Accelerated Adaptive Laboratory Evolution by Automated Repeated Batch Processes in Parallelized Bioreactors
Abstract
:1. Introduction
2. Material and Methods
2.1. Bacterial Strain
2.2. Media
2.3. Seed Culture
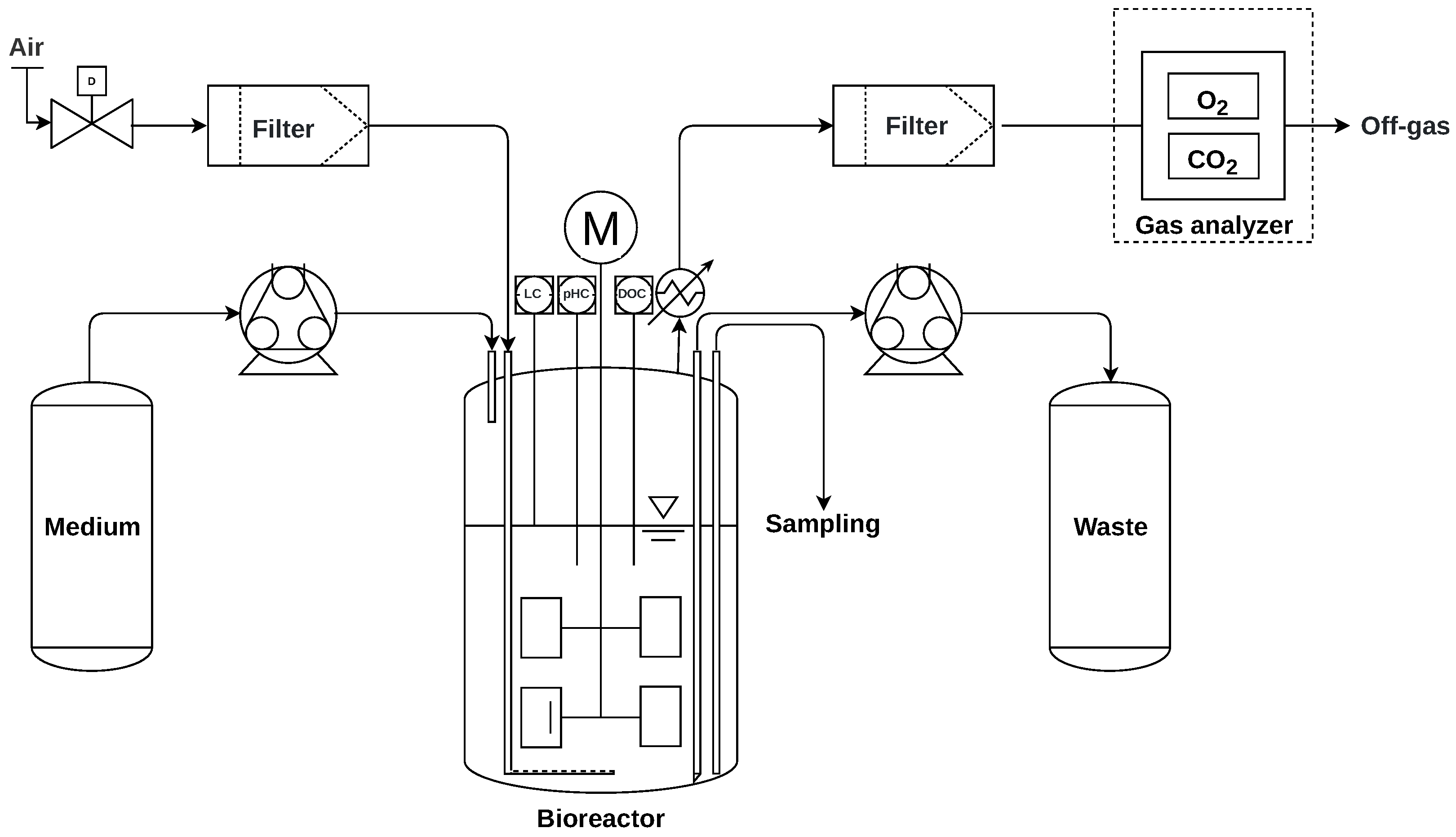
2.4. Experimental Setup and Process Control
2.4.1. Off-Gas Analysis
2.4.2. Peristaltic Pumps
2.4.3. Process Control Software
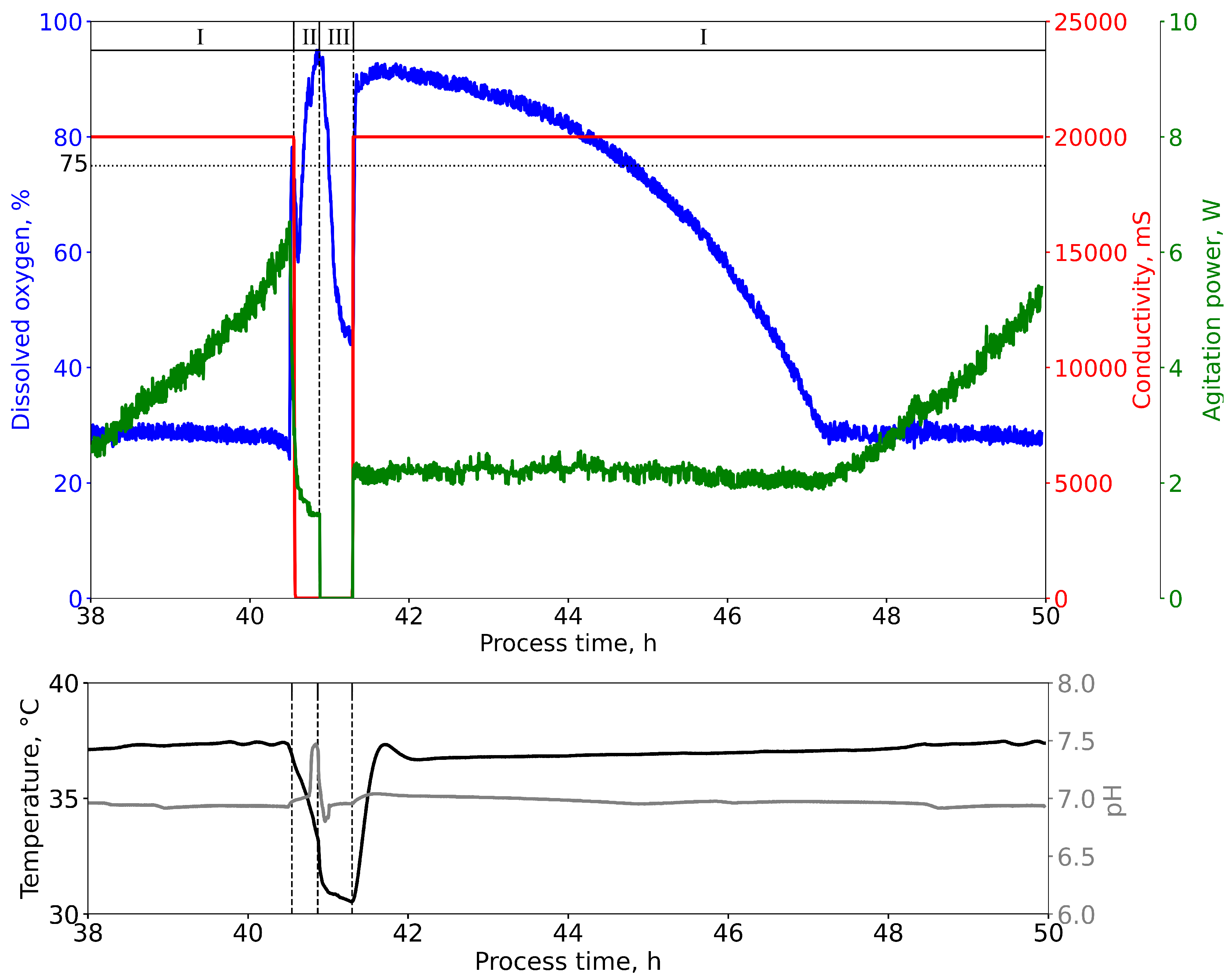
2.4.4. Repeated Batch Mode
2.5. Specific Growth Rate Estimation Using a Black-Box Model
2.5.1. Black Box Model
2.5.2. Oxygen Uptake Rate and Carbon Emission Rate
2.5.3. Biomass and Substrate Conversion Rate
2.5.4. Cumulative Number of Cell Divisions
2.5.5. Preprocessing
3. Results
3.1. Process Characteristics and Duration
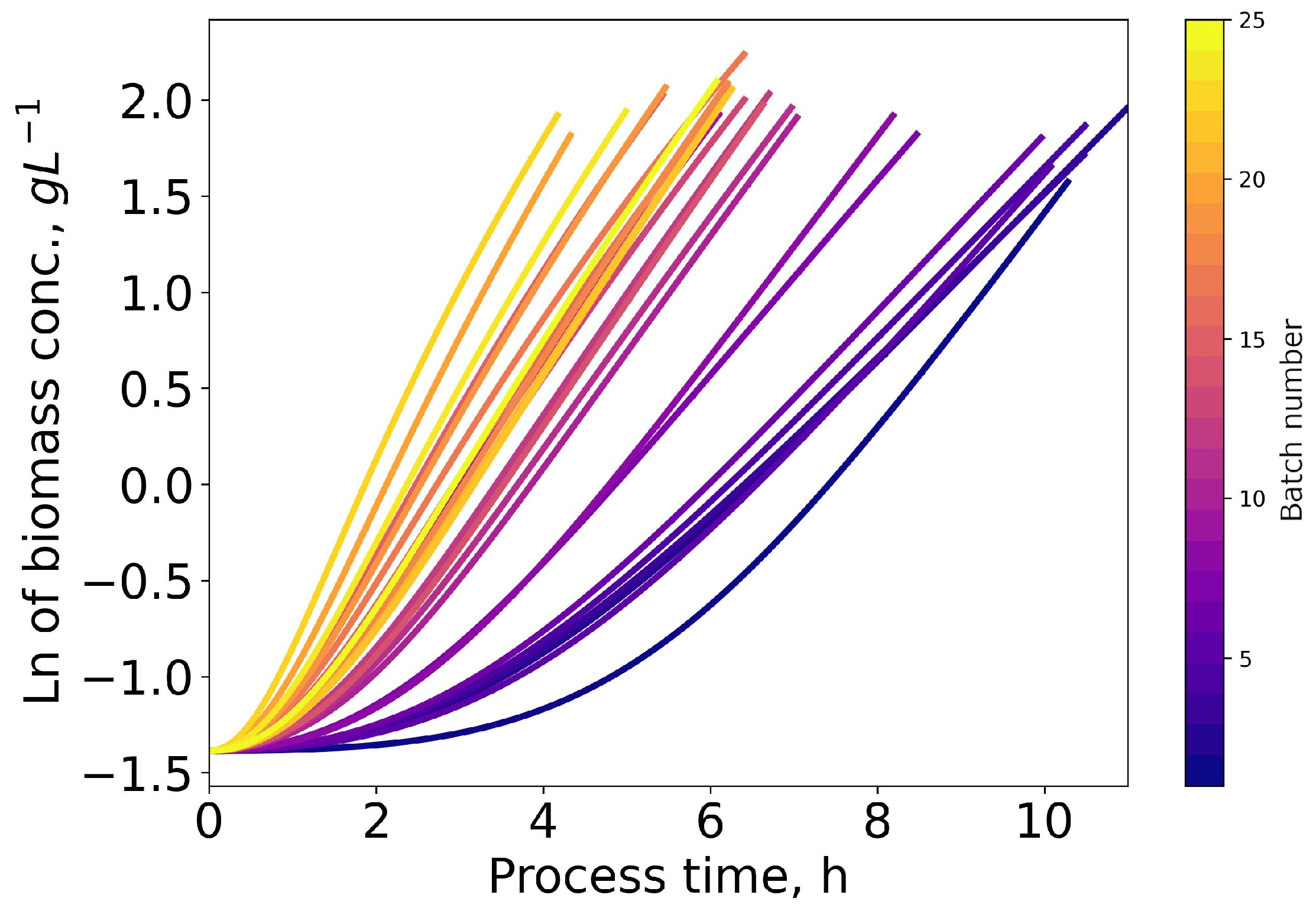
3.2. Model Predictions of Specific Growth Rate and Biomass Concentrations
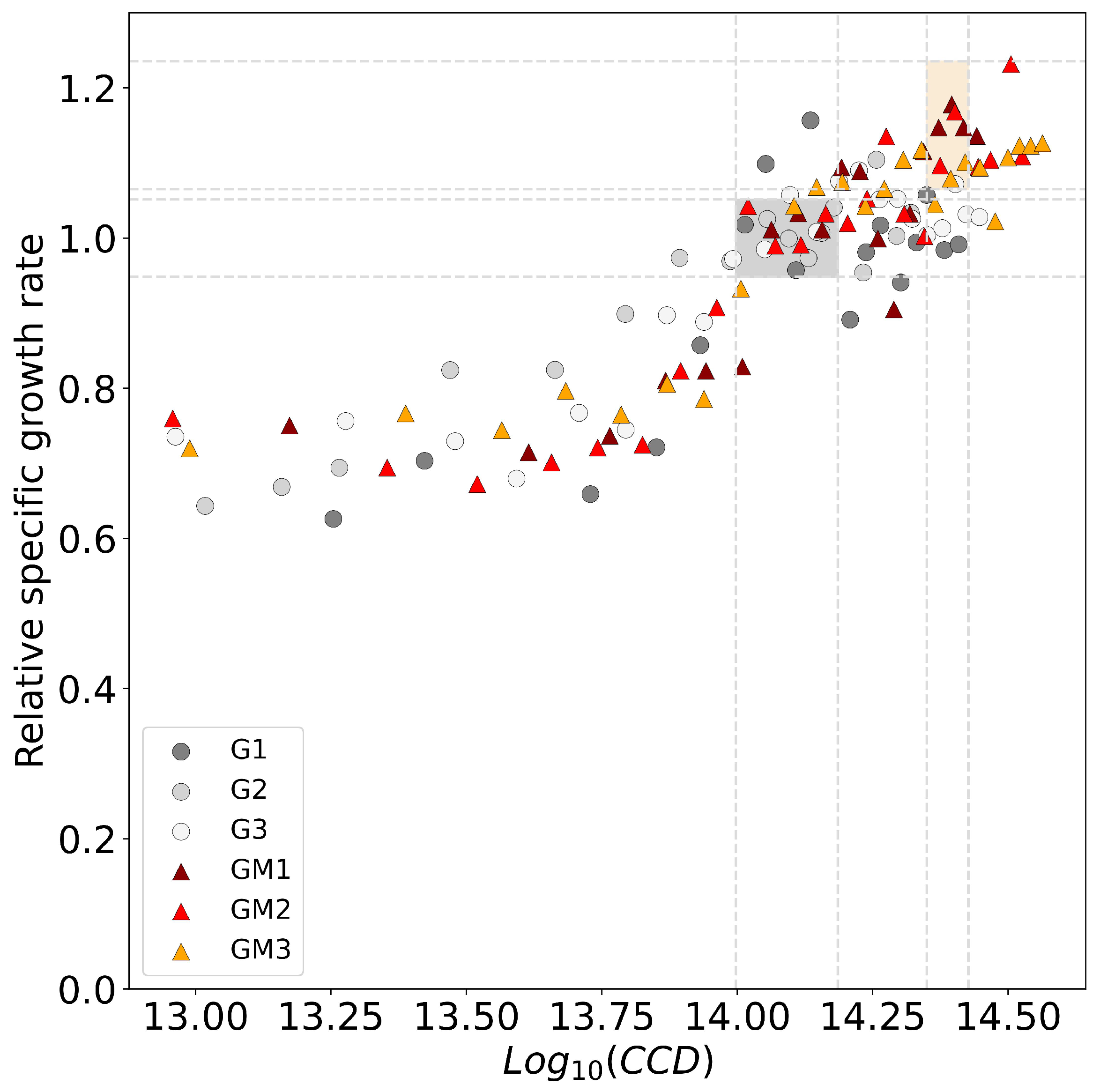
3.3. Comparison of Serial Passaging and Repeated Batch Process
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
Abbreviations
| ALE | Adaptive laboratory evolution |
| CCD | Cumulative number of cell divisions |
| CER | Carbon emission rate |
| DO | Dissolved oxygen |
| DoR | Degree of reduction |
| DCW | Dry cell weight |
| GR | Growth rate |
| LB | Lysogeny broth |
| NTG | N-methyl-N’nitro-N-nitrosoguanidine |
| OUR | Oxygen uptake rate |
| RB | Riesennberg |
| SiLA | Standardization in laboratory automation |
| WT | Wild type |
References
- Mavrommati, M.; Daskalaki, A.; Papanikolaou, S.; Aggelis, G. Adaptive laboratory evolution principles and applications in industrial biotechnology. Biotechnol. Adv. 2022, 54, 107795. [Google Scholar] [CrossRef] [PubMed]
- Herring, C.D.; Raghunathan, A.; Honisch, C.; Patel, T.; Applebee, M.K.; Joyce, A.R.; Albert, T.J.; Blattner, F.R.; Van den Boom, D.; Cantor, C.R. Comparative genome sequencing of Escherichia coli allows observation of bacterial evolution on a laboratory timescale. Nat. Genet. 2006, 38, 1406–1412. [Google Scholar] [CrossRef]
- Hua, Q.; Joyce, A.R.; Fong, S.S.; Palsson, B.Ø. Metabolic analysis of adaptive evolution for in silico-designed lactate-producing strains. Biotechnol. Bioeng. 2006, 95, 992–1002. [Google Scholar] [CrossRef]
- Stanek, M.T.; Cooper, T.F.; Lenski, R.E. Identification and dynamics of a beneficial mutation in a long-term evolution experiment with Escherichia coli. BMC Evol. Biol. 2009, 9, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Conrad, T.M.; Lewis, N.E.; Palsson, B.Ø. Microbial laboratory evolution in the era of genome-scale science. Mol. Syst. Biol. 2011, 7, 509. [Google Scholar] [CrossRef] [PubMed]
- Portnoy, V.A.; Bezdan, D.; Zengler, K. Adaptive laboratory evolution—harnessing the power of biology for metabolic engineering. Curr. Opin. Biotechnol. 2011, 22, 590–594. [Google Scholar] [CrossRef] [PubMed]
- Dragosits, M.; Mattanovich, D. Adaptive laboratory evolution – principles and applications for biotechnology. Microb. Cell Factories 2013, 12, 64. [Google Scholar] [CrossRef] [Green Version]
- Sandberg, T.E.; Salazar, M.J.; Weng, L.L.; Palsson, B.Ø; Feist, A.M. The emergence of adaptive laboratory evolution as an efficient tool for biological discovery and industrial biotechnology. Metab. Eng. 2019, 56, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Stella, R.G.; Wiechert, J.; Noack, S.; Frunzke, J. Evolutionary engineering of Corynebacterium glutamicum. Biotechnol. J. 2019, 14, 1800444. [Google Scholar] [CrossRef] [Green Version]
- Radek, A.; Tenhaef, N.; Müller, M.F.; Brüsseler, C.; Wiechert, W.; Marienhagen, J.; Polen, T.; Noack, S. Miniaturized and automated adaptive laboratory evolution: Evolving Corynebacterium glutamicum towards an improved d-xylose utilization. Bioresour. Technol. 2017, 245, 1377–1385. [Google Scholar] [CrossRef]
- Si, T.; Chao, R.; Min, Y.; Wu, Y.; Ren, W.; Zhao, H. Automated multiplex genome-scale engineering in yeast. Nat. Commun. 2017, 8, 15187. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Jian, X.; Xing, X.H.; Zhang, C.; Fei, Q. Empowering a Methanol-Dependent Escherichia coli via Adaptive Evolution Using a High-Throughput Microbial Microdroplet Culture System. Front. Bioeng. Biotechnol. 2020, 8. [Google Scholar] [CrossRef] [PubMed]
- Jian, X.; Guo, X.; Wang, J.; Tan, Z.L.; Xing, X.h.; Wang, L.; Zhang, C. Microbial microdroplet culture system (MMC): An integrated platform for automated, high-throughput microbial cultivation and adaptive evolution. Biotechnol. Bioeng. 2020, 117, 1724–1737. [Google Scholar] [CrossRef] [PubMed]
- Wong, B.G.; Mancuso, C.P.; Kiriakov, S.; Bashor, C.J.; Khalil, A.S. Precise, automated control of conditions for high-throughput growth of yeast and bacteria with eVOLVER. Nat. Biotechnol. 2018, 36, 614–623. [Google Scholar] [CrossRef]
- Ekkers, D.M.; Branco dos Santos, F.; Mallon, C.A.; Bruggeman, F.; van Doorn, G.S. The omnistat: A flexible continuous-culture system for prolonged experimental evolution. Methods Ecol. Evol. 2020, 11, 932–942. [Google Scholar] [CrossRef]
- LaCroix, R.A.; Palsson, B.Ø; Feist, A.M. A Model for Designing Adaptive Laboratory Evolution Experiments. Appl. Environ. Microbiol. 2017, 83, e03115-16. [Google Scholar] [CrossRef] [Green Version]
- Ji, X.J.; Huang, H.; Ouyang, P.K. Microbial 2,3-butanediol production: A state-of-the-art review. Biotechnol. Adv. 2011, 29, 351–364. [Google Scholar] [CrossRef]
- LaCroix, R.A.; Sandberg, T.E.; O’Brien, E.J.; Utrilla, J.; Ebrahim, A.; Guzman, G.I.; Szubin, R.; Palsson, B.Ø; Feist, A.M. Use of Adaptive Laboratory Evolution To Discover Key Mutations Enabling Rapid Growth of Escherichia coli K-12 MG1655 on Glucose Minimal Medium. Appl. Environ. Microbiol. 2015, 81, 17–30. [Google Scholar] [CrossRef] [Green Version]
- Wiser, M.J.; Lenski, R.E. A comparison of methods to measure fitness in Escherichia coli. PLoS ONE 2015, 10, e0126210. [Google Scholar] [CrossRef] [Green Version]
- Bacun-Druzina, V.; Cagalj, Z.; Gjuracic, K. The growth advantage in stationary-phase (GASP) phenomenon in mixed cultures of enterobacteria. FEMS Microbiol. Lett. 2007, 266, 119–127. [Google Scholar] [CrossRef]
- Sandberg, T.E.; Pedersen, M.; LaCroix, R.A.; Ebrahim, A.; Bonde, M.; Herrgard, M.J.; Palsson, B.Ø; Sommer, M.; Feist, A.M. Evolution of Escherichia coli to 42 °C and Subsequent Genetic Engineering Reveals Adaptive Mechanisms and Novel Mutations. Mol. Biol. Evol. 2014, 31, 2647–2662. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bertrand, R.L. Lag Phase Is a Dynamic, Organized, Adaptive, and Evolvable Period That Prepares Bacteria for Cell Division. J. Bacteriol. 2019, 201, e00697-18. [Google Scholar] [CrossRef] [PubMed]
- Augustin, J.C.; Brouillaud-Delattre, A.; Rosso, L.; Carlier, V. Significance of inoculum size in the lag time of Listeria monocytogenes. Appl. Environ. Microbiol. 2000, 66, 1706–1710. [Google Scholar] [CrossRef] [Green Version]
- Robinson, T.P.; Aboaba, O.O.; Kaloti, A.; Ocio, M.J.; Baranyi, J.; Mackey, B.M. The effect of inoculum size on the lag phase of Listeria monocytogenes. Int. J. Food Microbiol. 2001, 70, 163–173. [Google Scholar] [CrossRef]
- Lee, D.H.; Feist, A.M.; Barrett, C.L.; Palsson, B.Ø. Cumulative number of cell divisions as a meaningful timescale for adaptive laboratory evolution of Escherichia coli. PLoS ONE 2011, 6, e26172. [Google Scholar] [CrossRef] [Green Version]
- Fong, S.S.; Joyce, A.R.; Palsson, B.Ø. Parallel adaptive evolution cultures of Escherichia coli lead to convergent growth phenotypes with different gene expression states. Genome Res. 2005, 15, 1365–1372. [Google Scholar] [CrossRef] [Green Version]
- Riesenberg, D.; Schulz, V.; Knorre, W.A.; Pohl, H.D.; Korz, D.; Sanders, E.A.; Ross, A.; Deckwer, W.D. High cell density cultivation of Escherichia coli at controlled specific growth rate. J. Biotechnol. 1991, 20, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Kangwa, M.; Yelemane, V.; Polat, A.N.; Gorrepati, K.D.D.; Grasselli, M.; Fernández-Lahore, M. High-level fed-batch fermentative expression of an engineered Staphylococcal protein A based ligand in E. coli: Purification and characterization. AMB Express 2015, 5, 70. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bromig, L.; Leiter, D.; Mardale, A.V.; von den Eichen, N.; Bieringer, E.; Weuster-Botz, D. The SiLA 2 Manager for rapid device integration and workflow automation. SoftwareX 2022, 17, 100991. [Google Scholar] [CrossRef]
- SiLA 2 Community. Sila 2 python reference implementation.
- Roels, J.A. Application of macroscopic principles to microbial metabolism. Biotechnol. Bioeng. 1980, 22, 2457–2514. [Google Scholar] [CrossRef]
- Villadsen, J.; Nielsen, J.; Lidén, G. Elemental and Redox Balances. In Bioreaction Engineering Principles; Springer: Berlin/Heidelberg, Germany, 2011; pp. 63–118. [Google Scholar] [CrossRef]
- Neidhardt, F.C. Escherichia Coli and Salmonella: Typhimurium Cellular and Molecular Biology; American Society for Microbiology: Washington, DC, USA, 1987. [Google Scholar] [CrossRef]
- Herwig, C.; Marison, I.; Von Stockar, U. On-line stoichiometry and identification of metabolic state under dynamic process conditions. Biotechnol. Bioeng. 2001, 75, 345–354. [Google Scholar] [CrossRef] [PubMed]
- Sagmeister, P.; Wechselberger, P.; Jazini, M.; Meitz, A.; Langemann, T.; Herwig, C. Soft sensor assisted dynamic bioprocess control: Efficient tools for bioprocess development. Chem. Eng. Sci. 2013, 96, 190–198. [Google Scholar] [CrossRef]
- Campos, P.R.A.; Wahl, L.M. The Effects of Population Bottlenecks on Clonal Interference, and the Adaptation Effective Population Size. Evolution 2009, 63, 950–958. [Google Scholar] [CrossRef]
- Wielgoss, S.; Barrick, J.E.; Tenaillon, O.; Wiser, M.J.; Dittmar, W.J.; Cruveiller, S.; Chane-Woon-Ming, B.; Médigue, C.; Lenski, R.E.; Schneider, D. Mutation rate dynamics in a bacterial population reflect tension between adaptation and genetic load. Proc. Natl. Acad. Sci. USA 2013, 110, 222–227. [Google Scholar] [CrossRef] [Green Version]
- Swings, T.; Van den Bergh, B.; Wuyts, S.; Oeyen, E.; Voordeckers, K.; Verstrepen, K.J.; Fauvart, M.; Verstraeten, N.; Michiels, J. Adaptive tuning of mutation rates allows fast response to lethal stress in Escherichia coli. Elife 2017, 6, e22939. [Google Scholar] [CrossRef]
- Sprouffske, K.; Aguilar-Rodríguez, J.; Sniegowski, P.; Wagner, A. High mutation rates limit evolutionary adaptation in Escherichia coli. PLoS Genet. 2018, 14, e1007324. [Google Scholar] [CrossRef] [Green Version]
- Von den Eichen, N.; Bromig, L.; Sidarava, V.; Marienberg, H.; Weuster-Botz, D. Automated multi-scale cascade of parallel stirred-tank bioreactors for fast protein expression studies. J. Biotechnol. 2021, 332, 103–113. [Google Scholar] [CrossRef]
- Bromig, L.; von den Eichen, N.; Weuster-Botz, D. Control of parallelized bioreactors I: Dynamic scheduling software for efficient bioprocess management in high-throughput systems. Bioprocess Biosyst. Eng. 2022, 45, 1927–1937. [Google Scholar] [CrossRef] [PubMed]
- Osthege, M.; Dölle, M.; Bromig, L.; Wiechert, W.; Oldiges, M.; Weuster-Botz, D. Control of parallelized bioreactors II: Probabilistic quantification of carboxylic acid reductase activity for bioprocess optimization. Bioprocess Biosyst. Eng. 2022, 45, 1939–1954. [Google Scholar] [CrossRef]

| Evolution | Method | Replicates | GR (h) | CCD | Ref. |
|---|---|---|---|---|---|
| WT | mL scale, manual | 5 (GA, GB, GC, GD, GE) | [2,26] | ||
| mL scale, automated | 6 (1% passage size) | [16], suppl. | |||
| L scale, automated | 3 (G1, G2, G3) | this study | |||
| WT + NTG | mL scale, manual | 2 (GM1, GM2) | [25] | ||
| L scale, automated | 3 (GM1, GM2, GM3) | this study |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bromig, L.; Weuster-Botz, D. Accelerated Adaptive Laboratory Evolution by Automated Repeated Batch Processes in Parallelized Bioreactors. Microorganisms 2023, 11, 275. https://doi.org/10.3390/microorganisms11020275
Bromig L, Weuster-Botz D. Accelerated Adaptive Laboratory Evolution by Automated Repeated Batch Processes in Parallelized Bioreactors. Microorganisms. 2023; 11(2):275. https://doi.org/10.3390/microorganisms11020275
Chicago/Turabian StyleBromig, Lukas, and Dirk Weuster-Botz. 2023. "Accelerated Adaptive Laboratory Evolution by Automated Repeated Batch Processes in Parallelized Bioreactors" Microorganisms 11, no. 2: 275. https://doi.org/10.3390/microorganisms11020275
APA StyleBromig, L., & Weuster-Botz, D. (2023). Accelerated Adaptive Laboratory Evolution by Automated Repeated Batch Processes in Parallelized Bioreactors. Microorganisms, 11(2), 275. https://doi.org/10.3390/microorganisms11020275






