The Immune, Inflammatory and Hematological Response in COVID-19 Patients, According to the Severity of the Disease
Abstract
:1. Introduction
2. Material and Methods
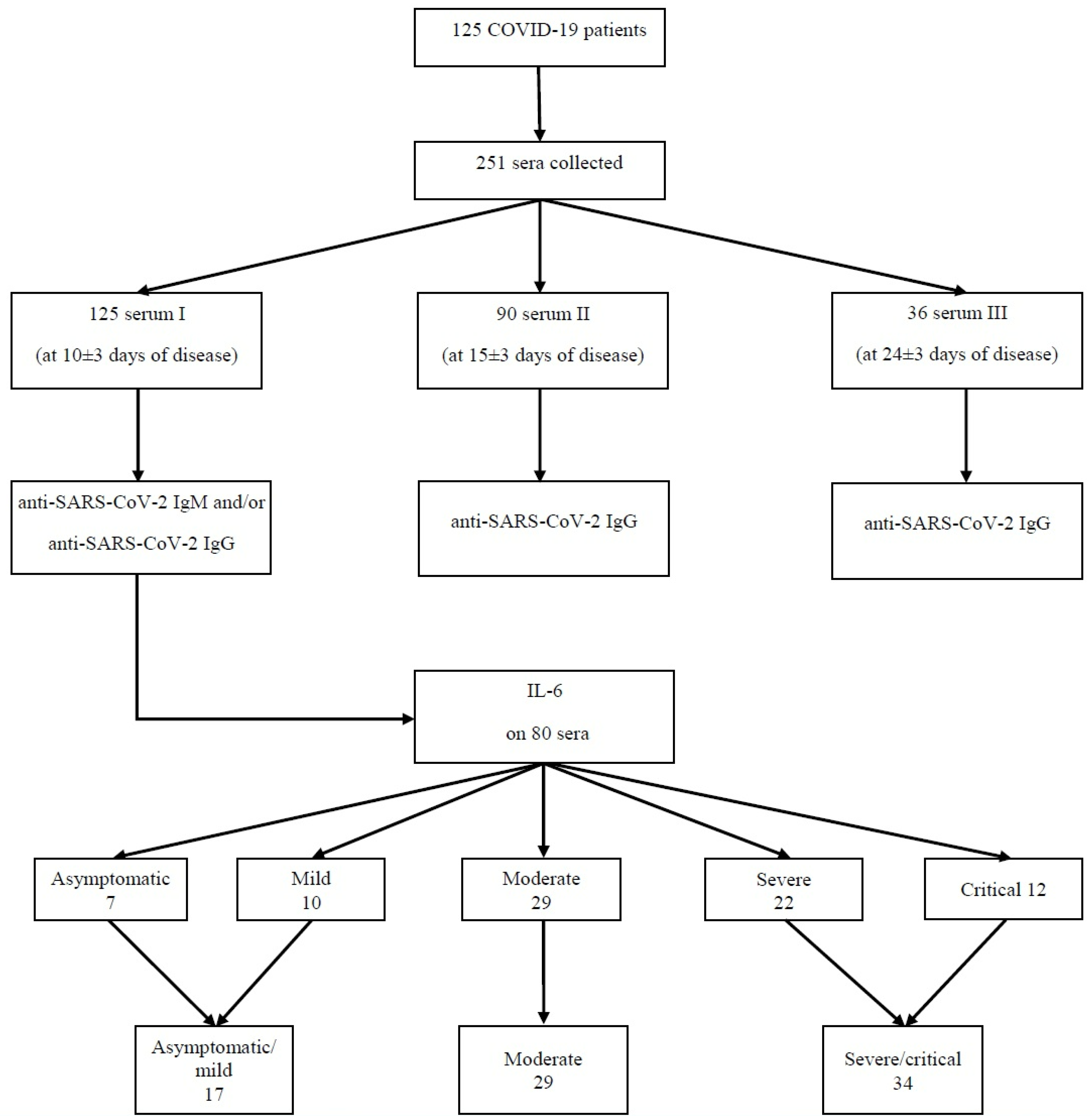
2.1. Study Design and Participants
2.2. Collection of Samples
2.3. Analysis of Samples
2.4. Ethical Principles
2.5. Statistical Analysis
3. Results
4. Discussion
4.1. Strengths of the Study
4.2. Limitations of the Study
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Guo, L.; Ren, L.; Yang, S.; Xiao, M.; Chang, D.; Yang, F.; Dela Cruz, C.S.; Wang, Y.; Wu, C.; Xiao, Y.; et al. Profiling Early Humoral Response to Diagnose Novel Coronavirus Disease (COVID-19). Clin. Infect. Dis. 2020, 71, 778–785. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, W.; Xu, Y.; Gao, R.; Lu, R.; Han, K.; Wu, G.; Tan, W. Detection of SARS-CoV-2 in Different Types of Clinical Specimens. JAMA 2020, 323, 1843–1844. [Google Scholar] [CrossRef] [Green Version]
- Bawiskar, N.; Talwar, D.; Acharya, S.; Kumar, S. Hematological Manifesta-tions of COVID-19 and Their Prognostic Significance in an Intensive Care Unit: A Cross-Sectional Study. Cureus 2021, 13, e19887. [Google Scholar] [CrossRef] [PubMed]
- Theel, E.S.; Slev, P.; Wheeler, S.; Couturier, M.R.; Wong, S.J.; Kadkhoda, K. The Role of Antibody Testing for SARS-CoV-2: Is There One? J. Clin. Microbiol. 2020, 58, e00797-20. [Google Scholar] [CrossRef] [PubMed]
- European Centre for Disease Prevention and Control (ECDC). Immune Responses and Immunity to SARS-CoV-2. Available online: https://www.ecdc.europa.eu/en/covid-19/latest-evidence/immune-responses (accessed on 18 July 2022).
- Ma, H.; Zeng, W.; He, H.; Zhao, D.; Jiang, D.; Zhou, P.; Cheng, L.; Li, Y.; Ma, X.; Jin, T. Serum IgA, IgM, and IgG responses in COVID-19. Cel. Molec. Immunol. 2020, 17, 773–775. [Google Scholar] [CrossRef] [PubMed]
- Wang, P. Significance of IgA antibody testing for early detection of SARS-CoV-2. J. Med. Virol. 2021, 93, 1888–1889. [Google Scholar] [CrossRef]
- Infantino, M.; Manfredi, M.; Grossi, V.; Lari, B.; Fabbri, S.; Benucci, M.; Fortini, A.; Damiani, A.; Mobilia, E.M.; Panciroli, M.; et al. Closing the serological gap in the diagnostic testing for COVID-19: The value of anti-SARS-CoV-2 IgA antibodies. J. Med. Virol. 2021, 93, 1436–1442. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.Q.; Sun, B.Q.; Fang, Z.F.; Zhao, J.C.; Liu, X.Y.; Li, Y.M.; Sun, X.Z.; Liang, H.F.; Zhong, B.; Huang, Z.F.; et al. Distinct features of SARS-CoV-2-specific IgA response in COVID-19 patients. Europ. Resp. J. 2020, 56, 2001526. [Google Scholar] [CrossRef]
- Sabaka, P.; Koščálová, A.; Straka, I.; Hodosy, J.; Lipták, R.; Kmotorková, B.; Kachlíková, M.; Kušnírová, A. Role of interleukin 6 as a predictive factor for a severe course of COVID-19: Retrospective data analysis of patients from a long-term care facility during COVID-19 outbreak. BMC Infect. Dis. 2021, 21, 308. [Google Scholar] [CrossRef] [PubMed]
- Herold, T.; Jurinovic, V.; Arnreich, C.; Lipworth, B.J.; Hellmuth, J.C.; von Bergwelt-Baildon, M.; Klein, M.; Weinberger, T. Elevated levels of IL-6 and CRP predict the need for mechanical ventilation in COVID-19. J. Allergy Clin. Immunol. 2020, 146, 128–136.e4. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Hao, Y.; Ou, W.; Ming, F.; Liang, G.; Qian, Y.; Cai, Q.; Dong, S.; Hu, S.; Wang, W.; et al. Serum interleukin-6 is an indicator for severity in 901 patients with SARS-CoV-2 infection: A cohort study. J. Transl. Med. 2020, 18, 406. [Google Scholar] [CrossRef]
- Velavan, T.P.; Meyer, C.G. Mild versus severe COVID-19: Laboratory markers. Int. J. Infect. Dis. 2020, 95, 304–307. [Google Scholar] [CrossRef]
- Hachim, M.Y.; Hachim, I.Y.; Naeem, K.B.; Hannawi, H.; Salmi, I.A.; Hannawi, S. D-dimer, Troponin, and Urea Level at Presentation With COVID-19 can Predict ICU Admission: A Single Centered Study. Front. Med. 2020, 7, 585003. [Google Scholar] [CrossRef] [PubMed]
- Man, M.A.; Rajnoveanu, R.M.; Motoc, N.S.; Bondor, C.I.; Chis, A.F.; Lesan, A.; Puiu, R.; Lucaciu, S.R.; Dantes, E.; Gergely-Domokos, B.; et al. Neutrophil-to-lymphocyte ratio, platelets-to-lymphocyte ratio, and eosinophils correlation with high-resolution computer tomography severity score in COVID-19 patients. PloS ONE 2021, 16, e0252599. [Google Scholar] [CrossRef] [PubMed]
- Hulkoti, V.S.; Acharya, S.; Kumar, S.; Talwar, D.; Khanna, S.; Annadatha, A.; Madaan, S.; Verma, V.; Sagar, V.V.S.S. Association of serum ferritin with COVID-19 in a cross-sectional study of 200 intensive care unit patients in a rural hospital: Is ferritin the forgotten biomarker of mortality in severe COVID-19? J. Fam. Med. Prim. Care 2022, 11, 2045–2050. [Google Scholar] [CrossRef]
- World Health Organization (WHO). Coronavirus Disease (COVID-19) Pandemic. Available online: https://www.who.int/emergencies/diseases/novel-coronavirus-2019 (accessed on 22 March 2021).
- National Institute for Health and Care Research (NIHR). Living with COVID-19—Second Review. 2021. Available online: https://evidence.nihr.ac.uk/themedreview/living-with-covid19-second-review/ (accessed on 25 March 2022).
- Petersen, L.R.; Sami, S.; Vuong, N.; Pathela, P.; Weiss, D.; Morgenthau, B.M.; Henseler, R.A.; Daskalakis, D.C.; Atas, J.; Patel, A.; et al. Lack of Antibodies to Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) in a Large Cohort of Previously Infected Persons. Clin. Infect.Dis. 2021, 73, e3066–e3073. [Google Scholar] [CrossRef]
- Malkova, A.; Kudryavtsev, I.; Starshinova, A.; Kudlay, D.; Zinchenko, Y.; Glushkova, A.; Yablonskiy, P.; Shoenfeld, Y. Post COVID-19 Syndrome in Patients with Asymptomatic/Mild Form. Pathogens 2021, 10, 1408. [Google Scholar] [CrossRef]
- Carod-Artal, F.J. Post-COVID-19 syndrome: Epidemiology, diagnostic criteria and pathogenic mechanisms involved. Rev. Neurol. 2021, 72, 384–396. [Google Scholar] [CrossRef] [PubMed]
- Ladds, E.; Rushforth, A.; Wieringa, S.; Taylor, S.; Rayner, C.; Husain, L.; Greenhalgh, T. Persistent symptoms after COVID-19: Qualitative study of 114 “long Covid” patients and draft quality principles for services. BMC Health Serv. Res. 2020, 20, 1144. [Google Scholar] [CrossRef]
- Office of National Statistics (ONS). Coronavirus (COVID-19) Infection Survey, UK Statistical Bulletins. Available online: https://www.ons.gov.uk/peoplepopulationandcommunity/healthandsocialcare/conditionsanddiseases/bulletins/coronaviruscovid19infectionsurveypilot/previousReleases (accessed on 25 March 2022).
- Covino, M.; De Matteis, G.; Polla, D.A.D.; Santoro, M.; Burzo, M.L.; Torelli, E.; Simeoni, B.; Russo, A.; Sandroni, C.; Gasbarrini, A.; et al. Predictors of in-hospital mortality AND death RISK STRATIFICATION among COVID-19 PATIENTS aged ≥ 80 YEARs OLD. Arch. Geront. Geriat. 2021, 95, 104383. [Google Scholar] [CrossRef] [PubMed]
- Fagard, K.; Gielen, E.; Deschodt, M.; Devriendt, E.; Flamaing, J. Risk factors for severe COVID-19 disease and death in patients aged 70 and over: A retrospective observational cohort study. Acta Clin. Belg. 2022, 77, 487–494. [Google Scholar] [CrossRef] [PubMed]
- Kompaniyets, L.; Goodman, A.B.; Belay, B.; Freedman, D.S.; Sucosky, M.S.; Lange, S.J.; Gundlapalli, A.V.; Boehmer, T.K.; Blanck, H.M. Body Mass Index and Risk for COVID-19-Related Hospitalization, Intensive Care Unit Admission, Invasive Mechanical Ventilation, and Death–United States, March-December 2020. MMWR. Morb. Mortal. Wkly. Rep. 2021, 70, 355–361. [Google Scholar] [CrossRef] [PubMed]
- Peckham, H.; de Gruijter, N.M.; Raine, C.; Radziszewska, A.; Ciurtin, C.; Wedderburn, L.R.; Rosser, E.C.; Webb, K.; Deakin, C.T. Male sex identified by global COVID-19 meta-analysis as a risk factor for death and ITU admission. Nat. Commun. 2021, 11, 6317. [Google Scholar] [CrossRef] [PubMed]
- Silva, I.; Faria, N.C.; Ferreira, Á.R.S.; Anastácio, L.R.; Ferreira, L.G. Risk factors for critical illness and death among adult Brazilians with COVID-19. Rev. Soc. Bras. Med. Trop. 2021, 54, e0014-2021. [Google Scholar] [CrossRef] [PubMed]
- Telle, K.E.; Grøsland, M.; Helgeland, J.; Håberg, S.E. Factors associated with hospitalization, invasive mechanical ventilation treatment and death among all confirmed COVID-19 cases in Norway: Prospective cohort study. Scand. J. Pubic Health 2021, 49, 41–47. [Google Scholar] [CrossRef]
- Grasselli, G.; Greco, M.; Zanella, A.; Albano, G.; Antonelli, M.; Bellani, G.; Bonanomi, E.; Cabrini, L.; Carlesso, E.; COVID-19 Lombardy ICU Network; et al. Risk Factors Associated With Mortality Among Patients With COVID-19 in Intensive Care Units in Lombardy, Italy. JAMA Intern. Med. 2020, 180, 1345–1355. [Google Scholar] [CrossRef]
- Sakowicz, A.; Ayala, A.E.; Ukeje, C.C.; Witting, C.S.; Grobman, W.A.; Miller, E.S. Risk factors for severe acute respiratory syndrome coronavirus 2 infection in pregnant women. Am. J. Obstet. Gynecol. MFM 2020, 2, 100198. [Google Scholar] [CrossRef]
- Mathur, R.; Rentsch, C.T.; Morton, C.E.; Hulme, W.J.; Schultze, A.; MacKenna, B.; Eggo, R.M.; Bhaskaran, K.; Wong, A.Y.S.; Williamson, E.J.; et al. Ethnic differences in SARS-CoV-2 infection and COVID-19-related hospitalisation, intensive care unit admission, and death in 17 million adults in England: An observational cohort study using the OpenSAFELY platform. Lancet 2021, 397, 1711–1724. [Google Scholar] [CrossRef]
- Choi, Y.J.; Park, J.Y.; Lee, H.S.; Suh, J.; Song, J.Y.; Byun, M.K.; Cho, J.H.; Kim, H.J.; Lee, J.H.; Park, J.W.; et al. Effect of asthma and asthma medication on the prognosis of patients with COVID-19. Eur. Resp. J. 2021, 57, 2002226. [Google Scholar] [CrossRef]
- Khunti, K.; Knighton, P.; Zaccardi, F.; Bakhai, C.; Barron, E.; Holman, N.; Kar, P.; Meace, C.; Sattar, N.; Sharp, S.; et al. Prescription of glucose-lowering therapies and risk of COVID-19 mortality in people with type 2 diabetes: A nationwide observational study in England. Lancet Diabetes Endocrinol. 2021, 9, 293–303. [Google Scholar] [CrossRef]
- Xia, Y.; Jin, R.; Zhao, J.; Li, W.; Shen, H. Risk of COVID-19 for patients with cancer. Lancet Oncol. 2020, 21, e180. [Google Scholar] [CrossRef] [PubMed]
- Rüthrich, M.M.; Giessen-Jung, C.; Borgmann, S.; Classen, A.Y.; Dolff, S.; Grüner, B.; Hanses, F.; Isberner, N.; Köhler, P.; Lanznaster, J.; et al. COVID-19 in cancer patients: Clinical characteristics and outcome-an analysis of the LEOSS registry. Ann. Hematol. 2021, 100, 383–393. [Google Scholar] [CrossRef]
- Pilgram, L.; Eberwein, L.; Wille, K.; Koehler, F.C.; Stecher, M.; Rieg, S.; Kielstein, J.T.; Jakob, C.E.M.; Rüthrich, M.; Burst, V.; et al. Clinical course and predictive risk factors for fatal outcome of SARS-CoV-2 infection in patients with chronic kidney disease. Infection 2021, 49, 725–737. [Google Scholar] [CrossRef]
- Huber, M.K.; Raichle, C.; Lingor, P.; Synofzik, M.; Borgmann, S.; Erber, J.; Tometten, L.; Rimili, W.; Dolff, S.; Wille, K.; et al. Outcomes of SARS-CoV-2 Infections in Patients with Neurodegenerative Diseases in the LEOSS Cohort. Mov. Disord. 2021, 36, 791–793. [Google Scholar] [CrossRef]
- Wehbe, Z.; Hammoud, S.H.; Yassine, H.M.; Fardoun, M.; El-Yazbi, A.F.; Eid, A.H. Molecular and Biological Mechanisms Underlying Gender Differences in COVID-19 Severity and Mortality. Front. Immunol. 2021, 12, 659339. [Google Scholar] [CrossRef]
- Wanschel, A.C.B.A.; Moretti, A.I.S.; Ouimet, M. Editorial: COVID-19 Mechanisms on Cardio-Vascular Dysfunction: From Membrane Receptors to Immune Response. Front. Cardiovasc. Med. 2021, 8, 686495. [Google Scholar] [CrossRef] [PubMed]
- Pesce, M.; Agostoni, P.; Bøtker, H.E.; Brundel, B.; Davidson, S.M.; Caterina, R.; Ferdinandy, P.; Girao, H.; Gyöngyösi, M.; Hulot, J.S.; et al. COVID-19-related cardiac complications from clinical evidences to basic mechanisms: Opinion paper of the ESC Working Group on Cellular Biology of the Heart. Cardiovasc. Resp. 2021, 117, 2148–2160. [Google Scholar] [CrossRef]
- Sun, B.; Feng, Y.; Mo, X.; Zheng, P.; Wang, Q.; Li, P.; Peng, P.; Liu, X.; Chen, Z.; Huang, H.; et al. Kinetics of SARS-CoV-2 specific IgM and IgG responses in COVID-19 patients. Emerg. Microbes. Infect. 2020, 9, 940–948. [Google Scholar] [CrossRef]
- Hambali, N.L.; Mohd Noh, M.; Paramasivam, S.; Chua, T.H.; Hayati, F.; Payus, A.O.; Tee, T.Y.; Rosli, K.T.; Abd Rachman Isnadi, M.F.; Manin, B.O. A Non-severe Coronavirus Disease 2019 Patient With Persistently High Interleukin-6 Level. Front. Public Health 2020, 8, 584552. [Google Scholar] [CrossRef]
- Gu, X.; Sha, L.; Zhang, S.; Shen, D.; Zhao, W.; Yi, Y. Neutrophils and Lymphocytes Can Help Distinguish Asymptomatic COVID-19 From Moderate COVID-19. Front. Cell. Infect Microbiol. 2021, 11, 654272. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Boon, A.; Michelson, A.P.; Foraker, R.E.; Zhan, M.; Payne, P. Estrogen Hormone Is an Essential Sex Factor Inhibiting Inflammation and Immune Response in COVID-19. Res. Sq. 2021, preprint, rs.3.rs-936900. [Google Scholar] [CrossRef]
- Hou, H.; Zhang, B.; Huang, H.; Luo, Y.; Wu, S.; Tang, G.; Liu, W.; Mao, L.; Mao, L.; Wang, F.; et al. Using IL-2R/lymphocytes for predicting the clinical progression of patients with COVID-19. Clin. Exp. Immunol. 2020, 201, 76–84. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.S.; Shu, T.; Kang, L.; Wu, D.; Zhou, X.; Liao, B.W.; Sun, X.L.; Zhou, X.; Wang, Y.Y. Temporal profiling of plasma cytokines, chemokines and growth factors from mild, severe and fatal COVID-19 patients. Signal Transduct Target Ther. 2020, 5, 100. [Google Scholar] [CrossRef] [PubMed]
- Talwar, D.; Kumar, S.; Acharya, S.; Raisinghani, N.; Madaan, S.; Hulkoti, V.; Akhilesh, A.; Khanna, S.; Shah, D.; Nimkar, S. Interleukin 6 and Its Correlation with COVID-19 in Terms of Outcomes in an Intensive Care Unit of a Rural Hospital:A Cross-sectional Study. Indian J. Crit. Care. Med. 2022, 26, 39–42. [Google Scholar] [CrossRef] [PubMed]
- Ding, R.; Yang, Z.; Huang, D.; Wang, Y.; Li, X.; Zhou, X.; Yan, L.; Lu, W.; Zhang, Z. Identification of parameters in routine blood and coagulation tests related to the severity of COVID-19. Int. J. Med. Sci. 2021, 18, 1207–1215. [Google Scholar] [CrossRef] [PubMed]
- Mo, P.; Xing, Y.; Xiao, Y.; Deng, L.; Zhao, Q.; Wang, H.; Xiong, Y.; Cheng, Z.; Gao, S.; Liang, K.; et al. Clinical Characteristics of Refractory Coronavirus Disease 2019 in Wuhan, China. Clin. Infect. Dis. 2021, 73, e4208–e4213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef] [Green Version]
- Wu, C.; Chen, X.; Cai, Y.; Xia, J.; Zhou, X.; Xu, S.; Huang, H.; Zhang, L.; Zhou, X.; Du, C.; et al. Risk Factors Associated With Acute Respiratory Distress Syndrome and Death in Patients With Coronavirus Disease 2019 Pneumonia in Wuhan, China. JAMA Intern. Med. 2020, 180, 934–943. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, L.; Liu, H.G.; Liu, W.; Liu, J.; Liu, K.; Shang, J.; Deng, Y.; Wei, S. [Analysis of clinical features of 29 patients with 2019 novel coronavirus pneumonia]. Zhonghua Jie He He Hu Xi Za Zhi Chin. J. Tuberc. Respir. Dis. 2020, 43, E005. [Google Scholar] [CrossRef]
- Gao, Y.; Li, T.; Han, M.; Li, X.; Wu, D.; Xu, Y.; Zhu, Y.; Liu, Y.; Wang, X.; Wang, L. Diagnostic utility of clinical laboratory data determinations for patients with the severe COVID-19. J. Med. Virol. 2020, 92, 791–796. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.J.; Dong, X.; Cao, Y.Y.; Yuan, Y.D.; Yang, Y.B.; Yan, Y.Q.; Akdis, C.A.; Gao, Y.D. Clinical characteristics of 140 patients infected with SARS-CoV-2 in Wuhan, China. Allergy 2020, 75, 1730–1741. [Google Scholar] [CrossRef] [PubMed]
- Li, L.Q.; Huang, T.; Wang, Y.Q.; Wang, Z.P.; Liang, Y.; Huang, T.B.; Zhang, H.Y.; Sun, W.; Wang, Y. COVID-19 patients’ clinical characteristics, discharge rate, and fatality rate of meta-analysis. J. Med. Virol. 2020, 92, 577–583. [Google Scholar] [CrossRef]
- Li, Y.Y.; Wang, W.N.; Lei, Y.; Zhang, B.; Yang, J.; Hu, J.W.; Ren, Y.L.; Lu, Q.F. Comparison of the clinical characteristics between RNA positive and negative patients clinically diagnosed with coronavirus disease 2019. Zhonghua Jie He He Hu Xi Za Zhi Chin. J. Tuberc. Respir. Dis. 2020, 43, 427–430. [Google Scholar] [CrossRef]
- Xia, T.; Zhang, W.; Xu, Y.; Wang, B.; Yuan, Z.; Wu, N.; Xiang, Y.; Li, C.; Shan, Y.; Xie, W.; et al. Early kidney injury predicts disease progression in patients with COVID-19: A cohort study. BMC Infect. Dis. 2021, 21, 1012. [Google Scholar] [CrossRef] [PubMed]
- Xiang, H.X.; Fei, J.; Xiang, Y.; Xu, Z.; Zheng, L.; Li, X.Y.; Fu, L.; Zhao, H. Renal dysfunction and prognosis of COVID-19 patients: A hospital-based retrospective cohort study. BMC Infect. Dis. 2021, 21, 158. [Google Scholar] [CrossRef]
- Sanyaolu, A.; Okorie, C.; Marinkovic, A.; Patidar, R.; Younis, K.; Desai, P.; Hosein, Z.; Padda, I.; Mangat, J.; Altaf, M. Comorbidity and its Impact on Patients with COVID-19. SN Compr. Clin. Med. 2020, 2, 1069–1076. [Google Scholar] [CrossRef] [PubMed]
- Franki, R. Comorbidities the Rule in New York’s COVID-19 Deaths. Hospitalist. 2020. Available online: https://www.the-hospitalist.org/hospitalist/article/220457/coronavirus-updates/comorbidities-rule-new-yorks-covid-19-deaths (accessed on 12 January 2021).
| Severity of Disease/ Blood Parameter | Unit | Mild/ Asymptomatic | Moderate | Severe/ Critical | Reference Interval |
|---|---|---|---|---|---|
| IL-6 | pg/mL | 19.57 | 73.26 | 149.36 | 0–7 |
| Anti-SARS-CoV-2 IgG | AU/mL | 25.82 | 36.81 | 64.17 | <1 |
| Leucocyte count | * 103/µL | 6.09 | 6.54 | 8.39 | 4–10 |
| Neutrophil count | * 103/µL | 3.64 | 4.66 | 6.77 | 2–8 |
| % neutrophils | 57.49 | 64.85 | 74.59 | 45–80 | |
| Lymphocyte count | * 103/µL | 1.82 | 1.29 | 1.01 | 1–4 |
| % lymphocytes | 30.79 | 25.28 | 16.09 | 20–45 | |
| Eosinophil count | * 103/µL | 0.09 | 0.05 | 0.03 | 0–0.5 |
| % eosinophils | 1.78 | 0.80 | 0.48 | 0–5 | |
| Hemoglobin | g/dL | 13.27 | 12.51 | 11.99 | 11.7–17.3 |
| Platelet count | * 103/µL | 247.41 | 221.59 | 234.53 | 150–380 |
| NLR | 2.32 | 4.01 | 8.26 | ||
| PLR | 153.60 | 185.72 | 257.77 | ||
| CRP | mg/L | 19.83 | 41.96 | 116.67 | 0–5 |
| Glucose | mg/dL | 143.06 | 115.66 | 142.32 | 70–115 |
| ALT | UI/L | 78.47 | 46.21 | 46.94 | 5–40 |
| AST | UI/L | 45.71 | 38.83 | 61 | 5–37 |
| Urea | mg/dL | 37.94 | 45.52 | 75.18 | 15–50 |
| Creatinine | mg/dL | 1.25 | 1.48 | 2.28 | 0.5–0.9 |
| eGFR | mL/min/1.73 m2 | 77.68 | 76.42 | 61.57 | 90–120 |
| LOS | days | 22.76 | 27.34 | 24.77 |
| Blood Parameters | Unit | Mean | Median | Standard Deviation | Variance | Range |
|---|---|---|---|---|---|---|
| IL-6 | pg/mL | 80.70 | 15.04 | 145.72 | 21234.75 | 499.50 |
| WBC count | * 103/µL | 7.23 | 5.69 | 4.96 | 24.64 | 33.20 |
| Neutrophils count | * 103/µL | 5.34 | 3.66 | 4.84 | 23.44 | 30.19 |
| % neutrophils | 67.43 | 66.75 | 15.49 | 239.93 | 62.30 | |
| Lymphocytes count | * 103/µL | 1.28 | 1.23 | 0.61 | 0.37 | 3.21 |
| % lymphocytes | 22.54 | 20.75 | 12.12 | 147.01 | 44.90 | |
| Eosinophils count | * 103/µL | 0.05 | 0.01 | 0.08 | 0.01 | 0.39 |
| % eosinophils | 0.87 | 0.25 | 1.39 | 1.93 | 7.00 | |
| Hemoglobin | g/dL | 12.45 | 12.70 | 2.01 | 4.05 | 9.30 |
| Platelets count | * 103/µL | 232.58 | 214.50 | 94.94 | 9013.99 | 488.00 |
| NLR | 5.46 | 3.22 | 6.17 | 38.12 | 29.50 | |
| PLR | 209.52 | 182.73 | 102.50 | 10507.04 | 469.16 | |
| LOS | 25.33 | 23.00 | 8.96 | 80.28 | 41.00 | |
| CRP | mg/L | 69.67 | 32.45 | 83.62 | 6993.11 | 365.78 |
| Glucose | mg/dL | 132.81 | 122.50 | 55.75 | 3108.43 | 411.00 |
| ALT | UI/L | 53.38 | 32.00 | 79.82 | 6371.25 | 548.00 |
| AST | UI/L | 49.71 | 33.00 | 50.53 | 2553.50 | 308.00 |
| Urea | mg/dL | 56.51 | 37.00 | 53.96 | 2911.87 | 314.00 |
| Creatinine | mg/dL | 1.77 | 0.95 | 2.14 | 4.60 | 10.50 |
| eGFR | mL/min/1.73 m2 | 70.30 | 77.72 | 33.00 | 1089.32 | 138.35 |
| Serum Parameters | p | r |
|---|---|---|
| IL-6 | <0.001 | 0.388 |
| IgG | 0.043 | 0.227 |
| neutrophil count | 0.005 | 0.312 |
| % neutrophils | <0.001 | 0.454 |
| NLR | <0.001 | 0.491 |
| PLR | <0.001 | 0.380 |
| CRP | <0.001 | 0.496 |
| AST | 0.012 | 0.281 |
| urea | 0.026 | 0.248 |
| lymphocyte count | <0.001 | −0.453 |
| % lymphocytes | <0.001 | −0.504 |
| eosinophil count | <0.001 | −0.400 |
| % eosinophils | <0.001 | −0.422 |
| hemoglobin | 0.018 | −0.265 |
| Serum Parameters | p | r |
|---|---|---|
| IgG | 0.045 | 0.216 |
| % neutrophils | 0.011 | 0.281 |
| NLR | 0.002 | 0.334 |
| PLR | 0.001 | 0.350 |
| CRP | <0.001 | 0.512 |
| lymphocyte count | 0.002 | −0.340 |
| % lymphocytes | 0.002 | −0.349 |
| hemoglobin | 0.003 | −0.329 |
| eGFR | 0.038 | −0.234 |
| Blood Parameters | p | r |
|---|---|---|
| leucocyte count | 0.002 | 0.340 |
| neutrophil count | <0.001 | 0.389 |
| % neutrophils | <0.001 | 0.389 |
| NLR | <0.001 | 0.423 |
| CRP | 0.004 | 0.324 |
| ALT | 0.003 | 0.324 |
| IL-6 | 0.045 | 0.216 |
| PLR | 0.028) | 0.245 |
| AST | 0.008 | 0.296 |
| % lymphocytes | <0.001 | −0.441 |
| Serum Parameters | p | |
|---|---|---|
| Asymptomatic/mild form | IL-6 | 0.002 |
| CRP | 0.016 | |
| IgG | 0.011 | |
| eosinophil count | 0.005 | |
| % eosinophils | 0.005 | |
| Moderate form | IL-6 | 0.009 |
| CRP | 0.002 | |
| IgG | <0.001 | |
| eosinophil count | <0.001 | |
| eGFR | 0.026 | |
| Severe/critical form | IgG | <0.001 |
| IL-6 | <0.001 | |
| CRP | <0.001 | |
| NLR | <0.001 | |
| PLR | 0.001 | |
| Glucose | 0.027 | |
| AST | 0.013 | |
| urea | 0.047 | |
| Creatinine | 0.006 | |
| eGFR | <0.001 | |
| eosinophil count | <0.001 | |
| Serum Parameters | p |
|---|---|
| IL-6 | 0.021 |
| CRP | <0.001 |
| % neutrophils | <0.001 |
| lymphocyte count | <0.001 |
| % lymphocytes | <0.001 |
| eosinophils | 0.038 |
| NLR | 0.001 |
| PLR | <0.001 |
| urea | 0.024 |
| Severe Form of COVID-19 | IL-6 Concentration | |
|---|---|---|
| Comorbidities | p = 0.014 r = 0.274 | p = 0.019 r = 0.262 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Trofin, F.; Nastase, E.-V.; Vâță, A.; Iancu, L.S.; Luncă, C.; Buzilă, E.R.; Vlad, M.A.; Dorneanu, O.S. The Immune, Inflammatory and Hematological Response in COVID-19 Patients, According to the Severity of the Disease. Microorganisms 2023, 11, 319. https://doi.org/10.3390/microorganisms11020319
Trofin F, Nastase E-V, Vâță A, Iancu LS, Luncă C, Buzilă ER, Vlad MA, Dorneanu OS. The Immune, Inflammatory and Hematological Response in COVID-19 Patients, According to the Severity of the Disease. Microorganisms. 2023; 11(2):319. https://doi.org/10.3390/microorganisms11020319
Chicago/Turabian StyleTrofin, Felicia, Eduard-Vasile Nastase, Andrei Vâță, Luminița Smaranda Iancu, Cătălina Luncă, Elena Roxana Buzilă, Mădălina Alexandra Vlad, and Olivia Simona Dorneanu. 2023. "The Immune, Inflammatory and Hematological Response in COVID-19 Patients, According to the Severity of the Disease" Microorganisms 11, no. 2: 319. https://doi.org/10.3390/microorganisms11020319
APA StyleTrofin, F., Nastase, E.-V., Vâță, A., Iancu, L. S., Luncă, C., Buzilă, E. R., Vlad, M. A., & Dorneanu, O. S. (2023). The Immune, Inflammatory and Hematological Response in COVID-19 Patients, According to the Severity of the Disease. Microorganisms, 11(2), 319. https://doi.org/10.3390/microorganisms11020319





