) |
| Komi, n = 130 | 26 (35.6) | 43 (58.9) | 2 (2.7) | 1 (1.4) | 1 (1.4) | 73 (56.2) |
| Murmansk, n = 67 | 7 (20.0) | 22 (62.9) | 1 (2.9) | 5 (14.3) | 0 | 35 (52.2) |
| Pskov, n = 78 | 8 (17.8) | 26 (57.8) | 2 (4.4) | 8 (17.8) | 1 (2.2) | 45 (57.7) |
| VNTR Locus | Locus Alias | Number of Alleles | Number of Repeats | HGI | Number of Alleles | Number of Repeats | HGI |
|---|---|---|---|---|---|---|---|
| Central Asian/Russian (n = 176) | B0/W148 (n = 77) | ||||||
| 154 | MIRU02 | 1 | 2 | 0 | 1 | 2 | 0 |
| 424 | Mtub04 | 4 | 2–5 | 0.109 | 2 | 2.4 | 0.026 |
| 577 | ETRC | 3 | 2.4.5 | 0.067 | 1 | 4 | 0 |
| 580 | MIRU04 | 1 | 2 | 0 | 1 | 2 | 0 |
| 802 | MIRU40 | 3 | 2–4 | 0.078 | 2 | 3–4 | 0.026 |
| 960 | MIRU10 | 2 | 2.3 | 0.011 | 1 | 3 | 0 |
| 1644 | MIRU16 | 1 | 3 | 0 | 1 | 3 | 0 |
| 1955 | Mtub21 | 5 | 1.3–6 | 0.120 | 2 | 4–5 | 0.051 |
| 2059 | MIRU20 | 1 | 2 | 0 | 1 | 2 | 0 |
| 2165 | ETRA | 4 | 1–4 | 0.224 | 1 | 4 | 0 |
| 2347 | Mtub29 | 3 | 2–4 | 0.077 | 1 | 4 | 0 |
| 2401 | Mtub30 | 2 | 2.4 | 0.034 | 2 | 2.4 | 0.026 |
| 2461 | ETRB | 1 | 2 | 0 | 1 | 2 | 0 |
| 2531 | MIRU23 | 1 | 5 | 0 | 1 | 5 | 0 |
| 2687 | MIRU24 | 1 | 1 | 0 | 1 | 1 | 0 |
| 2996 | MIRU26 | 4 | 4–6.9 | 0.161 | 3 | 4–6.7 | 0.077 |
| 3007 | MIRU27 | 1 | 3 | 0 | 1 | 3 | 0 |
| 3171 | Mtub34 | 2 | 2.3 | 0.011 | 1 | 3 | 0 |
| 3192 | MIRU31 | 3 | 4–6 | 0.149 | 2 | 4–5 | 0.123 |
| 3690 | Mtub39 | 2 | 3–4 | 0.011 | 3 | 2–4 | 0.052 |
| 4052 | QUB26 | 5 | 5–9 | 0.382 | 3 | 2.3.7 | 0.052 |
| 4156 | QUB4156 | 1 | 2 | 0 | 1 | 2 | 0 |
| 4348 | MIRU39 | 2 | 2.3 | 0.011 | 1 | 3 | 0 |
| 2163b | QUB11b | 5 | 2.5–8 | 0.170 | 2 | 5–6 | 0.076 |
| Beijing Subgroups | 24 Loci MIRU-VNTR Profile | MLVA MtbC 15-9 | 24-loci MIT | CC * | Number of Isolates |
|---|---|---|---|---|---|
| Central Asian/Russian n = 176 | 223325153533324682454433 | 9358-32 | 32 | CC1 | 2 |
| 223325153533424682444433 | 801-32 | Orphan | CC1 | 2 | |
| 223325153533422682454433 | 9357-32 | - | CC1 | 2 | |
| 223325153633422682454433 | 9356-32 | 71 | CC1 | 2 | |
| 223325153533424582454433 | 407-32 | - | CC4 * | 2 | |
| 223325153534424582454433 | 770-32 | - | CC4 * | 2 | |
| 223325153433424682254433 | ?-32 | - | - | 2 | |
| 223325143533424682454433 | 1068-32 | - | CC3 * | 3 | |
| 223325153533424682464433 | 96-32 | - | CC1 | 3 | |
| 223325153533424672452433 | 99-332 | - | CC1 | 4 | |
| 223325163533424682454433 | 1076-32 | Orphan | CC3 * | 5 | |
| 223325153533424672454433 | 99-32 | - | CC1 | 6 | |
| 223325153533324682454433 | 95-32 | 32 | CC1 | 12 | |
| 223325153533424662454433 | 1065-32 | 28 | CC1 | 17 | |
| 223325153533424682454433 | 94-32 | 25 | CC1 | 75 | |
| B0/W148 n = 77 | 223325173533424672444433 | 9378-32 | 35 | CC2 | 2 |
| 223325163533424672454433 | 1075-32 | - | CC2 | 2 | |
| 223325173433424672454433 | 4737-32 | Orphan | CC2 | 5 | |
| 223325173533424672454433 | 100-32 | 27 | CC2 | 59 | |
| Other modern Beijing, n = 25 | 244233352544425173353923 | 9380-32 | - | CC4 * | 2 |
| 244233352644425173353623 | 1066-32 | - | CC3 * | 2 | |
| 244233352644425173353823 | 1048-32 | - | CC3 * | 2 | |
| 244234352644425173353823 | 9391-32 | - | CC3 * | 9 | |
| Ancient Beijing, n = 8 | 244231342644425173353923 | 1071-32 | - | BL7 | 6 |
| Region | Number of Isolates | Number of VNTR Types | Number of Clusters | Cluster Size | Number of Clustered Isolates (%) | CR | HGI | Shannon Index |
|---|---|---|---|---|---|---|---|---|
| Vologda | 51 | 22 | 5 | 2–18 | 34 (66.7) | 0.57 | 0.852 | 2.48 |
| Kaliningrad | 46 | 24 | 6 | 2–13 | 28 (60.9) | 0.48 | 0.904 | 2.65 |
| Karelia | 36 | 18 | 3 | 3–9 | 21 (58.3) | 0.50 | 0.881 | 2.55 |
| Komi | 73 | 23 | 8 | 2–21 | 58 (79.5) | 0.68 | 0.854 | 2.28 |
| Murmansk | 35 | 13 | 4 | 2–15 | 26 (74.3) | 0.63 | 0.790 | 2.04 |
| Pskov | 45 | 23 | 6 | 2–10 | 28 (62.2) | 0.49 | 0.921 | 2.62 |
| Drug Resistance | Number of Isolates and % of All of This Subtype | p | ||||
|---|---|---|---|---|---|---|
| B0/W148, n = 77 | Central Asian/Russian (Other Than CAO), n = 157 | Central Asian/Russian CAO, n = 19 | Other Modern Beijing, n = 25 | Ancient Beijing, n = 8 | ||
| Susceptible. n = 69 | – | 67 (42.6) | – | 14 (56.0) | 2 (25) | <0.001 |
| Monoresistant/polyresistant n = 49 | 11 (14.3) | 30 (19.1) | 8 (42.1) | 6 (24.0) | – | |
| MDR. n = 143 | 66 (85.7) | 60 (38.2) | 11 (57.9) | 5 (20.0) | 6 (75) | |
| Beijing Clade | 24-MIRU-VNTR Type | Number of Strains and % of All in This Type | p | ||
|---|---|---|---|---|---|
| Susceptible | Monoresistant/Polyresistant | MDR | |||
| B0/W148 | 100-32 | 10 (16.9) | 49 (83.1) | p 100-32 vs. 1076-32 = 0.003 p 100-32 vs. 9391-32 < 0.001 p 100-32 vs. 94-32 < 0.001 p 100-32 vs. 95-32 = 0.008 p 100-32 vs. 99-32 < 0.001 p 1065-32 vs. 9391-32 < 0.001 p 1065-32 vs. 94-32 = 0.048 p 1065-32 vs. 99-32 = 0.010 p 1071-32 vs. 99-32 = 0.026 p 94-32 vs. 99-32 = 0.039 | |
| B0/W148 | 4737-32 | 5 (100) | |||
| C.A.R. | 94-32 | 33 (44.0) | 9 (12.0) | 33 (44.0) | |
| C.A.R. | 1065-32 | 2 (11.8) | 15 (88.2) | ||
| C.A.R. | 95-32 | 3 (25.0) | 3 (25.0) | 6 (50.0) | |
| C.A.R. | 99-32 | 2 (33.3) | 4 (66.7) | ||
| C.A.R. | 1076-32 | 1 (20.0) | 3 (60.0) | 1 (20.0) | |
| other | 9391-32 | 9 (100) | |||
| Ancient | 1071-32 | 6 (100) | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vyazovaya, A.; Gerasimova, A.; Mudarisova, R.; Terentieva, D.; Solovieva, N.; Zhuravlev, V.; Mokrousov, I. Genetic Diversity and Primary Drug Resistance of Mycobacterium tuberculosis Beijing Genotype Strains in Northwestern Russia. Microorganisms 2023, 11, 255. https://doi.org/10.3390/microorganisms11020255
Vyazovaya A, Gerasimova A, Mudarisova R, Terentieva D, Solovieva N, Zhuravlev V, Mokrousov I. Genetic Diversity and Primary Drug Resistance of Mycobacterium tuberculosis Beijing Genotype Strains in Northwestern Russia. Microorganisms. 2023; 11(2):255. https://doi.org/10.3390/microorganisms11020255
Chicago/Turabian StyleVyazovaya, Anna, Alena Gerasimova, Regina Mudarisova, Daria Terentieva, Natalia Solovieva, Viacheslav Zhuravlev, and Igor Mokrousov. 2023. "Genetic Diversity and Primary Drug Resistance of Mycobacterium tuberculosis Beijing Genotype Strains in Northwestern Russia" Microorganisms 11, no. 2: 255. https://doi.org/10.3390/microorganisms11020255
APA StyleVyazovaya, A., Gerasimova, A., Mudarisova, R., Terentieva, D., Solovieva, N., Zhuravlev, V., & Mokrousov, I. (2023). Genetic Diversity and Primary Drug Resistance of Mycobacterium tuberculosis Beijing Genotype Strains in Northwestern Russia. Microorganisms, 11(2), 255. https://doi.org/10.3390/microorganisms11020255





