Responses of Free-Living Planktonic Bacterial Communities to Experimental Acidification and Warming
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Setup and Sampling
2.2. pCO2 and Temperature Determination
2.3. DNA Extraction, Library Preparation, Sequencing and Sequence Processing
2.4. Determination of the Concentrations of Nitrate, Ammonium, Phosphate and Chlorophyll a
2.5. Determination of Bacterial Abundances
2.6. Statistics
3. Results and Discussion
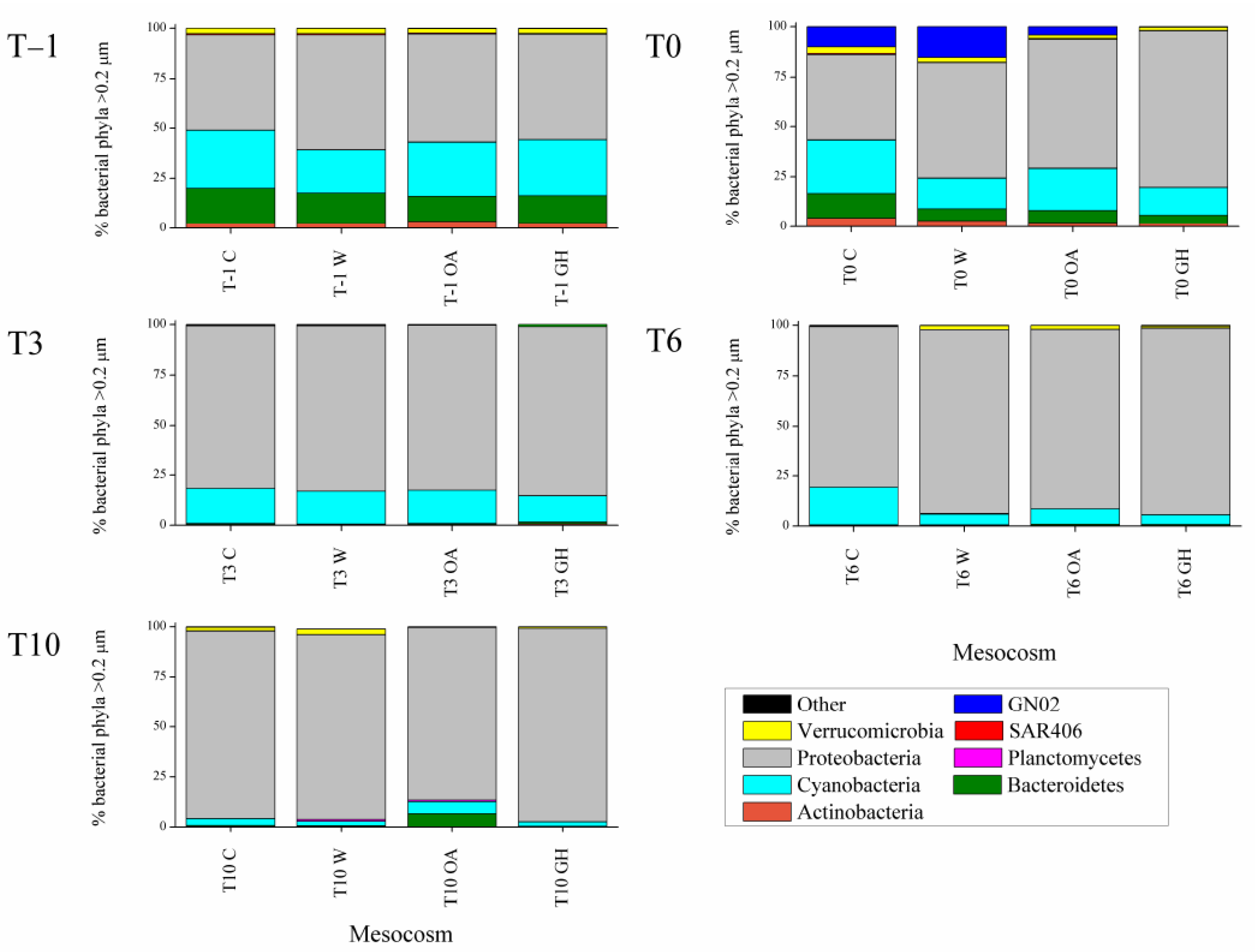
3.1. Immediate Treatment Effects at the >0.2 μm Bacterial Fraction (between T–1 and T0)
3.2. Short-Term Treatment Effects at the >0.2 μm Bacterial Fraction (between T3 and T10)
3.3. Short-Term Treatment Effects at the 0.1–0.2 μm Bacterial Fraction (between T3 and T10)

4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Halpern, B.S.; Frazier, M.; Potapenko, J.; Casey, K.S.; Koenig, K.; Longo, C.; Lowndes, J.S.; Rockwood, R.C.; Selig, E.R.; Selkoe, K.A.; et al. Spatial and Temporal Changes in Cumulative Human Impacts on the World’s Ocean. Nat. Commun. 2015, 6, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Cubash, U.; Wuebebles, D.; Chen, D.; Facchini, M.; Frame, D.; Mahowald, N.; Winther, J.-G. Climate Change 2013: The Physical Science Basis. Contribution of Working Group I to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change; Cambridge University Press: Cambridge, UK; New York, NY, USA, 2013. [Google Scholar]
- Gattuso, J.P.; Magnan, A.; Billé, R.; Cheung, W.W.L.; Howes, E.L.; Joos, F.; Allemand, D.; Bopp, L.; Cooley, S.R.; Eakin, C.M.; et al. Contrasting Futures for Ocean and Society from Different Anthropogenic CO2emissions Scenarios. Science 2015, 349. [Google Scholar] [CrossRef]
- IPCC. Climate Change 2013: The Physical Science Basis. Contribution of Working Group I to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change; Stocker, T.F., Qin, D., Plattner, G.K., Tignor, M.M.B., Allen, S.K., Boschung, J., Nauels, A., Xia, Y., Bex, V., Midgley, P.M., Eds.; Cambridge University Press: Cambridge, UK; New York, NY, USA, 2013; p. 1535. [Google Scholar] [CrossRef] [Green Version]
- Rasconi, S.; Gall, A.; Winter, K.; Kainz, M.J. Increasing Water Temperature Triggers Dominance of Small Freshwater Plankton. PLoS ONE 2015, 10, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Morán, X.A.G.; López-Urrutia, Á.; Calvo-Díaz, A.; LI, W.K.W. Increasing Importance of Small Phytoplankton in a Warmer Ocean. Glob. Chang. Biol. 2010, 16, 1137–1144. [Google Scholar] [CrossRef]
- Paul, C.; Matthiessen, B.; Sommer, U. Warming, but Not Enhanced CO2concentration, Quantitatively and Qualitatively Affects Phytoplankton Biomass. Mar. Ecol. Prog. Ser. 2015, 528, 39–51. [Google Scholar] [CrossRef] [Green Version]
- Tortell, P.D.; Payne, C.D.; Li, Y.; Trimborn, S.; Rost, B.; Smith, W.O.; Riesselman, C.; Dunbar, R.B.; Sedwick, P.; DiTullio, G.R. CO2 Sensitivity of Southern Ocean Phytoplankton. Geophys. Res. Lett. 2008, 35, 1–5. [Google Scholar] [CrossRef] [Green Version]
- Feng, Y.; Hare, C.E.; Leblanc, K.; Rose, J.M.; Zhang, Y.; DiTullio, G.R.; Lee, P.A.; Wilhelm, S.W.; Rowe, J.M.; Sun, J.; et al. Effects of Increased PCO2 and Temperature on the North Atlantic Spring Bloom. I. The Phytoplankton Community and Biogeochemical Response. Mar. Ecol. Prog. Ser. 2009, 388, 13–25. [Google Scholar] [CrossRef]
- Engel, A. Direct Relationship between CO2 Uptake and Transparent Exopolymer Particles Production in Natural Phytoplankton. J. Plankton Res. 2002, 24, 49–53. [Google Scholar] [CrossRef] [Green Version]
- Paulino, A.I.; Egge, J.K.; Larsen, A. Effects of Increased Atmospheric CO2 on Small and Intermediate Sized Osmotrophs during a Nutrient Induced Phytoplankton Bloom. Biogeosciences Discuss. 2007, 4, 4173–4195. [Google Scholar] [CrossRef]
- Newbold, L.K.; Oliver, A.E.; Booth, T.; Tiwari, B.; Desantis, T.; Maguire, M.; Andersen, G.; van der Gast, C.J.; Whiteley, A.S. The Response of Marine Picoplankton to Ocean Acidification. Environ. Microbiol. 2012, 14, 2293–2307. [Google Scholar] [CrossRef] [Green Version]
- Schulz, K.G.; Bellerby, R.G.J.; Brussaard, C.P.D.; Büdenbender, J.; Czerny, J.; Engel, a.; Fischer, M.; Koch-Klavsen, S.; Krug, S.A.; Lischka, S.; et al. Temporal Biomass Dynamics of an Arctic Plankton Bloom in Response to Increasing Levels of Atmospheric Carbon Dioxide. Biogeosciences 2013, 10, 161–180. [Google Scholar] [CrossRef] [Green Version]
- Taucher, J.; Boxhammer, T.; Bach, L.T.; Paul, A.J.; Schartau, M.; Stange, P.; Riebesell, U. Changing Carbon-to-Nitrogen Ratios of Organic-Matter Export under Ocean Acidification. Nat. Clim. Chang. 2021, 11, 52–57. [Google Scholar] [CrossRef]
- Grossart, H.P.; Allgaier, M.; Passow, U.; Riebesell, U. Testing the Effect of CO2 Concentration on the Dynamics of Marine Heterotrophic Bacterioplankton. Limnol. Oceanogr. 2006, 51, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Piontek, J.; Borchard, C.; Sperling, M.; Schulz, K.G.; Riebesell, U.; Engel, A. Response of Bacterioplankton Activity in an Arctic Fjord System to Elevated PCO2: Results from a Mesocosm Perturbation Study. Biogeosciences 2013, 10, 297–314. [Google Scholar] [CrossRef] [Green Version]
- Celussi, M.; Malfatti, F.; Franzo, A.; Gazeau, F.; Giannakourou, A.; Pitta, P.; Tsiola, A.; Del Negro, P. Ocean Acidification Effect on Prokaryotic Metabolism Tested in Two Diverse Trophic Regimes in the Mediterranean Sea. Estuar. Coast. Shelf Sci. 2015, 186, 125–138. [Google Scholar] [CrossRef]
- Tsiola, A.; Pitta, P.; Giannakourou, A.; Bourdin, G.; Marro, S.; Maugendre, L.; Pedrotti, M.L.; Gazeau, F. Ocean Acidification and Viral Replication Cycles: Frequency of Lytically Infected and Lysogenic Cells during a Mesocosm Experiment in the NW Mediterranean Sea. Estuar. Coast. Shelf Sci. 2017, 186, 139–151. [Google Scholar] [CrossRef]
- Cornwall, C.E. Hurd Experimental Design in Ocean Acidification Research: Problems and Solutions. ICES J. Mar. Sci. 2016, 73, 572–581. [Google Scholar] [CrossRef]
- Dutkiewicz, S.; Morris, J.J.; Follows, M.J.; Scott, J.; Levitan, O.; Dyhrman, S.T.; Berman-Frank, I. Impact of Ocean Acidification on the Structure of Future Phytoplankton Communities. Nat. Clim. Chang. 2015, 5, 1002–1006. [Google Scholar] [CrossRef]
- Sala, M.M.; Aparicio, F.L.; Balague, V.; Boras, J.A.; Borrull, E.; Cardelus, C.; Cros, L.; Gomes, A.; Lopez-Sanz, A.; Malits, A.; et al. Contrasting Effects of Ocean Acidification on the Microbial Food Web under Different Trophic Conditions. ICES J. Mar. Sci. 2016, 73, 670–679. [Google Scholar] [CrossRef]
- Maugendre, L.; Gattuso, J.P.; Poulton, A.J.; Dellisanti, W.; Gaubert, M.; Guieu, C.; Gazeau, F. No Detectable Effect of Ocean Acidification on Plankton Metabolism in the NW Oligotrophic Mediterranean Sea: Results from Two Mesocosm Studies. Estuar. Coast. Shelf Sci. 2017, 186, 89–99. [Google Scholar] [CrossRef] [Green Version]
- Mousing, E.A.; Ellegaard, M.; Richardson, K. Global Patterns in Phytoplankton Community Size Structure-Evidence for a Direct Temperature Effect. Mar. Ecol. Prog. Ser. 2014, 497, 25–38. [Google Scholar] [CrossRef] [Green Version]
- Brownman, H.I. Applying Organized Scepticism to Ocean Acidification Research. ICES J. Mar. Sci. 2016, 730, 529–536. [Google Scholar] [CrossRef] [Green Version]
- Fu, F.-X.; Warner, M.E.; Zhang, Y.; Feng, Y.; Hutchins, D.A. Effects of Increased Temperature and CO2 on Photosyntheis, Growth and Elemental Ratios in Marine Synechococcus and Prochlorococcus (Cyanobacteria). J. Phycol. 2007, 43, 485–496. [Google Scholar] [CrossRef]
- Basu, S.; Mackey, K.R.M. Effect of Rising Temperature and Carbon Dioxide on the Growth, Photophysiology, and Elemental Ratios of Marine Synechococcus: A Multistressor Approach. Sustainability 2022, 14, 9508. [Google Scholar] [CrossRef]
- Lindh, M.V.; Riemann, L.; Baltar, F.; Romero-Oliva, C.; Salomon, P.S.; Granéli, E.; Pinhassi, J. Consequences of Increased Temperature and Acidification on Bacterioplankton Community Composition during a Mesocosm Spring Bloom in the Baltic Sea. Environ. Microbiol. Rep. 2013, 5, 252–262. [Google Scholar] [CrossRef]
- Sommer, U.; Paul, C.; Moustaka-Gouni, M. Warming and Ocean Acidification Effects on Phytoplankton - From Species Shifts to Size Shifts within Species in a Mesocosm Experiment. PLoS ONE 2015, 10, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Hyun, B.; Kim, J.M.; Jang, P.G.; Jang, M.C.; Choi, K.H.; Lee, K.; Yang, E.J.; Noh, J.H.; Shin, K. The Effects of Ocean Acidification and Warming on Growth of a Natural Community of Coastal Phytoplankton. J. Mar. Sci. Eng. 2020, 8, 821. [Google Scholar] [CrossRef]
- Bergen, B.; Endres, S.; Engel, A.; Zark, M.; Dittmar, T. Acidification and Warming Affect Prominent Bacteria in Two Seasonal Phytoplankton Bloom Mesocosms. Env. Microbiol 2016, 18, 4579–4595. [Google Scholar] [CrossRef]
- Krom, M.D.; Kress, N.; Brenner, S.; Gordon, L.I. Phosphorus Limitation of Primary Productivity in the Eastern Mediterranean Sea. Limnol. Oceanogr. 1991, 36, 424–432. [Google Scholar] [CrossRef]
- Polovina, J.J.; Howell, E.A.; Abecassis, M. Ocean’s Least Productive Waters Are Expanding. Geophys. Res. Lett. 2008, 35, 2–6. [Google Scholar] [CrossRef] [Green Version]
- Bunse, C.; Lundin, D.; Karlsson, C.M.G.; Vila-Costa, M.; Palovaara, J.; Akram, N.; Svensson, L.; Holmfeldt, K.; González, J.M.; Calvo, E.; et al. Response of Marine Bacterioplankton PH Homeostasis Gene Expression to Elevated CO2. Nat. Clim. Chang. 2016, 6, 483–487. [Google Scholar] [CrossRef]
- Bach, L.T.; Alvarez-Fernandez, S.; Hornick, T.; Stuhr, A.; Riebesell, U. Simulated Ocean Acidification Reveals Winners and Losers in Coastal Phytoplankton. PLoS ONE 2017, 12, 1–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palmieri, J.; Orr, J.C.; Dutay, J.-C.; Beranger, K.; Schneider, A.; Beuvier, J.; Somot, S. Simulated Anthropogenic CO 2 Storage and Acidification of The. Biogeosciences 2015, 12, 781–802. [Google Scholar] [CrossRef] [Green Version]
- Goyet, C.; El, A.; Hassoun, R.; Gemayel, E.; Touratier, F. Thermodynamic Forecasts of the Mediterranean Sea Acidification Thermodynamic Forecasts of the Mediterranean Sea Acidification. Mediterr. Mar. Sci. 2016, 17, 508–518. [Google Scholar] [CrossRef] [Green Version]
- Polimene, L.; Sailley, S.; Clark, D.; Mitra, A.; Allen, J.I. Biological or Microbial Carbon Pump? The Role of Phytoplankton Stoichiometry in Ocean Carbon Sequestration. J. Plankton Res. 2017, 39, 180–186. [Google Scholar] [CrossRef] [Green Version]
- Cavicchioli, R.; Ripple, W.J.; Timmis, K.N.; Azam, F.; Bakken, L.R.; Baylis, M.; Behrenfeld, M.J.; Boetius, A.; Boyd, P.W.; Classen, A.T.; et al. Scientists’ Warning to Humanity: Microorganisms and Climate Change. Nat. Rev. Microbiol. 2019, 17, 569–586. [Google Scholar] [CrossRef] [Green Version]
- D’Amario, B.; Pérez, C.; Grelaud, M.; Pitta, P.; Krasakopoulou, E.; Ziveri, P. Coccolithophore Community Response to Ocean Acidification and Warming in the Eastern Mediterranean Sea: Results from a Mesocosm Experiment. Sci. Rep. 2020, 10, 1–14. [Google Scholar] [CrossRef]
- Gazeau, F.; Sallon, A.; Maugendre, L.; Louis, J.; Dellisanti, W.; Gaubert, M.; Lejeune, P.; Gobert, S.; Borges, A.V.; Harlay, J.; et al. First Mesocosm Experiments to Study the Impacts of Ocean Acidification on Plankton Communities in the NW Mediterranean Sea (MedSeA Project). Estuar. Coast. Shelf Sci. 2017, 186, 11–29. [Google Scholar] [CrossRef] [Green Version]
- Dickson, A.G.; Sabine, C.L.; Christian, J.R. Guide to Best Practices for Ocean CO2 Measurements; North Pacific Marine Sci-ence Organization: Sidney, Australia, 2007; ISBN 1897176074. [Google Scholar]
- Gattuso, J.-P.P.; Epitalon, J.M.; Lavigne, H. Seacarb: Seawater Carbonate Chemistry. R Package Version 3.3.0. 2022. Available online: https://CRAN.R-project.org/package=seacarb (accessed on 8 November 2022).
- Orr, J.C.; Epitalon, J.M.; Dickson, A.G.; Gattuso, J.P. Routine Uncertainty Propagation for the Marine Carbon Dioxide System. Mar. Chem. 2018, 207, 84–107. [Google Scholar] [CrossRef]
- Winnepenninckx, B.T.; Backeljau, T.; Wachter, R. De Extraction of High Molecular Weight DNA from Molluscs. Trends Genet. 1993, 9, 407. [Google Scholar] [CrossRef]
- Tsiola, A.; Toncelli, C.; Fodelianakis, S.; Michoud, G.; Bucheli, T.D.; Gavriilidou, A.; Kagiorgi, M.; Kalantzi, I.; Knauer, K.; Kotoulas, G.; et al. Low-Dose Addition of Silver Nanoparticles Stresses Marine Plankton Communities. Environ. Sci. Nano 2018, 5, 1965–1980. [Google Scholar] [CrossRef]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of General 16S Ribosomal RNA Gene PCR Primers for Classical and Next-Generation Sequencing-Based Diversity Studies. Nucleic Acids Res. 2013, 41, 1–11. [Google Scholar] [CrossRef]
- Apprill, A.; McNally, S.; Parsons, R.; Weber, L. Minor Revision to V4 Region SSU RRNA 806R Gene Primer Greatly Increases Detection of SAR11 Bacterioplankton. Aquat. Microb. Ecol. 2015, 75, 129–137. [Google Scholar] [CrossRef] [Green Version]
- Rimmelin, P.; Moutin, T. Re-Examination of the MAGIC Method to Determine Low Orthophosphate Concentration in Seawater. Anal. Chim. Acta 2005, 548, 174–182. [Google Scholar] [CrossRef]
- Strickland, J.; Parsons, T. Determination of Phosphorus. In A Practical Handbook of Sewater Analysis; Fisheries Research Board of Canada: Ottawa, QC, Canada, 1972; Volume Fisheries, p. 167. [Google Scholar]
- Ivancic, I.; Degobbis, D. An Optimal Manual Procedure for Ammonia Analysis in Natural Waters by the Indophenol Blue Method. Water Res. 1984, 18, 1143–1147. [Google Scholar] [CrossRef]
- Yentsch, C.S.; Menzel, D.W. A Method for the Determination of Phytoplankton Chlorophyll and Phaeophytin by Fluorescence. Deep. Res. Part I Oceanogr. Res. Pap. 1963, 10, 221–231. [Google Scholar] [CrossRef]
- Marie, D.; Partensky, F.; Jacquet, S.; Vaulot, D. Enumeration and Cell Cycle Analysis of Natural Populations of Marine Picoplankton by Flow Cytometry Using the Nucleic Acid Stain SYBR Green I. Appl. Environ. Microbiol. 1997, 63, 186–193. [Google Scholar] [CrossRef] [Green Version]
- Clarke, K.R.; Ainsworth, M. A Method of Linking Multivaritate Community Structure to Environmental Variable. Mar. Ecol. Prog. Ser. 1993, 92, 205–219. [Google Scholar] [CrossRef]
- Clarke, K.R.; Warwick, R.M. Change in Marine Communities: An Approach to Statistical Analysis and Interpretation; Primer-E Ltd: Plymouth, UK, 1994. [Google Scholar]
- Roy, A.-S.; Gibbons, S.M.; Schunck, H.; Owens, S.; Caporaso, J.G.; Sperling, M.; Nissimov, J.I.; Romac, S.; Bittner, L.; Riebesell, U.; et al. Ocean Acidification Shows Negligible Impacts on High-Latitude Bacterial Community Structure in Coastal Pelagic Mesocosms. Biogeosciences Discuss. 2012, 9, 13319–13349. [Google Scholar] [CrossRef]
- Zhang, R.; Xia, X.; Lau, S.C.K.; Motegi, C.; Weinbauer, M.G.; Jiao, N. Response of Bacterioplankton Community Structure to an Artificial Gradient of PCO2 in the Arctic Ocean. Biogeosciences Discuss. 2013, 10, 3679–3689. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Zhang, R.; Zheng, Q.; Deng, Y.; van Nostrand, J.D.; Zhou, J.; Jiao, N. Bacterioplankton Community Resilience to Ocean Acidificaiton: Evidence from Microbial Network Analysis. ICES J. Mar. Sci. 2016, 73, 865–875. [Google Scholar] [CrossRef]
- Lin, X.; Huang, R.; Li, Y.; Li, F.; Wu, Y.; Hutchins, D.A.; Dai, M.; Gao, K. Interactive Network Configuration Maintains Bacterioplankton Community Structure under Elevated CO2 in a Eutrophic Coastal Mesocosm Experiment. Biogeosciences 2018, 15, 551–565. [Google Scholar] [CrossRef] [Green Version]
- Aguayo, P.; Campos, V.L.; Henríquez, C.; Olivares, F.; De Ia Iglesia, R.; Ulloa, O.; Vargas, C.A. The Influence of PCO2-Driven Ocean Acidification on Open Ocean Bacterial Communities during a Short-Term Microcosm Experiment in the Eastern Tropical South Pacific (ETSP) off Northern Chile. Microorganisms 2020, 8, 1924. [Google Scholar] [CrossRef] [PubMed]
- Baltar, F.; Palovaara, J.; Vila-Costa, M.; Salazar, G.; Calvo, E.; Pelejero, C.; Marrasé, C.; Gasol, J.M.; Pinhassil, J. Response of Rare, Common and Abundant Bacterioplankton to Anthropogenic Perturbations in a Mediterranean Coastal Site. FEMS Microbiol. Ecol. 2015, 91, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Weels, S.S.L.; Welz, P.J.; Prins, A.; Le Roes-Hill, M. Impact of Physicochemical Parameters on the Diversity and Distribution of Microbial Communities Associated with Three South African Peatlands. Microorganisms 2022, 10, 2103. [Google Scholar] [CrossRef]
- Brown, M.V.; Lauro, F.M.; Demaere, M.Z.; Muir, L.; Wilkins, D.; Thomas, T.; Riddle, M.J.; Fuhrman, J.A.; Andrews-Pfannkoch, C.; Hoffman, J.M.; et al. Global Biogeography of SAR11 Marine Bacteria. Mol. Syst. Biol. 2012, 8, 1–13. [Google Scholar] [CrossRef]
- Sharma, A.K.; Becker, J.W.; Ottesen, E.A.; Bryant, J.A.; Duhamel, S.; Karl, D.M.; Cordero, O.X.; Repeta, D.J.; Delong, E.F. Distinct Dissolved Organic Matter Sources Induce Rapid Transcriptional Responses in Coexisting Populations of Prochlorococcus, Pelagibacter and the OM60 Clade. Environ. Microbiol. 2014, 16, 2815–2830. [Google Scholar] [CrossRef] [Green Version]
- Herlemann, D.P.R.; Woelk, J.; Labrenz, M.; Jürgens, K. Diversity and Abundance of “Pelagibacterales” (SAR11) in the Baltic Sea Salinity Gradient. Syst. Appl. Microbiol. 2014, 37, 601–604. [Google Scholar] [CrossRef] [Green Version]
- Hartmann, M.; Hill, P.G.; Tynan, E.; Achterberg, E.P.; Leakey, R.J.G.; Zubkov, M.V. Resilience of SAR11 Bacteria to Rapid Acidification in the High-Latitude Open Ocean. FEMS Microbiol. Ecol. 2016, 92, 1–10. [Google Scholar] [CrossRef]
- Sieber, C.M.K.; Paul, B.G.; Castelle, C.J.; Hu, P.; Tringe, S.G.; Valentine, D.L.; Andersen, G.L.; Banfield, F. Unusual Metabolism and Hypervariation in the Genome of a Gracilibacterium (BD1-5) from an Oil-Degrading Community. MBio 2019, 10, e02128-19. [Google Scholar] [CrossRef] [Green Version]
- Monier, A.; Findlay, H.S.; Charvet, S.; Lovejoy, C.; Steen, I.H. Late Winter under Ice Pelagic Microbial Communities in the High Arctic Ocean and the Impact of Short-Term Exposure to Elevated CO2 Levels. Front. Microbiol. 2014, 5, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Obayashi, Y.; Suzuki, S. High Growth Potential and Activity of 0.2 Μm Filterable Bacteria Habitually Present in Coastal Seawater. Biogeosciences Discuss 2017. [Google Scholar] [CrossRef]


| Mesocosm | Temperature | pCO2 | pH | NO3– | NH4+ | PO43– | Chl | Shannon Index |
|---|---|---|---|---|---|---|---|---|
| T–1 C1 | 25.10 | nd | 8.11 | 0.13 | 0.10 | 1.9 | 0.04 | 3.5 |
| T–1 C2 | 25.10 | 369 | 8.11 | 0.12 | 0.05 | 1.6 | 0.04 | 3.6 |
| T–1 W1 | 27.06 | nd | 8.11 | 0.06 | 0.28 | 3.1 | 0.05 | 3.5 |
| T–1 W2 | 27.06 | 364 | 8.11 | 0.04 | 0.14 | 4.1 | 0.04 | 3.5 |
| T–1 OA1 | 25.12 | 371 | 8.11 | 0.05 | 0.04 | 1.1 | 0.03 | 3.8 |
| T–1 OA2 | 25.12 | 366 | 8.11 | 0.08 | 0.08 | 1.6 | 0.04 | 3.6 |
| T–1 GH1 | 27.04 | 362 | 8.11 | 0.08 | 0.16 | 7.5 | 0.05 | 3.5 |
| T–1 GH2 | 27.04 | 371 | 8.10 | 0.48 | 0.15 | 6.2 | 0.05 | 3.6 |
| T0 C1 | 25.03 | 418 | 8.07 | 0.13 | 0.02 | 5.0 | 0.05 | 3.6 |
| T0 C2 | 25.03 | 403 | 8.08 | 0.14 | 0.03 | 9.9 | 0.06 | 3.6 |
| T0 W1 | 27.46 | 425 | 8.06 | 0.14 | 0.04 | 4.6 | 0.05 | 3.3 |
| T0 W2 | 27.40 | 420 | 8.06 | 0.11 | 0.02 | 5.3 | 0.05 | 3.4 |
| T0 OA1 | 25.02 | 549 | 7.97 | 0.03 | 0.011 | 3.9 | 0.06 | 3.4 |
| T0 OA2 | 25.02 | 533 | 7.98 | 0.12 | 0.06 | 3.8 | 0.06 | 3.5 |
| T0 GH1 | 27.35 | 612 | 7.93 | 0.07 | 0.04 | 4.9 | 0.06 | 3.5 |
| T0 GH2 | 27.34 | 611 | 7.93 | 0.18 | 0.05 | 7.3 | 0.06 | 3.4 |
| T3 C1 | 25.04 | 416 | 8.07 | 0.01 | 0.08 | 1.8 | 0.07 | 3.6 |
| T3 C2 | 25.01 | 420 | 8.07 | 0.13 | 0.06 | 3.1 | 0.07 | 3.3 |
| T3 W1 | 28.27 | 397 | 8.08 | 0.10 | 0.03 | 4.1 | 0.07 | 3.4 |
| T3 W2 | 28.25 | 411 | 8.07 | 0.03 | 0.04 | 3.5 | 0.07 | 3.3 |
| T3 OA1 | 25.05 | 794 | 7.84 | 0.07 | 0.06 | 3.1 | 0.09 | 2.9 |
| T3 OA2 | 25.02 | 768 | 7.85 | 0.06 | 0.04 | 3.0 | 0.10 | 3.1 |
| T3 GH1 | 28.24 | 803 | 7.83 | 0.05 | 0.00 | 5.6 | 0.10 | 3.5 |
| T3 GH2 | 28.25 | 872 | 7.80 | 0.06 | 0.00 | 5.5 | 0.10 | 3.5 |
| T6 C1 | 25.16 | 451 | 8.04 | 0.02 | 0.03 | 2.6 | 0.04 | 3.4 |
| T6 C2 | 25.09 | 447 | 8.04 | 0.02 | 0.03 | 1.8 | 0.05 | 3.5 |
| T6 W1 | 28.41 | 442 | 8.05 | 0.06 | 0.01 | 2.6 | 0.04 | 3.6 |
| T6 W2 | 28.32 | 476 | 8.02 | 0.06 | 0.00 | 2.1 | 0.04 | 3.7 |
| T6 OA1 | 25.27 | 805 | 7.83 | 0.02 | 0.01 | 3.0 | 0.05 | 3.6 |
| T6 OA2 | 25.02 | 816 | 7.83 | 0.03 | 0.02 | 1.9 | 0.05 | nd |
| T6 GH1 | 28.26 | 874 | 7.80 | 0.05 | 0.00 | 1.6 | 0.04 | 3.9 |
| T6 GH2 | 28.41 | 887 | 7.80 | 0.04 | 0.00 | 2.4 | 0.04 | 3.5 |
| T10 C1 | 25.26 | 438 | 8.05 | 0.02 | 0.00 | 4.4 | 0.04 | 3.8 |
| T10 C2 | 25.23 | 442 | 8.05 | 0.06 | 0.00 | 2.6 | 0.05 | 3.4 |
| T10 W1 | 27.86 | 437 | 8.05 | 0.05 | 0.00 | 3.6 | 0.05 | 3.3 |
| T10 W2 | 27.92 | 456 | 8.04 | 0.00 | 0.00 | 3.9 | 0.04 | 3.8 |
| T10 OA1 | 25.29 | 785 | 7.84 | 0.02 | 0.00 | 3.4 | 0.05 | 3.7 |
| T10 OA2 | 25.22 | 793 | 7.84 | 0.03 | 0.00 | 2.6 | 0.06 | nd |
| T10 GH1 | 27.90 | 812 | 7.83 | 0.03 | 0.00 | 5.5 | 0.06 | 3.5 |
| T10 GH2 | 27.96 | 816 | 7.83 | 0.05 | 0.00 | 4.4 | 0.07 | 3.2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tsiola, A.; Krasakopoulou, E.; Daffonchio, D.; Frangoulis, C.; Tsagaraki, T.M.; Fodelianakis, S.; Pitta, P. Responses of Free-Living Planktonic Bacterial Communities to Experimental Acidification and Warming. Microorganisms 2023, 11, 273. https://doi.org/10.3390/microorganisms11020273
Tsiola A, Krasakopoulou E, Daffonchio D, Frangoulis C, Tsagaraki TM, Fodelianakis S, Pitta P. Responses of Free-Living Planktonic Bacterial Communities to Experimental Acidification and Warming. Microorganisms. 2023; 11(2):273. https://doi.org/10.3390/microorganisms11020273
Chicago/Turabian StyleTsiola, Anastasia, Evangelia Krasakopoulou, Daniele Daffonchio, Constantin Frangoulis, Tatiana M. Tsagaraki, Stilianos Fodelianakis, and Paraskevi Pitta. 2023. "Responses of Free-Living Planktonic Bacterial Communities to Experimental Acidification and Warming" Microorganisms 11, no. 2: 273. https://doi.org/10.3390/microorganisms11020273
APA StyleTsiola, A., Krasakopoulou, E., Daffonchio, D., Frangoulis, C., Tsagaraki, T. M., Fodelianakis, S., & Pitta, P. (2023). Responses of Free-Living Planktonic Bacterial Communities to Experimental Acidification and Warming. Microorganisms, 11(2), 273. https://doi.org/10.3390/microorganisms11020273






