Comparative Proteomic Analysis of Protein Patterns of Stenotrophomonas maltophilia in Biofilm and Planktonic Lifestyles
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strain and Growth Conditions
2.2. Collection of Planktonic Cells
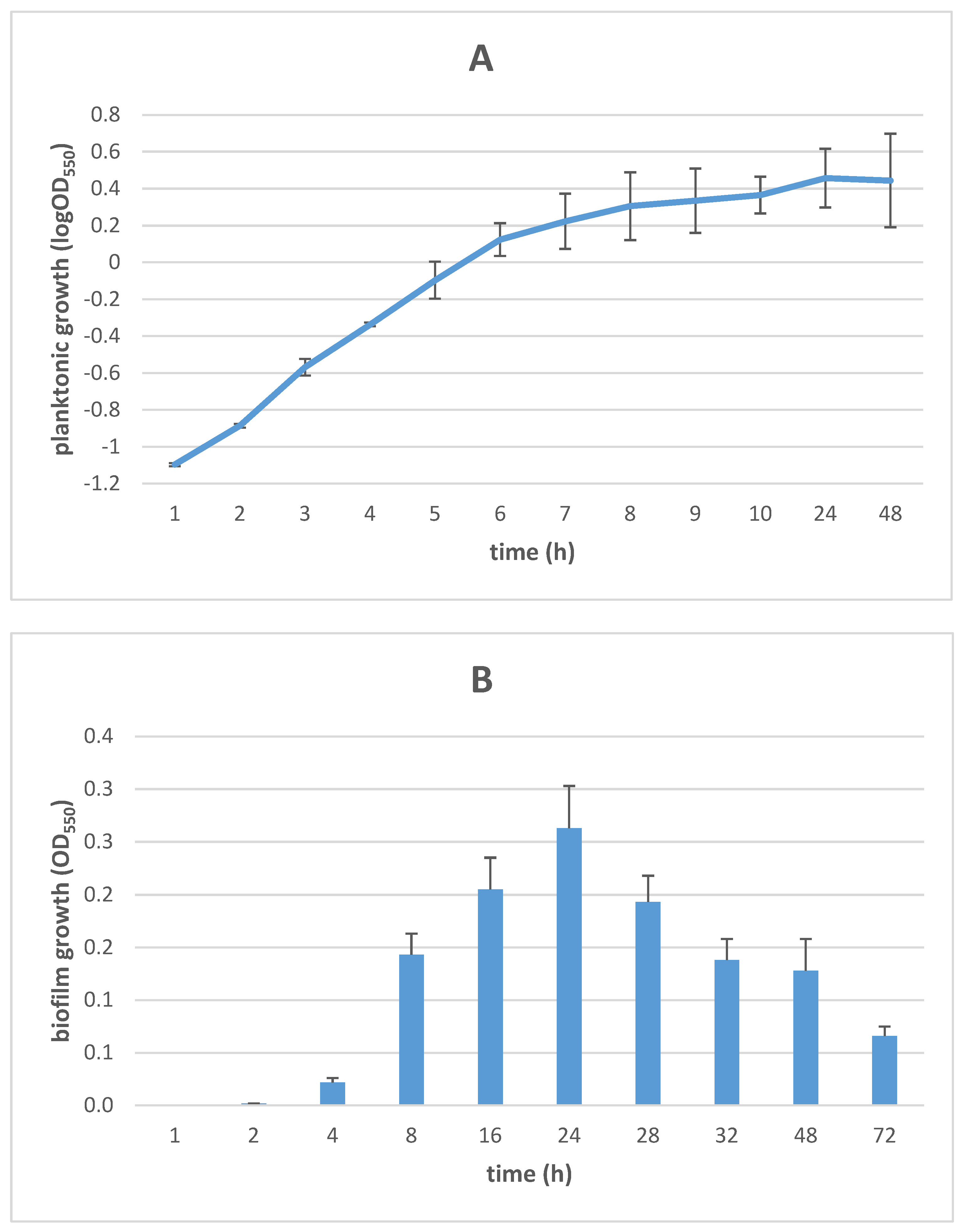
2.3. Collection of Biofilm Cells and Kinetics of Biofilm Formation
2.4. Bacterial Lysis and Protein Extraction
2.5. 2-D Electrophoresis
2.6. Analysis of Protein Patterns
2.7. Protein Identification by MALDI-TOF-MS and N-Terminal Sequencing
2.8. Scanning Electron Microscopy
3. Results and Discussion
3.1. Growth Kinetics of Planktonic and Biofilm S. maltophilia Cells
3.2. Proteomic Analysis: Biofilm versus Planktonic Lifestyles
3.3. Proteins and Related Pathways Expressed Only in Biofilm Cells
3.3.1. Quorum Sensing-Mediated Intercellular Communication
3.3.2. General Stress Response and Nutrient Starvation
Putative Heat Shock Protein (B6109 Protein)
Acetyl-CoA Synthetase (B3712 Protein)
Chorismate-Synthetase (B3102 Protein) and Polyribonucleotide phosphorylase (B3808 Protein)
3.3.3. Carbohydrate Metabolism and Energy Production
3.3.4. Other Pathways
3.4. Proteins Significantly Upregulated in Biofilm Cells
3.4.1. Putative Urocanate Hydratase
3.4.2. NADP-Dependent Alcohol Dehydrogenase
3.4.3. Serine Hydroxymethyltransferase
3.4.4. Acetyl-CoA Carboxylase Biotin Carboxylase Subunit
3.4.5. Aminotransferase
3.5. Proteins Significantly Downregulated in Biofilm Cells
3.5.1. OmpA/MotB Proteins
3.5.2. Thioester Dehydrase
3.5.3. Thioredoxin
3.5.4. Cold Shock Protein
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- An, S.Q.; Berg, G. Stenotrophomonas maltophilia. Trends Microbiol. 2018, 26, 637–638. [Google Scholar] [CrossRef]
- Insuwanno, W.; Kiratisin, P.; Jitmuang, A. Stenotrophomonas maltophilia infections: Clinical characteristics and factors associated with mortality of hospitalized patients. Infect. Drug. Resist. 2020, 13, 1559–1566. [Google Scholar] [CrossRef]
- Brooke, J.S. Advances in the microbiology of Stenotrophomonas maltophilia. Clin. Microbiol. Rev. 2021, 34, e0003019. [Google Scholar] [CrossRef]
- Francis, F.; Enaud, R.; Soret, P.; Lussac-Sorton, F.; Avalos-Fernandez, M.; MucoFong Investigation Group; Bui, S.; Fayon, M.; Thiébaut, R.; Delhaes, L. New Insights in Microbial Species Predicting Lung Function Decline in CF: Lessons from the MucoFong Project. J. Clin. Med. 2021, 10, 3725. [Google Scholar] [CrossRef]
- Trifonova, A.; Strateva, T. Stenotrophomonas maltophilia—A low-grade pathogen with numerous virulence factors. Infect. Dis. 2019, 51, 168–178. [Google Scholar] [CrossRef] [PubMed]
- Flores-Treviño, S.; Bocanegra-Ibarias, P.; Camacho-Ortiz, A.; Morfín-Otero, R.; Salazar-Sesatty, H.A.; Garza-González, E. Stenotrophomonas maltophilia biofilm: Its role in infectious diseases. Expert. Rev. Anti. Infect. Ther. 2019, 17, 877–893. [Google Scholar] [CrossRef]
- Pompilio, A.; Ranalli, M.; Piccirilli, A.; Perilli, M.; Vukovic, D.; Savic, B.; Krutova, M.; Drevinek, P.; Jonas, D.; Fiscarelli, E.V.; et al. Biofilm Formation among Stenotrophomonas maltophilia Isolates Has Clinical Relevance: The ANSELM Prospective Multicenter Study. Microorganisms 2020, 9, 49. [Google Scholar] [CrossRef]
- Pompilio, A.; Crocetta, V.; Confalone, P.; Nicoletti, M.; Petrucca, A.; Guarnieri, S.; Fiscarelli, E.; Savini, V.; Piccolomini, R.; Di Bonaventura, G. Adhesion to and biofilm formation on IB3-1 bronchial cells by Stenotrophomonas maltophilia isolates from cystic fibrosis patients. BMC Microbiol. 2010, 10, 102. [Google Scholar] [CrossRef]
- Pompilio, A.; Piccolomini, R.; Picciani, C.; D’Antonio, D.; Savini, V.; Di Bonaventura, G. Factors associated with adherence to and biofilm formation on polystyrene by Stenotrophomonas maltophilia: The role of cell surface hydrophobicity and motility. FEMS Microbiol. Lett. 2008, 287, 41–47. [Google Scholar] [CrossRef] [PubMed]
- An, S.Q.; Tang, J.I. The Ax21 protein influences virulence and biofilm formation in Stenotrophomonas maltophilia. Arch Microbiol. 2018, 200, 183–187. [Google Scholar] [CrossRef] [PubMed]
- An, S.; Tang, J. Diffusible signal factor signaling regulates multiple functions in the opportunistic pathogen Stenotrophomonas maltophilia. BMC Res. Notes 2018, 11, 569. [Google Scholar] [CrossRef] [PubMed]
- Alio, I.; Gudzuhn, M.; Pérez García, P.; Danso, D.; Schoelmerich, M.C.; Mamat, U.; Schaible, U.E.; Steinmann, J.; Yero, D.; Gibert, I.; et al. Phenotypic and Transcriptomic Analyses of Seven Clinical Stenotrophomonas maltophilia Isolates Identify a Small Set of Shared and Commonly Regulated Genes Involved in the Biofilm Lifestyle. Appl. Environ. Microbiol. 2020, 86, e02038-20. [Google Scholar] [CrossRef] [PubMed]
- Esposito, A.; Pompilio, A.; Bettua, C.; Crocetta, V.; Giacobazzi, E.; Fiscarelli, E.; Jousson, O.; Di Bonaventura, G. Evolution of Stenotrophomonas maltophilia in Cystic Fibrosis Lung over Chronic Infection: A Genomic and Phenotypic Population Study. Front. Microbiol. 2017, 8, 1590. [Google Scholar] [CrossRef] [PubMed]
- Pompilio, A.; Crocetta, V.; Scocchi, M.; Pomponio, S.; Di Vincenzo, V.; Mardirossian, M.; Gherardi, G.; Fiscarelli, E.; Dicuonzo, G.; Gennaro, R.; et al. Potential novel therapeutic strategies in cystic fibrosis: Antimicrobial and anti-biofilm activity of natural and designed α-helical peptides against Staphylococcus aureus, Pseudomonas aeruginosa, and Stenotrophomonas maltophilia. BMC Microbiol. 2012, 12, 145. [Google Scholar] [CrossRef] [PubMed]
- Karp, N.A.; Lilley, K.S. Investigating sample pooling strategies for DIGE experiments to address biological variability. Proteomics 2009, 9, 388–397. [Google Scholar] [CrossRef]
- Crossman, L.C.; Gould, V.C.; Dow, J.M.; Vernikos, G.S.; Okazaki, A.; Sebaihia, M.; Saunders, D.; Arrowsmith, C.; Carver, T.; Peters, N.; et al. The complete genome, comparative and functional analysis of Stenotrophomonas maltophilia reveals an organism heavily shielded by drug resistance determinants. Genome Biol. 2008, 9, R74. [Google Scholar] [CrossRef]
- Wang, Y.; Bian, Z.; Wang, Y. Biofilm formation and inhibition mediated by bacterial quorum sensing. Appl. Microbiol. Biotechnol. 2022, 106, 6365–6381. [Google Scholar] [CrossRef]
- Simanek, K.A.; Paczkowski, J.E. Resistance Is Not Futile: The Role of Quorum Sensing Plasticity in Pseudomonas aeruginosa Infections and Its Link to Intrinsic Mechanisms of Antibiotic Resistance. Microorganisms 2022, 10, 1247. [Google Scholar] [CrossRef]
- Huedo, P.; Yero, D.; Martinez-Servat, S.; Ruyra, À.; Roher, N.; Daura, X.; Gibert, I. Decoding the genetic and functional diversity of the DSF quorum-sensing system in Stenotrophomonas maltophilia. Front. Microbiol. 2015, 6, 761. [Google Scholar] [CrossRef]
- Pompilio, A.; Crocetta, V.; De Nicola, S.; Verginelli, F.; Fiscarelli, E.; Di Bonaventura, G. Cooperative pathogenicity in cystic fibrosis: Stenotrophomonas maltophilia modulates Pseudomonas aeruginosa virulence in mixed biofilm. Front. Microbiol. 2015, 6, 951. [Google Scholar] [CrossRef] [PubMed]
- Martínez, P.; Huedo, P.; Martinez-Servat, S.; Planell, R.; Ferrer-Navarro, M.; Daura, X.; Yero, D.; Gilbert, I. Stenotrophomonas maltophilia responds to exogenous AHL signals through the LuxR solo SmoR (Smlt1839). Front. Cell. Infect. Microbiol. 2015, 5, 41. [Google Scholar] [CrossRef]
- Givskov, M.; Rasmussen, B.T. Quorum sensing inhibitors: A bargain of effects. Microbiology 2006, 152, 895–904. [Google Scholar]
- Givskov, M.R.; De Nys, M.; Manefield, L.; Gram, R.; Maximilien, L.; Eberl, S.; Molin, P.; Steinberg, D.; Kjelleberg, S. Eukaryotic interference with homoserine lactone-mediated prokaryotic signalling. J. Bacteriol. 1996, 178, 6618–6622. [Google Scholar] [CrossRef]
- Uroz, S.; Oger, P.M.; Chapelle, E.; Adeline, M.T.; Faure, D.; Dessaux, Y. A Rhodococcus qsdA-encoded enzyme defines a novel class of large-spectrum quorum-quenching lactonases. Appl. Environ. Microbiol. 2008, 74, 1357–1366. [Google Scholar] [CrossRef] [PubMed]
- Park, S.Y.; Kang, H.O.; Jang, H.S.; Lee, J.K.; Koo, B.T.; Yum, D.Y. Identification of extracellular N-acylhomoserine lactone acylase from a Streptomyces sp and its application to quorum quenching. Appl. Environ. Microbiol. 2005, 71, 2632–2641. [Google Scholar] [CrossRef] [PubMed]
- Webb, K.; Cámara, M.; Zain, N.M.M.; Halliday, N.; Bruce, K.D.; Nash, E.F.; Whitehouse, J.L.; Knox, A.; Forrester, D.; Smyth, A.R.; et al. Novel detection of specific bacterial quorum sensing molecules in saliva: Potential non-invasive biomarkers for pulmonary Pseudomonas aeruginosa in cystic fibrosis. J. Cyst. Fibros. 2022, 21, 626–629. [Google Scholar] [CrossRef]
- O’Connor, A.; McClean, S. The role of universal stress proteins in bacterial infections. Curr. Med. Chem. 2017, 24, 3970–3976. [Google Scholar] [CrossRef]
- Zeng, B.; Wang, C.; Zhang, P.; Guo, Z.; Chen, L.; Duan, K. Heat Shock Protein DnaJ in Pseudomonas aeruginosa Affects Biofilm Formation via Pyocyanin Production. Microorganisms 2020, 8, 395. [Google Scholar] [CrossRef] [PubMed]
- Wolfe, A.J. The acetate switch. Microbiol. Mol. Biol. Rev. 2005, 69, 12–50. [Google Scholar] [CrossRef] [PubMed]
- Pru¨ß, B.M.; Wolfe, A.J. Regulation of acetyl phosphate synthesis and degradation, and the control of flagellar expression in Escherichia coli. Mol. Microbiol. 1994, 12, 973–984. [Google Scholar] [PubMed]
- Wolfe, A.J.; Chang, D.E.; Walker, J.D.; Seitz-Partridge, J.E.; Vidaurri, M.D.; Lange, C.F.; Pru¨ß, B.M.; Henk, M.C.; Larkin, J.C.; Conway, T. Evidence that acetyl phosphate functions as a global signal during biofilm development. Mol. Microbiol. 2003, 48, 977–988. [Google Scholar] [CrossRef] [PubMed]
- Anderson, G.G.; Palermo, J.J.; Schilling, J.D.; Roth, R.; Heuser, J.; Hultgren, S.J. Intracellular bacterial biofilm-like pods in urinary tract infections. Science 2003, 301, 105–107. [Google Scholar] [CrossRef] [PubMed]
- Sharif, D.I.; Gallon, J.; Smith, C.J.; Dudley, E. Quorum sensing in Cyanobacteria: N-octanoyl-homoserine lactone release and response, by the epilithic colonial cyanobacterium. Gloeothece PCCISME J. 2008, 2, 1171–1182. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, D.; Fisher, P.B. Polynucleotide phosphorylase: An evolutionary conserved gene with an expanding repertoire of functions. Pharmacol. Ther. 2006, 112, 243–263. [Google Scholar]
- Ladjouzi, R.; Duban, M.; Lucau-Danila, A.; Drider, D. The absence of PNPase activity in Enterococcus faecalis results in alterations of the bacterial cell-wall but induces high proteolytic and adhesion activities. Gene 2022, 833, 146610. [Google Scholar] [CrossRef]
- Rouf, S.F.; Ahmad, I.; Anwar, N.; Vodnala, S.K.; Kader, A.; Römling, U.; Rhen, M. Opposing contributions of polynucleotide phosphorylase and the membrane protein NlpI to biofilm formation by Salmonella enterica serovar Typhimurium. J. Bacteriol. 2011, 193, 580–582. [Google Scholar] [CrossRef]
- Carzaniga, T.; Antoniani, D.; Dehò, G.; Briani, F.; Landini, P. The RNA processing enzyme polynucleotide phosphorylase negatively controls biofilm formation by repressing poly-N-acetylglucosamine (PNAG) production in Escherichia coli C. BMC Microbiol. 2012, 12, 270. [Google Scholar] [CrossRef]
- Ulett, G.C.; Webb, R.I.; Schembri, M.A. Antigen-43-mediated autoaggregation impairs motility in Escherichia coli. Microbiology 2006, 152, 2101–2110. [Google Scholar] [CrossRef]
- Ramos-Hegazy, L.; Chakravarty, S.; Anderson, G.G. Phosphoglycerate mutase affects Stenotrophomonas maltophilia attachment to biotic and abiotic surfaces. Microbes Infect. 2020, 22, 60–64. [Google Scholar] [CrossRef] [PubMed]
- Isom, C.M.; Fort, B.; Anderson, G.G. Evaluating Metabolic Pathways and Biofilm Formation in Stenotrophomonas maltophilia. J. Bacteriol. 2022, 204, e0039821. [Google Scholar] [CrossRef]
- Xu, K.D.; Stewart, P.S.; Xia, F.; Huang, C.T.; McFeters, G.A. Spatial physiological heterogeneity in Pseudomonas aeruginosa biofilm is determined by oxygen availability. Appl. Environ. Microbiol. 1998, 64, 4035–4039. [Google Scholar] [CrossRef]
- Prigent-Combaret, C.; Vidal, O.; Dorel, C.; Lejeune, P. Abiotic surface sensing and biofilm-dependent regulation of gene expression in Escherichia coli. J. Bacteriol. 1999, 181, 5993–6002. [Google Scholar] [CrossRef]
- Pancholi, V.; Fischetti, A.V. a-Enolase, a novel strong plasmin(ogen) binding protein on the surface of pathogenic streptococci. J. Biol. Chem. 1998, 273, 14503–14515. [Google Scholar] [CrossRef]
- Zhao, L.; Liu, Q.; Huang, Q.; Liu, F.; Liu, H.; Wang, G. Isocitrate dehydrogenase of Bacillus cereus is involved in biofilm formation. World J. Microbiol. Biotechnol. 2021, 37, 207. [Google Scholar] [CrossRef]
- Prasad, U.V.; Vasu, D.; Yeswanth, S.; Swarupa, V.; Sunitha, M.M.; Choudhary, A.; Sarma, P.V. Phosphorylation controls the functioning of Staphylococcus aureus isocitrate dehydrogenase--favours biofilm formation. J. Enzyme Inhib. Med. Chem. 2015, 30, 655–661. [Google Scholar] [CrossRef]
- Keegan, R.; Waterman, D.G.; Hopper, D.J.; Coates, L.; Taylor, G.; Guo, J.; Coker, A.R.; Erskine, P.T.; Wood, S.P.; Cooper, J.B. The 1.1 Å resolution structure of a periplasmic phosphate-binding protein from Stenotrophomonas maltophilia: A crystallization contaminant identified by molecular replacement using the entire Protein Data Bank. Acta Crystallogr. D. Struct. Biol. 2016, 72, 933–943. [Google Scholar] [CrossRef] [PubMed]
- Neznansky, A.; Blus-Kadosh, I.; Yerushalmi, G.; Banin, E.; Opatowsky, Y. The Pseudomonas aeruginosa phosphate transport protein PstS plays a phosphate-independent role in biofilm formation. FASEB J. 2014, 28, 5223–5233. [Google Scholar] [CrossRef] [PubMed]
- Beliaeva, M.A.; Atac, R.; Seebeck, F.P. Bacterial Degradation of Nτ-Methylhistidine. ACS Chem. Biol. 2022, 17, 1989–1995. [Google Scholar] [CrossRef]
- Shibamura-Fujiogi, M.; Wang, X.; Maisat, W.; Koutsogiannaki, S.; Li, Y.; Chen, Y.; Lee, J.C.; Yuki, K. GltS regulates biofilm formation in methicillin-resistant Staphylococcus aureus. Commun. Biol. 2022, 5, 1284. [Google Scholar] [CrossRef]
- Hassanov, T.; Karunker, I.; Steinberg, N.; Erez, A.; Kolodkin-Gal, I. Novel antibiofilm chemotherapies target nitrogen from glutamate and glutamine. Sci. Rep. 2018, 8, 7097. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Yang, X.; Yang, J.; Qiao, X.; Li, F.; Liu, X.; Wei, J.; Wang, L. Alcohol dehydrogenase modulates quorum sensing in biofilm formations of Acinetobacter baumannii. Microb. Pathog. 2020, 148, 104451. [Google Scholar] [CrossRef] [PubMed]
- Becker, P.; Hufnagle, W.; Peters, G.; Herrmann, M. Detection of differential gene expression in biofilm-forming versus planktonic populations of Staphylococcus aureus using micro-representational-difference analysis. Appl. Environ. Microbiol. 2001, 67, 2958–2965. [Google Scholar] [CrossRef] [PubMed]
- Pu, M.; Sheng, L.; Song, S.; Gong, T.; Wood, T.K. Serine Hydroxymethyltransferase ShrA (PA2444) Controls Rugose Small-Colony Variant Formation in Pseudomonas aeruginosa. Front. Microbiol. 2018, 9, 315. [Google Scholar] [CrossRef] [PubMed]
- Choi-Rhee, E.; Cronan, J.E. The biotin carboxylase-biotin carboxyl carrier protein complex of Escherichia coli acetyl-CoA carboxylase. J. Biol. Chem. 2003, 278, 30806–30812. [Google Scholar] [CrossRef] [PubMed]
- Huedo, P.; Coves, X.; Daura, X.; Gibert, I.; Yero, D. Quorum Sensing Signaling and Quenching in the Multidrug-Resistant Pathogen Stenotrophomonas maltophilia. Front. Cell. Infect. Microbiol. 2018, 8, 122. [Google Scholar] [CrossRef] [PubMed]
- Alcaraz, E.; García, C.; Friedman, L.; de Rossi, B.P. The rpf/DSF signalling system of Stenotrophomonas maltophilia positively regulates biofilm formation, production of virulence-associated factors and β-lactamase induction. FEMS Microbiol. Lett. 2019, 366, fnz069. [Google Scholar] [CrossRef] [PubMed]
- Twomey, K.B.; O’Connell, O.J.; McCarthy, Y.; Dow, J.M.; O’Toole, G.A.; Plant, B.J.; Ryan, R.P. Bacterial cis-2-unsaturated fatty acids found in the cystic fibrosis airway modulate virulence and persistence of Pseudomonas aeruginosa. ISME J. 2012, 6, 939–950. [Google Scholar] [CrossRef] [PubMed]
- Weiland-Bräuer, N.; Kisch, M.J.; Pinnow, N.; Liese, A.; Schmitz, R.A. Highly Effective Inhibition of Biofilm Formation by the First Metagenome-Derived AI-2 Quenching Enzyme. Front. Microbiol. 2016, 7, 1098. [Google Scholar] [CrossRef]
- Ma, F.; Wang, T.; Ma, X.; Wang, P. Identification and characterization of protein encoded by orf382 as L-threonine dehydrogenase. J. Microbiol. Biotechnol. 2014, 24, 748–755. [Google Scholar] [CrossRef]
- Mougous, J.D.; Cuff, M.E.; Raunser, S.; Shen, A.; Zhou, M.; Gifford, C.A.; Goodman, A.L.; Joachimiak, G.; Ordoñez, C.L.; Lory, S.; et al. A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science 2006, 312, 1526–1530. [Google Scholar] [CrossRef]
- Liao, C.H.; Chang, C.L.; Huang, H.H.; Lin, Y.T.; Li, L.H.; Yang, T.C. Interplay between OmpA and RpoN Regulates Flagellar Synthesis in Stenotrophomonas maltophilia. Microorganisms 2021, 9, 1216. [Google Scholar] [CrossRef]
- Di Bonaventura, G.; Prosseda, G.; Del Chierico, F.; Cannavacciuolo, S.; Cipriani, P.; Petrucca, A.; Superti, F.; Ammendolia, M.G.; Concato, C.; Fiscarelli, E.; et al. Molecular characterization of virulence determinants of Stenotrophomonas maltophilia strains isolated from patients affected by cystic fibrosis. Int. J. Immunopathol. Pharmacol. 2007, 20, 529–537. [Google Scholar] [CrossRef]
- D’Arpa, P.; Karna, S.L.R.; Chen, T.; Leung, K.P. Pseudomonas aeruginosa transcriptome adaptations from colonization to biofilm infection of skin wounds. Sci. Rep. 2021, 11, 20632. [Google Scholar] [CrossRef]
- Cheng, C.; Dong, Z.; Han, X.; Wang, H.; Jiang, L.; Sun, J.; Yang, Y.; Ma, T.; Shao, C.; Wang, X.; et al. Thioredoxin A is essential for motility and contributes to host infection of Listeria monocytogenes via redox interactions. Front. Cell. Infect. Microbiol. 2017, 7, 287. [Google Scholar] [CrossRef]
- Windle, H.J.; Fox, A.; Ni Eidhin, D.; Kelleher, D. The thioredoxin system of Helicobacter pylori. J. Biol. Chem. 2000, 275, 5081–5089. [Google Scholar] [CrossRef]
- May, H.C.; Yu, J.J.; Zhang, H.; Wang, Y.; Cap, A.P.; Chambers, J.P.; Guentzel, M.N.; Arulanandam, B.P. Thioredoxin-A is a virulence factor and mediator of the type IV pilus system in Acinetobacter baumannii. PLoS ONE 2019, 14, e0218505. [Google Scholar] [CrossRef]
- Gilmore, K.S.; Srinivas, P.; Akins, D.R.; Hatter, K.L.; Gilmore, M.S. Growth, development, and gene expression in a persistent Streptococcus gordonii biofilm. Infect. Immun. 2003, 71, 4759–4766. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Petrova, O.E.; Su, S.; Lau, G.W.; Panmanee, W.; Na, R.; Hassett, D.J.; Davies, D.G.; Sauer, K. BdlA, DipA and induced dispersion contribute to acute virulence and chronic persistence of Pseudomonas aeruginosa. PLoS Pathog. 2014, 10, e1004168. [Google Scholar] [CrossRef]
- Hamilton, S.; Bongaerts, R.J.; Mulholland, F.; Cochrane, B.; Porter, J.; Lucchini, S.; Lappin-Scott, H.M.; Hinton, J.C. The transcriptional program of Salmonella enterica serovar Typhimurium reveals a key role for tryptophan metabolism in biofilms. BMC Genomics 2009, 10, 599. [Google Scholar] [CrossRef] [PubMed]
- Folkesson, A.; Jelsbak, L.; Yang, L.; Johansen, H.K.; Ciofu, O.; Høiby, N.; Molin, S. Adaptation of Pseudomonas aeruginosa to the cystic fibrosis airway: An evolutionary perspective. Nat. Rev. Microbiol. 2012, 10, 841–851. [Google Scholar] [CrossRef] [PubMed]
- Pompilio, A.; Crocetta, V.; Ghosh, D.; Chakrabarti, M.; Gherardi, G.; Vitali, L.A.; Fiscarelli, E.; Di Bonaventura, G. Stenotrophomonas maltophilia Phenotypic and Genotypic Diversity during a 10-year Colonization in the Lungs of a Cystic Fibrosis Patient. Front. Microbiol. 2016, 7, 1551. [Google Scholar] [CrossRef] [PubMed]
- Yu, T.; Keto-Timonen, R.; Jiang, X.; Virtanen, J.P.; Korkeala, H. Insights into the phylogeny and evolution of cold shock proteins: From enteropathogenic Yersinia and Escherichia coli to Eubacteria. Int. J. Mol. Sci. 2019, 20, 4059. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Sawyer, T.; Burbank, L. Csp1, a Cold Shock Protein Homolog in Xylella fastidiosa Influences Cell Attachment, Pili Formation, and Gene Expression. Microbiol. Spectr. 2021, 9, e0159121. [Google Scholar] [CrossRef] [PubMed]
- Muchaamba, F.; Stephan, R.; Tasara, T. Listeria monocytogenes Cold Shock Proteins: Small Proteins with A Huge Impact. Microorganisms 2021, 9, 1061. [Google Scholar] [CrossRef] [PubMed]
- Ray, S.; Da Costa, R.; Thakur, S.; Nandi, D. Salmonella Typhimurium encoded cold shock protein E is essential for motility and biofilm formation. Microbiology 2020, 166, 460–473. [Google Scholar] [CrossRef]


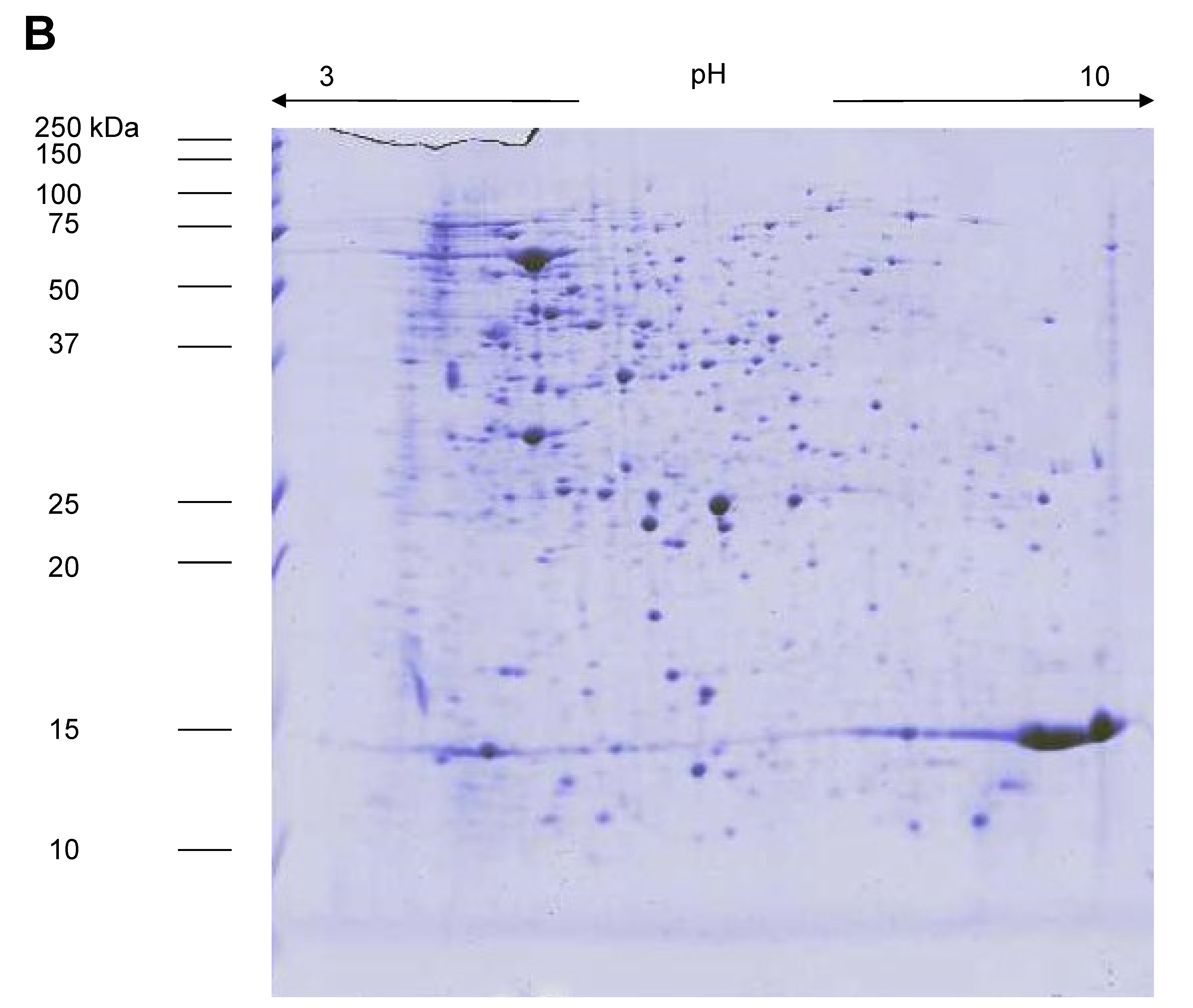
| Spot No. | Protein | Biological Process | Molecular Function | Peptides Matched a | Sequence Coverage b | pI c | MW d | GI e |
|---|---|---|---|---|---|---|---|---|
| B1305 | regulatory protein | Not known | Not known | 6 | 33 | 4.83/5.96 | 36,096/25,543 | 197784663 |
| B5613 | putative amidohydrolase | Nitrogen compound metabolic process | Hydrolase activity | 11 | 31 | 5.65/6.38 | 49,616/52,720 | 190575191 |
| B6708 | putative electron transfer flavoprotein-ubiquinone oxidoreductase | Energy production and conversion | Electron-transferring-flavoprotein dehydrogenase activity; Metal ion binding | 11 | 29 | 5.89/6.60 | 61,233/72,075 | 190572277 |
| B8509 | putative 3-ketoacyl-CoA thiolase | Fatty acid beta-oxidation | Acetyl-CoA C-acyltransferase | 14 | 43 | 7.15/8.66 | 46,154/45,239 | 190572243 |
| B7007 | hypothetical protein Smal | Not known | Not known | 8 | 48 | 6.09/7.19 | 16,432/11,577 | 194367327 |
| B4409 | putative isocitrate/isopropylmalate dehydrogenase | Leucine biosynthesis | Oxidoreductase activity; Mg and NAD binding | 16 | 57 | 5.61/6.17 | 35,841/39,353 | 190573015 |
| B3102 | chorismate synthase | Aromatic amino acid family biosynthetic process | Lyase chorismate synthase activity | 13 | 45 | 5.41/5.56 | 42,701/20,525 | 191161246 |
| B3712 | putative acetyl-coenzyme A synthetase | Acetyl-CoA biosynthesis from acetate | Metal ion binding; AMP/ATP binding; Acetate-CoA ligase activity | 17 | 28 | 5.55/5.61 | 72,107/67,921 | 190576408 |
| B6109 | putative heat shock protein | Not known | Not known | 10 | 74 | 5.74/6.61 | 19,550/17,758 | 190575907 |
| B8812 | conserved hypothetical exported protein | Not known | Catalytic activity | 28 | 43 | 8.76/8.73 | 77,975/83,683 | 190576412 |
| B3808 | putative polyribonucleotide phosphorylase | RNA processing; mRNA catabolic processing; 3′-5-exoribonuclease activity; RNA binding; Polyribonucleotide nucleotidyl-transferase | Nucleotidyl-transferase | 20 | 34 | 5.42/5.63 | 75,448/79,698 | 190575258 |
| B7203 | phosphoglycerate mutase 1 family | Gluconeogenesis; Glycolysis | Isomerase | 9 | 54 | 6.51/7.24 | 28,036/26,920 | 194364976 |
| B5617 | putative 4-aminobutyrate-2-oxoglutarate aminotransferase | Alanine, glutamate, and glutamine metabolism | Pyridoxal phosphate binding; 4-amino-butyrate:2-oxoglutarate transaminase activity | 9 | 26 | 5.82/6.58 | 52,261/54,309 | 190573417 |
| B8206 | 2,3,4,5-tetrahydropyridine-2,6-dicarboxylate N-succinyltransferase | Lysine and diamonipimelate biosynthesis | 2,3,4,5-tetrahydropyridine-2,6-dicarboxylate N-succinyltransferase activity | 8 | 18 | 9.71/8.76 | 37,654/29,196 | 190573512 |
| B9202 | putative dihydrolipoamide succinyltransferase, E2 component | Tricarboxylic acid cycle | dihydrolipoyllysine-residue succinyltransferase activity | 9 | 18 | 5.71/9.39 | 41,992/26,817 | 190575085 |
| B6314 | 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase | Gluconeogenesis; Glycolysis | 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity | 11 | 61 | 6.32/6.93 | 28,062/27,123 | 190573432 |
| Spot No. | Protein | Biological Process | Molecular Function | Fold Up-Regulation a | Peptides Matched b | Sequence Coverage c | pI d | MW e | GI f |
|---|---|---|---|---|---|---|---|---|---|
| MB4713 | Putative urocanate hydratase | Aminoacid degradation | Conversion of urocanate to 4-imidazolone-5-propionate | 7.8 | 13 | 30 | 5.58/6.06 | 60,368/69,065 | 190575003 |
| MB4807 | Putative zinc metallopeptidase | Cell division; Protein catabolysis; Proteolysis | Metallopeptidase activity; Zinc ion binding | 10.9 | 15 | 25 | 5.73/6.05 | 76,512/82,630 | 190572608 |
| MB1806 | Putative 30S ribosomal protein | Protein synthesis | Protein synthesis | 3.0 | 4.44/4.79 | 66,234/72,144 | 1702195087 | ||
| MB6403 | Putative aminotransferase | Protein metabolism; Gluconeogenesis | Transamination | 4.7 | 7 | 31 | 6.07/6.91 | 42,450/43,319 | 190575152 |
| MB6405 | Putative NADP-dependent alcohol dehydrogenase | Response to reactive oxygen species | Metal ion binding; Butanol dehydrogenase activity | 4.3 | 4 | 17 | 5.71/6.93 | 38,658/41,493 | 190575829 |
| MB7303 | Putative threonine 3-dehydrogenase | Threonine catabolism | L-threonine 3-dehydrogenase activity | 3.5 | 20 | 47 | 6.21/7.26 | 37,502/35,401 | 190572994 |
| MB7502 | Putative serine hydroxymethyltransferase | Glycine biosynthesis from serine | Glycine hydroxymethyltransferase activity | 10.1 | 9 | 27 | 6.12/7.44 | 45,122/49,476 | 190572766 |
| MB8406 | Conserved hypothetical protein | Not known | Not known | 7.8 | 22 | 59 | 7.15/5.99 | 43,882/85,923 | 190576469 |
| MB8602 | Putative biotin carboxylase | Fatty acid biosynthesis | ATP binding; Metal ion binding | 5.2 | 13 | 34 | 6.55/7.97 | 49,609/60,821 | 190576068 |
| Spot No. | Protein | Biological Process | Molecular Function | Fold Up-Regulation a | Peptides Matched b | Sequence Coverage c | pI d | MW e | GI f |
|---|---|---|---|---|---|---|---|---|---|
| MP0304 | OmpA/MotB domain-containing protein | Ion transport | Porin activity | 19.0 | 11 | 43 | 4.80/4.36 | 39,148/37,464 | 194364578 |
| MP4106 | Hypothetical protein Smal_0271 | Not known | Not known | 13.5 | 9 | 44 | 5.80/5.97 | 21,071/16,406 | 194364049 |
| MP8009 | Thioester dehydrase family protein | Fatty acid biosynthetic process | Dehydrase activity | 3.6 | 8 | 43 | 4.90/8.21 | 17,497/7237 | 223039807 |
| MP0011 | Ribosomal protein L7/L12 | Cytoplasmatic translation | Structural constituent of ribosome | 4.0 | 4 | 50 | 4.79/4.51 | 12,589/10,677 | 254524344 |
| MP0201 | Thioredoxin | Removal of superoxide radicals | Detoxification | 4.4 | 3 | 14 | 4.57/4.34 | 30,871/31,825 | 194366686 |
| MP1006 | 50S ribosomal protein L9 | Cytoplasmatic translation | Large ribosomal subunit rRNA binding | 10.3 | 6 | 36 | 5.24/4.9 | 15,719/8238 | 190575039 |
| MP8002 | Putative sigma-54 modulation protein | Translation regulation | Translation regulation | 5.5 | 2 | 26 | 6.64/8.17 | 11,739/10,127 | 190573137 |
| MP8004 | Cold shock protein | Response to stress | Nucleic acid binding | 9.8 | 3 | 40 | 8.15/8.80 | 7573/7464 | 190573988 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Di Bonaventura, G.; Picciani, C.; Lupetti, V.; Pompilio, A. Comparative Proteomic Analysis of Protein Patterns of Stenotrophomonas maltophilia in Biofilm and Planktonic Lifestyles. Microorganisms 2023, 11, 442. https://doi.org/10.3390/microorganisms11020442
Di Bonaventura G, Picciani C, Lupetti V, Pompilio A. Comparative Proteomic Analysis of Protein Patterns of Stenotrophomonas maltophilia in Biofilm and Planktonic Lifestyles. Microorganisms. 2023; 11(2):442. https://doi.org/10.3390/microorganisms11020442
Chicago/Turabian StyleDi Bonaventura, Giovanni, Carla Picciani, Veronica Lupetti, and Arianna Pompilio. 2023. "Comparative Proteomic Analysis of Protein Patterns of Stenotrophomonas maltophilia in Biofilm and Planktonic Lifestyles" Microorganisms 11, no. 2: 442. https://doi.org/10.3390/microorganisms11020442
APA StyleDi Bonaventura, G., Picciani, C., Lupetti, V., & Pompilio, A. (2023). Comparative Proteomic Analysis of Protein Patterns of Stenotrophomonas maltophilia in Biofilm and Planktonic Lifestyles. Microorganisms, 11(2), 442. https://doi.org/10.3390/microorganisms11020442








