The Contribution of the Human Oral Microbiome to Oral Disease: A Review
Abstract
:1. Introduction
2. The Human Microbiome Analysis
3. The Human Oral Microbiome
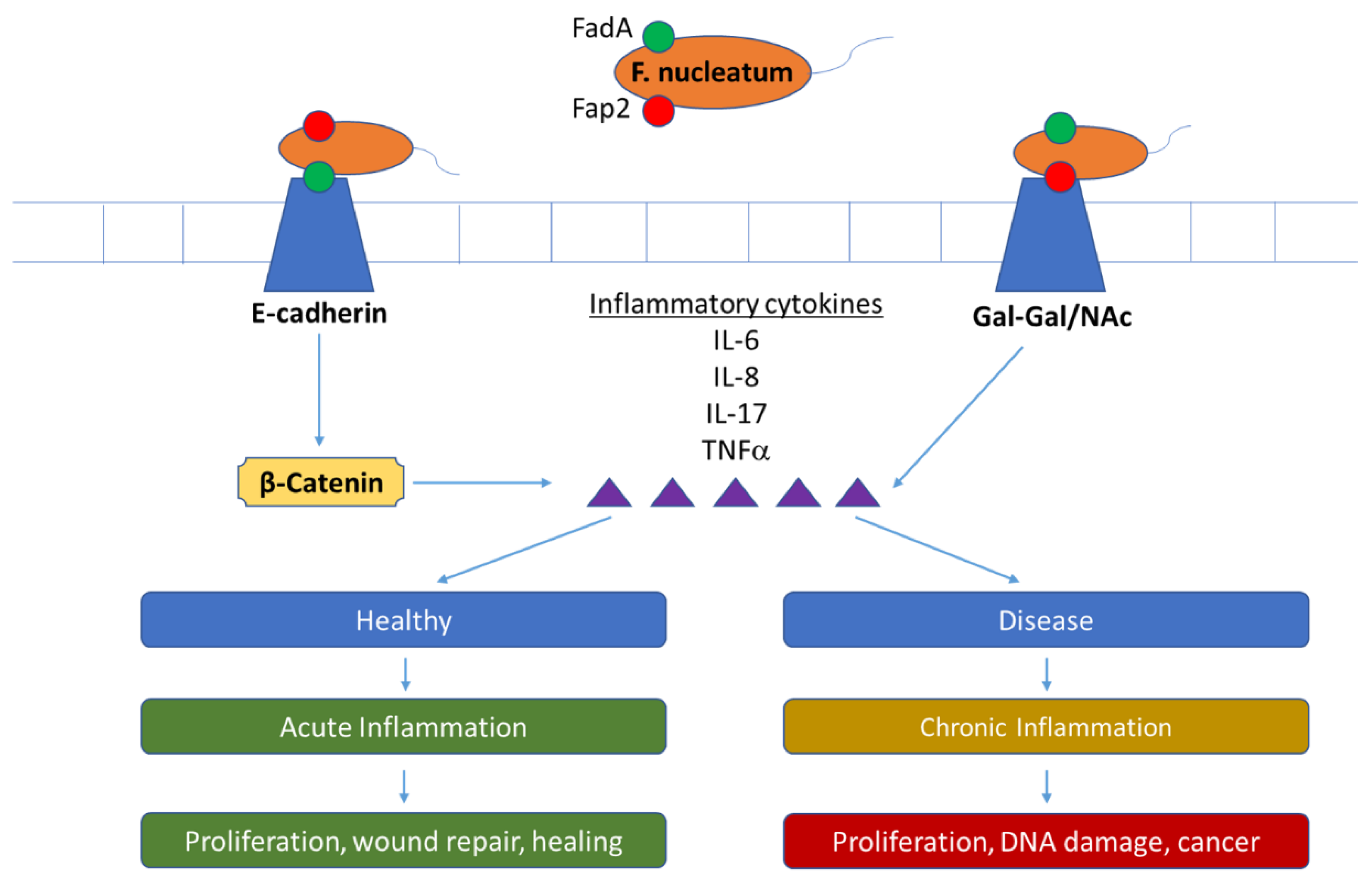
4. Key Bacteria Involved in Oral Disease
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kilian, M.; Chapple, I.L.; Hannig, M.; Marsh, P.D.; Meuric, V.; Pedersen, A.M.; Tonetti, M.S.; Wade, W.G.; Zaura, E. The oral microbiome-an update for oral healthcare professionals. Br. Dent. J. 2016, 221, 657–666. [Google Scholar] [CrossRef] [PubMed]
- Sender, R.; Fuchs, S.; Milo, R. Are We Really Vastly Outnumbered? Revisiting the Ratio of Bacterial to Host Cells in Humans. Cell 2016, 164, 337–340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manos, J. The human microbiome in disease and pathology. APMIS 2022, 130, 690–705. [Google Scholar] [CrossRef] [PubMed]
- Gomaa, E.Z. Human gut microbiota/microbiome in health and diseases: A review. Antonie Van Leeuwenhoek 2020, 113, 2019–2040. [Google Scholar] [CrossRef] [PubMed]
- Yoo, W.; Byndloss, M.X. How to thrive in the inflamed gut. Nat. Microbiol. 2020, 5, 10–11. [Google Scholar] [CrossRef]
- Herremans, K.M.; Riner, A.N.; Cameron, M.E.; McKinley, K.L.; Triplett, E.W.; Hughes, S.J.; Trevino, J.G. The oral microbiome, pancreatic cancer and human diversity in the age of precision medicine. Microbiome 2022, 10, 93. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.; Cheng, L.; You, Y.; Tang, C.; Ren, B.; Li, Y.; Xu, X.; Zhou, X. Oral microbiota in human systematic diseases. Int. J. Oral Sci. 2022, 14, 14. [Google Scholar] [CrossRef]
- Frankel, T.L.; Pasca di Magliano, M. Immune sensing of microbial metabolites: Action at the tumor. Immunity 2022, 55, 192–194. [Google Scholar] [CrossRef]
- Hezaveh, K.; Shinde, R.S.; Klotgen, A.; Halaby, M.J.; Lamorte, S.; Ciudad, M.T.; Quevedo, R.; Neufeld, L.; Liu, Z.Q.; Jin, R.; et al. Tryptophan-derived microbial metabolites activate the aryl hydrocarbon receptor in tumor-associated macrophages to suppress anti-tumor immunity. Immunity 2022, 55, 324–340.e28. [Google Scholar] [CrossRef]
- Nasidze, I.; Li, J.; Quinque, D.; Tang, K.; Stoneking, M. Global diversity in the human salivary microbiome. Genome Res. 2009, 19, 636–643. [Google Scholar] [CrossRef] [Green Version]
- Deo, P.N.; Deshmukh, R. Oral microbiome: Unveiling the fundamentals. J. Oral Maxillofac. Pathol. 2019, 23, 122–128. [Google Scholar] [CrossRef] [PubMed]
- Lane, N. The unseen world: Reflections on Leeuwenhoek (1677) ‘Concerning little animals’. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2015, 370, 20140344. [Google Scholar] [CrossRef] [PubMed]
- Wade, W.G. The oral microbiome in health and disease. Pharmacol Res. 2013, 69, 137–143. [Google Scholar] [CrossRef]
- Huttenhower, C.; Gevers, D.; Knight, R.; Abubucker, S.; Badger, J.H.; Chinwalla, A.T.; Creasy, H.H.; Earl, A.M.; FitzGerald, M.G.; Fulton, R.S.; et al. Structure, function and diversity of the healthy human microbiome. Nature 2012, 486, 207–214. [Google Scholar] [CrossRef] [Green Version]
- Tuominen, H.; Collado, M.C.; Rautava, J.; Syrjanen, S.; Rautava, S. Composition and maternal origin of the neonatal oral cavity microbiota. J. Oral Microbiol. 2019, 11, 1663084. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Collins, L.M.; Dawes, C. The surface area of the adult human mouth and thickness of the salivary film covering the teeth and oral mucosa. J. Dent. Res. 1987, 66, 1300–1302. [Google Scholar] [CrossRef] [PubMed]
- Xie, H.; Cook, G.S.; Costerton, J.W.; Bruce, G.; Rose, T.M.; Lamont, R.J. Intergeneric communication in dental plaque biofilms. J. Bacteriol. 2000, 182, 7067–7069. [Google Scholar] [CrossRef] [Green Version]
- D’Argenio, V. Human microbiome acquisition and bioinformatic challenges in metagenomic studies. Int. J. Mol. Sci. 2018, 19, 383. [Google Scholar] [CrossRef] [Green Version]
- Dentino, A.; Lee, S.; Mailhot, J.; Hefti, A.F. Principles of periodontology. Periodontol 2000 2013, 61, 16–53. [Google Scholar] [CrossRef] [Green Version]
- Subbarao, K.C.; Nattuthurai, G.S.; Sundararajan, S.K.; Sujith, I.; Joseph, J.; Syedshah, Y.P. Gingival Crevicular Fluid: An Overview. J. Pharm. Bioallied. Sci. 2019, 11, S135–S139. [Google Scholar] [CrossRef]
- Genco, R.J.; Borgnakke, W.S. Risk factors for periodontal disease. Periodontol 2000 2013, 62, 59–94. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhang, D.; Jia, H.; Feng, Q.; Wang, D.; Liang, D.; Wu, X.; Li, J.; Tang, L.; Li, Y.; et al. The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat. Med. 2015, 21, 895–905. [Google Scholar] [CrossRef] [PubMed]
- Dhotre, S.; Jahagirdar, V.; Suryawanshi, N.; Davane, M.; Patil, R.; Nagoba, B. Assessment of periodontitis and its role in viridans streptococcal bacteremia and infective endocarditis. Indian Heart. J. 2018, 70, 225–232. [Google Scholar] [CrossRef]
- Ordovas, J.M.; Mooser, V. Metagenomics: The role of the microbiome in cardiovascular diseases. Curr. Opin. Lipidol. 2006, 17, 157–161. [Google Scholar] [CrossRef]
- Scannapieco, F.A.; Genco, R.J. Association of periodontal infections with atherosclerotic and pulmonary diseases. J. Periodontal. Res. 1999, 34, 340–345. [Google Scholar] [CrossRef]
- Bajaj, J.S.; Betrapally, N.S.; Hylemon, P.B.; Heuman, D.M.; Daita, K.; White, M.B.; Unser, A.; Thacker, L.R.; Sanyal, A.J.; Kang, D.J.; et al. Salivary microbiota reflects changes in gut microbiota in cirrhosis with hepatic encephalopathy. Hepatology 2015, 62, 1260–1271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meurman, J.H. Oral microbiota and cancer. J. Oral Microbiol. 2010, 2, 5195. [Google Scholar] [CrossRef] [Green Version]
- Bianchi, S.; Fantozzi, G.; Bernardi, S.; Antonouli, S.; Continenza, M.A.; Macchiarelli, G. Commercial oral hygiene products and implant collar surfaces: Scanning electron microscopy observations. Can. J. Dent. Hyg. 2020, 54, 26–31. [Google Scholar]
- Gholizadeh, P.; Eslami, H.; Yousefi, M.; Asgharzadeh, M.; Kafil, H.S. Role of oral microbiome on oral cancers, a review. Biomed Pharm. 2016, 84, 552–558. [Google Scholar] [CrossRef]
- Lamont, R.J.; Koo, H.; Hajishengallis, G. The oral microbiota: Dynamic communities and host interactions. Nat. Rev. Microbiol. 2018, 16, 745–759. [Google Scholar] [CrossRef]
- Seitz, H.K.; Becker, P. Alcohol metabolism and cancer risk. Alcohol. Res. Health 2007, 30, 38–41, 44–47. [Google Scholar] [PubMed]
- Rajakaruna, G.A.; Negi, M.; Uchida, K.; Sekine, M.; Furukawa, A.; Ito, T.; Kobayashi, D.; Suzuki, Y.; Akashi, T.; Umeda, M.; et al. Localization and density of Porphyromonas gingivalis and Tannerella forsythia in gingival and subgingival granulation tissues affected by chronic or aggressive periodontitis. Sci. Rep. 2018, 8, 9507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jemal, A.; Bray, F.; Center, M.M.; Ferlay, J.; Ward, E.; Forman, D. Global cancer statistics. CA Cancer J. Clin. 2011, 61, 69–90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morgan, X.C.; Huttenhower, C. Chapter 12: Human microbiome analysis. PLoS Comput. Biol. 2012, 8, e1002808. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olsen, G.J.; Lane, D.J.; Giovannoni, S.J.; Pace, N.R.; Stahl, D.A. Microbial ecology and evolution: A ribosomal RNA approach. Annu. Rev. Microbiol. 1986, 40, 337–365. [Google Scholar] [CrossRef] [PubMed]
- Kuczynski, J.; Lauber, C.L.; Walters, W.A.; Parfrey, L.W.; Clemente, J.C.; Gevers, D.; Knight, R. Experimental and analytical tools for studying the human microbiome. Nat. Rev. Genet. 2011, 13, 47–58. [Google Scholar] [CrossRef] [Green Version]
- Galloway-Pena, J.; Hanson, B. Tools for Analysis of the Microbiome. Dig. Dis. Sci. 2020, 65, 674–685. [Google Scholar] [CrossRef] [Green Version]
- Ni, J.; Yan, Q.; Yu, Y. How much metagenomic sequencing is enough to achieve a given goal? Sci. Rep. 2013, 3, 1968. [Google Scholar] [CrossRef] [Green Version]
- Quince, C.; Walker, A.W.; Simpson, J.T.; Loman, N.J.; Segata, N. Shotgun metagenomics, from sampling to analysis. Nat. Biotechnol. 2017, 35, 833–844. [Google Scholar] [CrossRef] [Green Version]
- Segata, N.; Waldron, L.; Ballarini, A.; Narasimhan, V.; Jousson, O.; Huttenhower, C. Metagenomic microbial community profiling using unique clade-specific marker genes. Nat. Methods 2012, 9, 811–814. [Google Scholar] [CrossRef]
- Huson, D.H.; Mitra, S.; Ruscheweyh, H.J.; Weber, N.; Schuster, S.C. Integrative analysis of environmental sequences using MEGAN4. Genome Res. 2011, 21, 1552–1560. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meyer, F.; Paarmann, D.; D’Souza, M.; Olson, R.; Glass, E.M.; Kubal, M.; Paczian, T.; Rodriguez, A.; Stevens, R.; Wilke, A.; et al. The metagenomics RAST server–a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinform. 2008, 9, 386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chakravorty, S.; Helb, D.; Burday, M.; Connell, N.; Alland, D. A detailed analysis of 16S ribosomal RNA gene segments for the diagnosis of pathogenic bacteria. J. Microbiol. Methods 2007, 69, 330–339. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vetrovsky, T.; Baldrian, P. The variability of the 16S rRNA gene in bacterial genomes and its consequences for bacterial community analyses. PLoS ONE 2013, 8, e57923. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xue, Z.; Kable, M.E.; Marco, M.L. Impact of DNA Sequencing and Analysis Methods on 16S rRNA Gene Bacterial Community Analysis of Dairy Products. mSphere 2018, 3, e00410-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, J.S.; Spakowicz, D.J.; Hong, B.Y.; Petersen, L.M.; Demkowicz, P.; Chen, L.; Leopold, S.R.; Hanson, B.M.; Agresta, H.O.; Gerstein, M.; et al. Evaluation of 16S rRNA gene sequencing for species and strain-level microbiome analysis. Nat. Commun. 2019, 10, 5029. [Google Scholar] [CrossRef] [Green Version]
- Park, S.C.; Won, S. Evaluation of 16S rRNA Databases for Taxonomic Assignments Using Mock Community. Genom. Inform. 2018, 16, e24. [Google Scholar] [CrossRef] [Green Version]
- Man, S.M.; Kaakoush, N.O.; Octavia, S.; Mitchell, H. The internal transcribed spacer region, a new tool for use in species differentiation and delineation of systematic relationships within the Campylobacter genus. Appl. Environ. Microbiol. 2010, 76, 3071–3081. [Google Scholar] [CrossRef] [Green Version]
- Nilsson, R.H.; Larsson, K.-H.; Taylor, A.F.S.; Bengtsson-Palme, J.; Jeppesen, T.S.; Schigel, D.; Kennedy, P.; Picard, K.; Glöckner, F.O.; Tedersoo, L.; et al. The UNITE database for molecular identification of fungi: Handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res. 2018, 47, D259–D264. [Google Scholar] [CrossRef]
- Sutton, T.D.S.; Clooney, A.G.; Ryan, F.J.; Ross, R.P.; Hill, C. Choice of assembly software has a critical impact on virome characterisation. Microbiome 2019, 7, 12. [Google Scholar] [CrossRef] [Green Version]
- Stern, J.; Miller, G.; Li, X.; Saxena, D. Virome and bacteriome: Two sides of the same coin. Curr. Opin. Virol. 2019, 37, 37–43. [Google Scholar] [CrossRef]
- Breitwieser, F.P.; Lu, J.; Salzberg, S.L. A review of methods and databases for metagenomic classification and assembly. Brief. Bioinform. 2019, 20, 1125–1136. [Google Scholar] [CrossRef] [PubMed]
- Wesolowska-Andersen, A.; Bahl, M.I.; Carvalho, V.; Kristiansen, K.; Sicheritz-Pontén, T.; Gupta, R.; Licht, T.R. Choice of bacterial DNA extraction method from fecal material influences community structure as evaluated by metagenomic analysis. Microbiome 2014, 2, 19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thurber, R.V.; Haynes, M.; Breitbart, M.; Wegley, L.; Rohwer, F. Laboratory procedures to generate viral metagenomes. Nat. Protoc. 2009, 4, 470. [Google Scholar] [CrossRef] [PubMed]
- Goodrich, J.K.; Di Rienzi, S.C.; Poole, A.C.; Koren, O.; Walters, W.A.; Caporaso, J.G.; Knight, R.; Ley, R.E. Conducting a microbiome study. Cell 2014, 158, 250–262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turner, P.; Turner, C.; Jankhot, A.; Helen, N.; Lee, S.J.; Day, N.P.; White, N.J.; Nosten, F.; Goldblatt, D. A longitudinal study of Streptococcus pneumoniae carriage in a cohort of infants and their mothers on the Thailand-Myanmar border. PLoS ONE 2012, 7, e38271. [Google Scholar] [CrossRef] [Green Version]
- Salter, S.J.; Cox, M.J.; Turek, E.M.; Calus, S.T.; Cookson, W.O.; Moffatt, M.F.; Turner, P.; Parkhill, J.; Loman, N.J.; Walker, A.W. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 2014, 12, 87. [Google Scholar] [CrossRef] [Green Version]
- Ross, M.G.; Russ, C.; Costello, M.; Hollinger, A.; Lennon, N.J.; Hegarty, R.; Nusbaum, C.; Jaffe, D.B. Characterizing and measuring bias in sequence data. Genome Biol. 2013, 14, R51. [Google Scholar] [CrossRef] [Green Version]
- Pérez-Cobas, A.E.; Gomez-Valero, L.; Buchrieser, C. Metagenomic approaches in microbial ecology: An update on whole-genome and marker gene sequencing analyses. Microb. Genom. 2020, 6, e000409. [Google Scholar] [CrossRef]
- Mulcahy-O’Grady, H.; Workentine, M.L. The challenge and potential of metagenomics in the clinic. Front. Immunol. 2016, 7, 29. [Google Scholar] [CrossRef] [Green Version]
- Abubucker, S.; Segata, N.; Goll, J.; Schubert, A.M.; Izard, J.; Cantarel, B.L.; Rodriguez-Mueller, B.; Zucker, J.; Thiagarajan, M.; Henrissat, B. Metabolic reconstruction for metagenomic data and its application to the human microbiome. PLoS Comput. Biol. 2012, 8, e1002358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, Y.; Leung, H.C.M.; Yiu, S.M.; Chin, F.Y.L. Meta-IDBA: A de Novo assembler for metagenomic data. In Bioinformatics; Oxford University Press: Oxford, UK, 2011; Volume 27, p. i94. Available online: http://bioinformatics.oxfordjournals.org/ (accessed on 9 December 2022).
- Alneberg, J.; Bjarnason, B.S.; De Bruijn, I.; Schirmer, M.; Quick, J.; Ijaz, U.Z.; Lahti, L.; Loman, N.J.; Andersson, A.F.; Quince, C. Binning metagenomic contigs by coverage and composition. Nat. Methods 2014, 11, 1144–1146. [Google Scholar] [CrossRef] [PubMed]
- Eren, A.M.; Esen, Ö.C.; Quince, C.; Vineis, J.H.; Morrison, H.G.; Sogin, M.L.; Delmont, T.O. Anvi’o: An advanced analysis and visualization platform for ‘omics data. PeerJ 2015, 3, e1319. [Google Scholar] [CrossRef] [Green Version]
- Ayling, M.; Clark, M.D.; Leggett, R.M. New approaches for metagenome assembly with short reads. Brief Bioinform 2020, 21, 584–594. [Google Scholar] [CrossRef] [Green Version]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef]
- Hu, X.; Haas, J.G.; Lathe, R. The electronic tree of life (eToL): A net of long probes to characterize the microbiome from RNA-seq data. BMC Microbiol. 2022, 22, 317. [Google Scholar] [CrossRef]
- Aguiar-Pulido, V.; Huang, W.; Suarez-Ulloa, V.; Cickovski, T.; Mathee, K.; Narasimhan, G. Metagenomics, Metatranscriptomics, and Metabolomics Approaches for Microbiome Analysis. Evol. Bioinform. 2016, 12, 5–16. [Google Scholar] [CrossRef] [Green Version]
- Zierer, J.; Jackson, M.A.; Kastenmuller, G.; Mangino, M.; Long, T.; Telenti, A.; Mohney, R.P.; Small, K.S.; Bell, J.T.; Steves, C.J.; et al. The fecal metabolome as a functional readout of the gut microbiome. Nat. Genet. 2018, 50, 790–795. [Google Scholar] [CrossRef]
- Roessner, U.; Bowne, J. What is metabolomics all about? Biotechniques 2009, 46, 363–365. [Google Scholar] [CrossRef]
- Blakeley-Ruiz, J.A.; Erickson, A.R.; Cantarel, B.L.; Xiong, W.; Adams, R.; Jansson, J.K.; Fraser, C.M.; Hettich, R.L. Metaproteomics reveals persistent and phylum-redundant metabolic functional stability in adult human gut microbiomes of Crohn’s remission patients despite temporal variations in microbial taxa, genomes, and proteomes. Microbiome 2019, 7, 18. [Google Scholar] [CrossRef] [Green Version]
- Shakya, M.; Lo, C.C.; Chain, P.S.G. Advances and Challenges in Metatranscriptomic Analysis. Front. Genet. 2019, 10, 904. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lagier, J.-C.; Dubourg, G.; Million, M.; Cadoret, F.; Bilen, M.; Fenollar, F.; Levasseur, A.; Rolain, J.-M.; Fournier, P.-E.; Raoult, D. Culturing the human microbiota and culturomics. Nat. Rev. Microbiol. 2018, 16, 540–550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gevers, D.; Knight, R.; Petrosino, J.F.; Huang, K.; McGuire, A.L.; Birren, B.W.; Nelson, K.E.; White, O.; Methe, B.A.; Huttenhower, C. The Human Microbiome Project: A community resource for the healthy human microbiome. PLoS Biol. 2012, 10, e1001377. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Proctor, L.M. The Human Microbiome Project in 2011 and beyond. Cell Host Microbe 2011, 10, 287–291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- iHMP Consortium. The Integrative Human Microbiome Project. Nature 2019, 569, 641–648. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verma, D.; Garg, P.K.; Dubey, A.K. Insights into the human oral microbiome. Arch. Microbiol. 2018, 200, 525–540. [Google Scholar] [CrossRef]
- Wang, S.; Song, F.; Wang, Y.; Huang, Y.; Xie, B.; Luo, H. High resolution melting analysis (HRM) based on 16SrRNA as a tool for personal identification with the human oral microbiome. Forensic Sci. Int. Genet. Suppl. Ser. 2019, 7, 161–163. [Google Scholar] [CrossRef]
- Abeles, S.R.; Pride, D.T. Molecular bases and role of viruses in the human microbiome. J. Mol. Biol. 2014, 426, 3892–3906. [Google Scholar] [CrossRef]
- Baker, J.L.; Bor, B.; Agnello, M.; Shi, W.; He, X. Ecology of the Oral Microbiome: Beyond Bacteria. Trends Microbiol. 2017, 25, 362–374. [Google Scholar] [CrossRef] [Green Version]
- Pride, D.T.; Salzman, J.; Haynes, M.; Rohwer, F.; Davis-Long, C.; White, R.A., 3rd; Loomer, P.; Armitage, G.C.; Relman, D.A. Evidence of a robust resident bacteriophage population revealed through analysis of the human salivary virome. ISME J. 2012, 6, 915–926. [Google Scholar] [CrossRef]
- Gruffaz, M.; Zhang, T.; Marshall, V.; Goncalves, P.; Ramaswami, R.; Labo, N.; Whitby, D.; Uldrick, T.S.; Yarchoan, R.; Huang, Y.; et al. Signatures of oral microbiome in HIV-infected individuals with oral Kaposi’s sarcoma and cell-associated KSHV DNA. PLoS Pathog. 2020, 16, e1008114. [Google Scholar] [CrossRef] [Green Version]
- Wood, L.F.; Chahroudi, A.; Chen, H.L.; Jaspan, H.B.; Sodora, D.L. The oral mucosa immune environment and oral transmission of HIV/SIV. Immunol. Rev. 2013, 254, 34–53. [Google Scholar] [CrossRef] [Green Version]
- Hayes, R.B.; Ahn, J.; Fan, X.; Peters, B.A.; Ma, Y.; Yang, L.; Agalliu, I.; Burk, R.D.; Ganly, I.; Purdue, M.P.; et al. Association of Oral Microbiome With Risk for Incident Head and Neck Squamous Cell Cancer. JAMA Oncol. 2018, 4, 358–365. [Google Scholar] [CrossRef]
- Gomez, A.; Espinoza, J.L.; Harkins, D.M.; Leong, P.; Saffery, R.; Bockmann, M.; Torralba, M.; Kuelbs, C.; Kodukula, R.; Inman, J.; et al. Host Genetic Control of the Oral Microbiome in Health and Disease. Cell Host Microbe 2017, 22, 269–278. [Google Scholar] [CrossRef] [Green Version]
- Matarazzo, F.; Ribeiro, A.C.; Feres, M.; Faveri, M.; Mayer, M.P. Diversity and quantitative analysis of Archaea in aggressive periodontitis and periodontally healthy subjects. J. Clin. Periodontol. 2011, 38, 621–627. [Google Scholar] [CrossRef]
- Liu, N.N.; Jiao, N.; Tan, J.C.; Wang, Z.; Wu, D.; Wang, A.J.; Chen, J.; Tao, L.; Zhou, C.; Fang, W.; et al. Multi-kingdom microbiota analyses identify bacterial-fungal interactions and biomarkers of colorectal cancer across cohorts. Nat. Microbiol. 2022, 7, 238–250. [Google Scholar] [CrossRef]
- Ghannoum, M.A.; Jurevic, R.J.; Mukherjee, P.K.; Cui, F.; Sikaroodi, M.; Naqvi, A.; Gillevet, P.M. Characterization of the oral fungal microbiome (mycobiome) in healthy individuals. PLoS Pathog. 2010, 6, e1000713. [Google Scholar] [CrossRef] [Green Version]
- Cheung, M.K.; Chan, J.Y.K.; Wong, M.C.S.; Wong, P.Y.; Lei, P.; Cai, L.; Lan, L.; Ho, W.C.S.; Yeung, A.C.M.; Chan, P.K.S.; et al. Determinants and Interactions of Oral Bacterial and Fungal Microbiota in Healthy Chinese Adults. Microbiol. Spectr. 2022, 10, e0241021. [Google Scholar] [CrossRef]
- Sultan, A.S.; Kong, E.F.; Rizk, A.M.; Jabra-Rizk, M.A. The oral microbiome: A Lesson in coexistence. PLoS Pathog. 2018, 14, e1006719. [Google Scholar] [CrossRef] [Green Version]
- Burgain, A.; Pic, É.; Markey, L.; Tebbji, F.; Kumamoto, C.A.; Sellam, A. A novel genetic circuitry governing hypoxic metabolic flexibility, commensalism and virulence in the fungal pathogen Candida albicans. PLoS Pathogens. 2019, 15, e1007823. [Google Scholar] [CrossRef] [Green Version]
- Vylkova, S.; Lorenz, M.C. Phagosomal neutralization by the fungal pathogen Candida albicans induces macrophage pyroptosis. Infect. Immun. 2017, 85, e00832-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Metwalli, K.H.; Khan, S.A.; Krom, B.P.; Jabra-Rizk, M.A. Streptococcus mutans, Candida albicans, and the human mouth: A sticky situation. PLoS Pathog. 2013, 9, e1003616. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krom, B.P.; Kidwai, S.; Ten Cate, J.M. Candida and other fungal species: Forgotten players of healthy oral microbiota. J. Dent. Res. 2014, 93, 445–451. [Google Scholar] [CrossRef]
- Morse, D.J.; Wilson, M.J.; Wei, X.; Bradshaw, D.J.; Lewis, M.A.O.; Williams, D.W. Modulation of Candida albicans virulence in in vitro biofilms by oral bacteria. Lett. Appl. Microbiol. 2019, 68, 337–343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ishijima, S.A.; Hayama, K.; Burton, J.P.; Reid, G.; Okada, M.; Matsushita, Y.; Abe, S. Effect of Streptococcus salivarius K12 on the in vitro growth of Candida albicans and its protective effect in an oral candidiasis model. Appl. Environ. Microbiol. 2012, 78, 2190–2199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vila, T.; Rizk, A.M.; Sultan, A.S.; Jabra-Rizk, M.A. The power of saliva: Antimicrobial and beyond. PLoS Pathog. 2019, 15, e1008058. [Google Scholar] [CrossRef]
- Verma, A.; Gaffen, S.L.; Swidergall, M. Innate Immunity to Mucosal Candida Infections. J. Fungi 2017, 3, 60. [Google Scholar] [CrossRef] [Green Version]
- Benn, A.; Heng, N.; Broadbent, J.; Thomson, W. Studying the human oral microbiome: Challenges and the evolution of solutions. Aust. Dent. J. 2018, 63, 14–24. [Google Scholar] [CrossRef] [Green Version]
- Dewhirst, F.E.; Chen, T.; Izard, J.; Paster, B.J.; Tanner, A.C.; Yu, W.H.; Lakshmanan, A.; Wade, W.G. The human oral microbiome. J. Bacteriol. 2010, 192, 5002–5017. [Google Scholar] [CrossRef] [Green Version]
- Perera, M.; Al-Hebshi, N.N.; Speicher, D.J.; Perera, I.; Johnson, N.W. Emerging role of bacteria in oral carcinogenesis: A review with special reference to perio-pathogenic bacteria. J. Oral Microbiol. 2016, 8, 32762. [Google Scholar] [CrossRef] [Green Version]
- Pushalkar, S.; Paul, B.; Li, Q.; Yang, J.; Vasconcelos, R.; Makwana, S.; Gonzalez, J.M.; Shah, S.; Xie, C.; Janal, M.N.; et al. Electronic Cigarette Aerosol Modulates the Oral Microbiome and Increases Risk of Infection. iScience 2020, 23, 100884. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ibrahim, M.; Subramanian, A.; Anishetty, S. Comparative pan genome analysis of oral Prevotella species implicated in periodontitis. Funct. Integr. Genom. 2017, 17, 513–536. [Google Scholar] [CrossRef] [PubMed]
- Eribe, E.R.K.; Olsen, I. Leptotrichia species in human infections II. J. Oral Microbiol. 2017, 9, 1368848. [Google Scholar] [CrossRef] [Green Version]
- Eribe, E.R.; Olsen, I. Leptotrichia species in human infections. Anaerobe 2008, 14, 131–137. [Google Scholar] [CrossRef]
- Langfeldt, D.; Neulinger, S.C.; Stiesch, M.; Stumpp, N.; Bang, C.; Schmitz, R.A.; Eberhard, J. Health- and disease-associated species clusters in complex natural biofilms determine the innate immune response in oral epithelial cells during biofilm maturation. FEMS Microbiol. Lett. 2014, 360, 137–143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gupta, V.K.; Chaudhari, N.M.; Iskepalli, S.; Dutta, C. Divergences in gene repertoire among the reference Prevotella genomes derived from distinct body sites of human. BMC Genom. 2015, 16, 153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Randis, T.M.; Ratner, A.J. Gardnerella and Prevotella: Co-conspirators in the Pathogenesis of Bacterial Vaginosis. J. Infect. Dis. 2019, 220, 1085–1088. [Google Scholar] [CrossRef]
- Lukens, J.R.; Gurung, P.; Vogel, P.; Johnson, G.R.; Carter, R.A.; McGoldrick, D.J.; Bandi, S.R.; Calabrese, C.R.; Vande Walle, L.; Lamkanfi, M.; et al. Dietary modulation of the microbiome affects autoinflammatory disease. Nature 2014, 516, 246–249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Brito, B.B.; da Silva, F.A.F.; Soares, A.S.; Pereira, V.A.; Santos, M.L.C.; Sampaio, M.M.; Neves, P.H.M.; de Melo, F.F. Pathogenesis and clinical management of Helicobacter pylori gastric infection. World J. Gastroenterol. 2019, 25, 5578–5589. [Google Scholar] [CrossRef]
- Yee, J.K.C. Are the view of Helicobacter pylori colonized in the oral cavity an illusion? Exp. Mol. Med. 2017, 49, e397. [Google Scholar] [CrossRef] [Green Version]
- Brown, L.M. Helicobacter pylori: Epidemiology and routes of transmission. Epidemiol. Rev. 2000, 22, 283–297. [Google Scholar] [CrossRef]
- Loster, B.W.; Majewski, S.W.; Czesnikiewicz-Guzik, M.; Bielanski, W.; Pierzchalski, P.; Konturek, S.J. The relationship between the presence of Helicobacter pylori in the oral cavity and gastric in the stomach. J. Physiol.Pharmacol. 2006, 57 (Suppl. 3), 91–100. [Google Scholar] [PubMed]
- Namiot, D.B.; Namiot, Z.; Kemona, A.; Bucki, R.; Gotebiewska, M. Oral health status and oral hygiene practices of patients with peptic ulcer and how these affect Helicobacter pylori eradication from the stomach. Helicobacter 2007, 12, 63–67. [Google Scholar] [CrossRef]
- Jia, C.L.; Jiang, G.S.; Li, C.H.; Li, C.R. Effect of dental plaque control on infection of Helicobacter pylori in gastric mucosa. Tex. Dent. J. 2012, 129, 1069–1073. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Regis, W.; Reis, A.; Rocha, F.; Guedes, S.; Maia, D.; Rodrigues, L. The Role of Candida Albicans and Streptococcus Mutans Spent Culture Supernatant in Single and Dual-Species Biofilm. J. Sci. Tech. Res. 2019, 14, 1–3. [Google Scholar]
- Falsetta, M.L.; Klein, M.I.; Colonne, P.M.; Scott-Anne, K.; Gregoire, S.; Pai, C.H.; Gonzalez-Begne, M.; Watson, G.; Krysan, D.J.; Bowen, W.H.; et al. Symbiotic relationship between Streptococcus mutans and Candida albicans synergizes virulence of plaque biofilms in vivo. Infect. Immun. 2014, 82, 1968–1981. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dashper, S.G.; Mitchell, H.L.; Le Cao, K.A.; Carpenter, L.; Gussy, M.G.; Calache, H.; Gladman, S.L.; Bulach, D.M.; Hoffmann, B.; Catmull, D.V.; et al. Temporal development of the oral microbiome and prediction of early childhood caries. Sci. Rep. 2019, 9, 19732. [Google Scholar] [CrossRef] [Green Version]
- Baker, J.L.; He, X.; Shi, W. Precision Reengineering of the Oral Microbiome for Caries Management. Adv. Dent. Res. 2019, 30, 34–39. [Google Scholar] [CrossRef]
- Krzysciak, W.; Koscielniak, D.; Papiez, M.; Vyhouskaya, P.; Zagorska-Swiezy, K.; Kolodziej, I.; Bystrowska, B.; Jurczak, A. Effect of a Lactobacillus Salivarius Probiotic on a Double-Species Streptococcus Mutans and Candida Albicans Caries Biofilm. Nutrients 2017, 9, 1242. [Google Scholar] [CrossRef] [Green Version]
- Surdu, A.E.; Popa, C.; Luchian, I. Identification of bacteria involved in periodontal disease using molecular biology techniques. Rev. Chim. 2017, 68, 2407–2412. [Google Scholar]
- Teodorescu, A.C.; Teslaru, S.; Solomon, S.M.; Zetu, L.; Luchian, I.; Sioustis, I.-A.; Martu, M.A.; Vasiliu, B.; Martu, S. Assessment of Bacterial Associations Involved in Periodontal Disease Using Crevicular Fluid. Rev. Chim. 2019, 70, 2145–2149. [Google Scholar] [CrossRef]
- Olsen, I.; Lambris, J.D.; Hajishengallis, G. Porphyromonas gingivalis disturbs host-commensal homeostasis by changing complement function. J. Oral Microbiol. 2017, 9, 1340085. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Irshad, M.; van der Reijden, W.A.; Crielaard, W.; Laine, M.L. In vitro invasion and survival of Porphyromonas gingivalis in gingival fibroblasts; role of the capsule. Arch. Immunol. Ther Exp. 2012, 60, 469–476. [Google Scholar] [CrossRef] [PubMed]
- How, K.Y.; Song, K.P.; Chan, K.G. Porphyromonas gingivalis: An Overview of Periodontopathic Pathogen below the Gum Line. Front. Microbiol. 2016, 7, 53. [Google Scholar] [CrossRef] [Green Version]
- Amano, A. Bacterial adhesins to host components in periodontitis. Periodontol 2000 2010, 52, 12–37. [Google Scholar] [CrossRef]
- Xiao, L.; Zhang, Q.; Peng, Y.; Wang, D.; Liu, Y. The effect of periodontal bacteria infection on incidence and prognosis of cancer: A systematic review and meta-analysis. Medicine 2020, 99, e19698. [Google Scholar] [CrossRef] [PubMed]
- Olsen, I.; Yilmaz, O. Possible role of Porphyromonas gingivalis in orodigestive cancers. J. Oral Microbiol. 2019, 11, 1563410. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, Y.W. Fusobacterium nucleatum: A commensal-turned pathogen. Curr. Opin. Microbiol. 2015, 23, 141–147. [Google Scholar] [CrossRef] [Green Version]
- Loozen, G.; Ozcelik, O.; Boon, N.; De Mol, A.; Schoen, C.; Quirynen, M.; Teughels, W. Inter-bacterial correlations in subgingival biofilms: A large-scale survey. J. Clin. Periodontol. 2014, 41, 12167. [Google Scholar] [CrossRef]
- Han, Y.W.; Wang, X. Mobile microbiome: Oral bacteria in extra-oral infections and inflammation. J. Dent. Res. 2013, 92, 485–491. [Google Scholar] [CrossRef] [Green Version]
- Bashir, A.; Miskeen, A.Y.; Hazari, Y.M.; Asrafuzzaman, S.; Fazili, K.M. Fusobacterium nucleatum, inflammation, and immunity: The fire within human gut. Tumor Biol. 2016, 37, 2805–2810. [Google Scholar] [CrossRef] [PubMed]
- Cao, P.; Chen, Y.; Chen, Y.; Su, W.; Zhan, N.; Dong, W. Fusobacterium nucleatum Activates Endoplasmic Reticulum Stress to Promote Crohn’s Disease Development via the Upregulation of CARD3 Expression. Front. Pharmacol. 2020, 11, 106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Corona, P.S.; Lung, M.; Rodriguez-Pardo, D.; Pigrau, C.; Soldado, F.; Amat, C.; Carrera, L. Acute periprosthetic joint infection due to Fusobacterium nucleatum in a non-immunocompromised patient. Failure using a Debridement, Antibiotics + Implant retention approach. Anaerobe 2018, 49, 116–120. [Google Scholar] [CrossRef]
- Hintze, T.; Steed, M.; Sievers, E.; Bagwell, J.T.; Selfa, N. Primary meningitis due to Fusobacterium nucleatum successfully treated with ceftriaxone in a healthy adult male. IDCases 2019, 18, e00616. [Google Scholar] [CrossRef] [PubMed]
- Hattori, T.; Yuasa, N.; Ikegami, S.; Nishiyama, H.; Takeuchi, E.; Miyake, H.; Kuno, R.; Miyata, K.; Fujino, M.; Minami, M. Culture-based bacterial evaluation of the appendix lumen in patients with and without acute appendicitis. J. Infect. Chemother. 2019, 25, 708–713. [Google Scholar] [CrossRef]
- Vander Haar, E.L.; So, J.; Gyamfi-Bannerman, C.; Han, Y.W. Fusobacterium nucleatum and adverse pregnancy outcomes: Epidemiological and mechanistic evidence. Anaerobe 2018, 50, 55–59. [Google Scholar] [CrossRef]
- Amer, A.; Galvin, S.; Healy, C.M.; Moran, G.P. The Microbiome of Potentially Malignant Oral Leukoplakia Exhibits Enrichment for Fusobacterium, Leptotrichia, Campylobacter, and Rothia Species. Front Microbiol. 2017, 8, 2391. [Google Scholar] [CrossRef] [Green Version]
- Brennan, C.A.; Garrett, W. Fusobacterium nucleatum-symbiont, opportunist and oncobacterium. Nat. Rev. Microbiol. 2019, 17, 156–166. [Google Scholar] [CrossRef]
- Coppenhagen-Glazer, S.; Sol, A.; Abed, J.; Naor, R.; Zhang, X.; Han, Y.W.; Bachrach, G. Fap2 of Fusobacterium nucleatum Is a Galactose-Inhibitable Adhesin Involved in Coaggregation, Cell Adhesion, and Preterm Birth. Infect. Immun. 2015, 83, 1104–1113. [Google Scholar] [CrossRef] [Green Version]
- Park, S.R.; Kim, D.J.; Han, S.H.; Kang, M.J.; Lee, J.Y.; Jeong, Y.J.; Lee, S.J.; Kim, T.H.; Ahn, S.G.; Yoon, J.H.; et al. Diverse Toll-like receptors mediate cytokine production by Fusobacterium nucleatum and Aggregatibacter actinomycetemcomitans in macrophages. Infect. Immun. 2014, 82, 1914–1920. [Google Scholar] [CrossRef] [Green Version]
- Chaushu, S.; Wilensky, A.; Gur, C.; Shapira, L.; Elboim, M.; Halftek, G.; Polak, D.; Achdout, H.; Bachrach, G.; Mandelboim, O. Direct recognition of Fusobacterium nucleatum by the NK cell natural cytotoxicity receptor NKp46 aggravates periodontal disease. PLoS Pathog. 2012, 8, e1002601. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, X.; Zhang, L.; Xu, L.; Meng, H.; Lu, R.; Chen, Z.; Shi, D.; Wang, X. Detection of eight periodontal microorganisms and distribution of Porphyromonas gingivalis fimA genotypes in Chinese patients with aggressive periodontitis. J. Periodontol. 2014, 85, 150–159. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Tan, L.; Wang, H.; Kou, Y.; Shi, X.; Zhang, S.; Pan, Y. Fusobacterium nucleatum Interaction with Pseudomonas aeruginosa Induces Biofilm-Associated Antibiotic Tolerance via Fusobacterium Adhesin A. ACS Infect. Dis. 2020, 6, 1686–1696. [Google Scholar] [CrossRef] [PubMed]
- Yu, T.; Guo, F.; Yu, Y.; Sun, T.; Ma, D.; Han, J.; Qian, Y.; Kryczek, I.; Sun, D.; Nagarsheth, N.; et al. Fusobacterium nucleatum Promotes Chemoresistance to Colorectal Cancer by Modulating Autophagy. Cell 2017, 170, 548–563.e16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Binder Gallimidi, A.; Fischman, S.; Revach, B.; Bulvik, R.; Maliutina, A.; Rubinstein, A.M.; Nussbaum, G.; Elkin, M. Periodontal pathogens Porphyromonas gingivalis and Fusobacterium nucleatum promote tumor progression in an oral-specific chemical carcinogenesis model. Oncotarget 2015, 6, 22613–22623. [Google Scholar] [CrossRef] [Green Version]
| Characterization Method | Description | References |
|---|---|---|
| Targeted Gene Analysis | Analysis involving the use of one or a couple hypervariable regions. This is the most common analysis and is best used for highly specific sequencing. | [26,27,28] |
| Shotgun Metagenomics | A widespread, untargeted approach that incorporates a wide variety of genetic information. This analysis is best used in the absence of reference genome. | [33,34,35] |
| Metabolomics | A comprehensive analysis which utilizes mass spectrometry for characterization. This is best used to gain information on the role of small metabolites in cell function. | [38,39] |
| Metraproteomics | Analysis using mass spectrometry to provide information on macromolecules. This should be used when inquiring about protein interaction with the system. | [26,40] |
| Metratransciptomics | An approach which utilizes microbes within their natural environment to provide additional information on the overall function of the community | [41] |
| Microbial Culturomics | A method used in conjunction with other characterization methods to provide information of unknown species through their culturing methods. Experimental design is critical. | [42] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Morrison, A.G.; Sarkar, S.; Umar, S.; Lee, S.T.M.; Thomas, S.M. The Contribution of the Human Oral Microbiome to Oral Disease: A Review. Microorganisms 2023, 11, 318. https://doi.org/10.3390/microorganisms11020318
Morrison AG, Sarkar S, Umar S, Lee STM, Thomas SM. The Contribution of the Human Oral Microbiome to Oral Disease: A Review. Microorganisms. 2023; 11(2):318. https://doi.org/10.3390/microorganisms11020318
Chicago/Turabian StyleMorrison, Austin Gregory, Soumyadev Sarkar, Shahid Umar, Sonny T. M. Lee, and Sufi Mary Thomas. 2023. "The Contribution of the Human Oral Microbiome to Oral Disease: A Review" Microorganisms 11, no. 2: 318. https://doi.org/10.3390/microorganisms11020318
APA StyleMorrison, A. G., Sarkar, S., Umar, S., Lee, S. T. M., & Thomas, S. M. (2023). The Contribution of the Human Oral Microbiome to Oral Disease: A Review. Microorganisms, 11(2), 318. https://doi.org/10.3390/microorganisms11020318







