SARS-CoV-2 Affects Both Humans and Animals: What Is the Potential Transmission Risk? A Literature Review
Abstract
:1. Introduction
2. Methods
3. Coronavirus Disease 2019 (COVID-19)
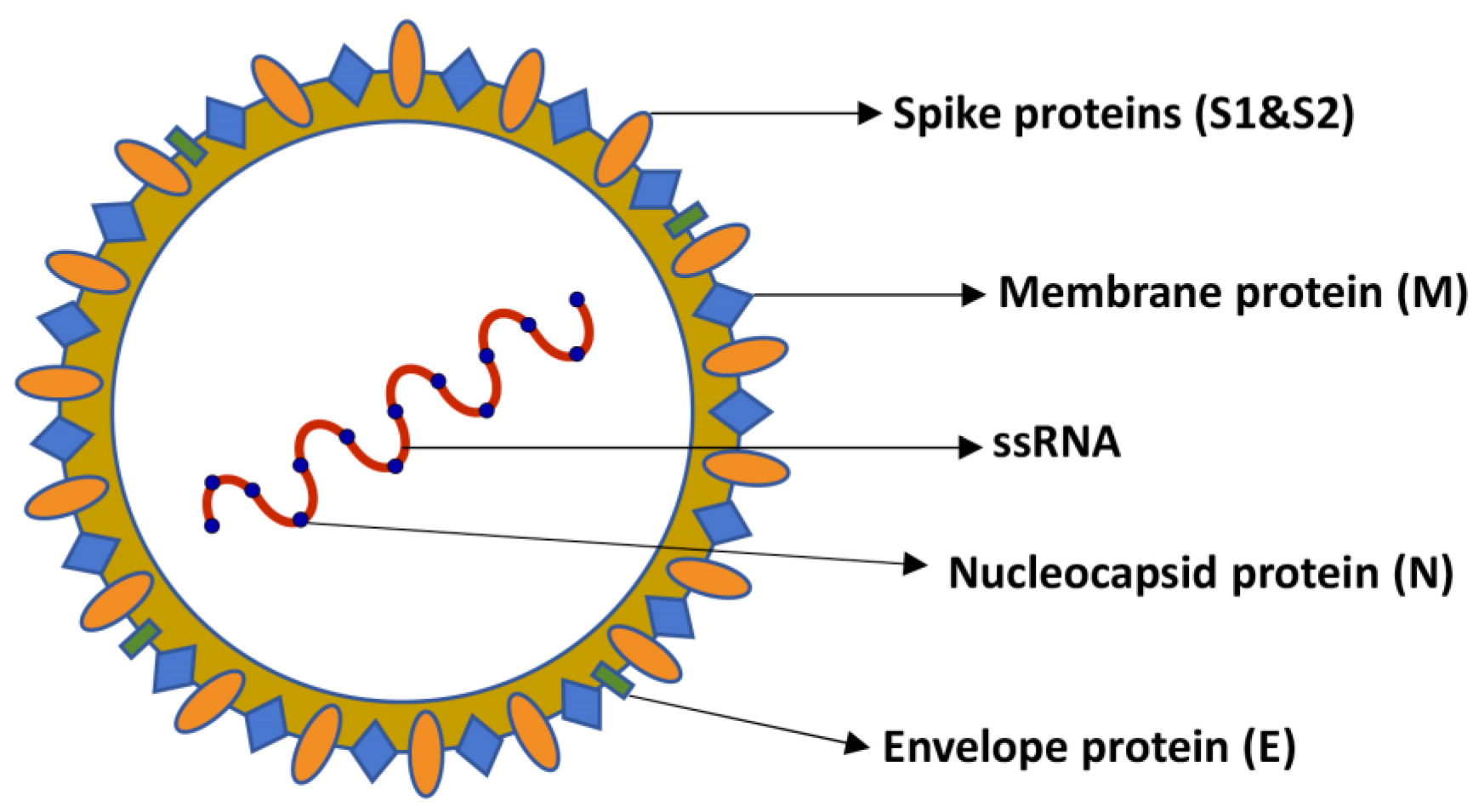
3.1. SARS-CoV-2 Virus
3.2. Infections in Animals
3.2.1. Cats
3.2.2. Dogs
3.2.3. Other Animal Species
3.3. Infections in Humans
4. Serological and Molecular Diagnosis
4.1. SARS-CoV-2 Testing in Humans
4.2. SARS-CoV-2 Testing in Animals
5. Potential Transmission Risk by SARS-CoV-2 in Human–Animal Relationship
6. Discussion
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Russotto, Y.; Micali, C.; Pellicano, G.F.; Nunnari, G.; Venanzi Rullo, E. HIV and Mediterranean Zoonoses: A Review of the Literature. Infect. Dis. Rep. 2022, 14, 694–709. [Google Scholar] [CrossRef] [PubMed]
- Lindahl, J.F.; Grace, D. The consequences of human actions on risks for infectious diseases: A review. Infect. Ecol. Epidemiol. 2015, 5, 30048. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wolfe, N.D.; Dunavan, C.P.; Diamond, J. Origins of major human infectious diseases. Nature 2007, 447, 279–283. [Google Scholar] [CrossRef] [PubMed]
- Salyer, S.J.; Silver, R.; Simone, K.; Barton Behravesh, C. Prioritizing Zoonoses for Global Health Capacity Building-Themes from One Health Zoonotic Disease Workshops in 7 Countries, 2014–2016. Emerg. Infect. Dis. 2017, 23, S55–S64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santaniello, A.; Cimmino, I.; Dipineto, L.; Agognon, A.L.; Beguinot, F.; Formisano, P.; Fioretti, A.; Menna, L.F.; Oriente, F. Zoonotic Risk of Encephalitozoon cuniculi in Animal-Assisted Interventions: Laboratory Strategies for the Diagnosis of Infections in Humans and Animals. Int. J. Environ. Res. Public Health 2021, 18, 9333. [Google Scholar] [CrossRef]
- Segal, E.; Elad, D. Human and Zoonotic Dermatophytoses: Epidemiological Aspects. Front. Microbiol. 2021, 12, 713532. [Google Scholar] [CrossRef]
- Butler, T. Plague history: Yersin’s discovery of the causative bacterium in 1894 enabled, in the subsequent century, scientific progress in understanding the disease and the development of treatments and vaccines. Clin. Microbiol. Infect. 2014, 20, 202–209. [Google Scholar] [CrossRef] [Green Version]
- Jefferson, T.; Demicheli, V.; Pratt, M. Vaccines for preventing plague. Cochrane Database Syst. Rev. 2000, 1998, CD000976. [Google Scholar] [CrossRef]
- Yousaf, M.Z.; Qasim, M.; Zia, S.; Khan, M.; Ashfaq, U.A.; Khan, S. Rabies molecular virology, diagnosis, prevention and treatment. Virol. J. 2012, 9, 50. [Google Scholar] [CrossRef] [Green Version]
- Goldman, J.S.; Vallabh, S.M. Genetic counseling for prion disease: Updates and best practices. Genet. Med. 2022, 24, 1993–2003. [Google Scholar] [CrossRef]
- Gallardo, M.J.; Delgado, F.O. Animal prion diseases: A review of intraspecies transmission. Open Vet. J. 2021, 11, 707–723. [Google Scholar] [CrossRef]
- Asjo, B.; Kruse, H. Zoonoses in the Emergence of Human Viral Diseases. Perspect. Med. Virol. 2006, 16, 15–41. [Google Scholar] [PubMed]
- Sharp, P.M.; Hahn, B.H. Origins of HIV and the AIDS pandemic. Cold Spring Harb. Perspect. Med. 2011, 1, a006841. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piot, P.; Bartos, M.; Ghys, P.D.; Walker, N.; Schwartlander, B. The global impact of HIV/AIDS. Nature 2001, 410, 968–973. [Google Scholar] [CrossRef] [PubMed]
- Ananworanich, J.; Mellors, J.W. A cure for HIV: What will it take? Curr. Opin. HIV AIDS 2015, 10, 1–3. [Google Scholar] [CrossRef]
- Eisinger, R.W.; Fauci, A.S. Ending the HIV/AIDS Pandemic(1). Emerg. Infect. Dis. 2018, 24, 413–416. [Google Scholar] [CrossRef] [Green Version]
- Kassutto, S.; Rosenberg, E.S. Primary HIV type 1 infection. Clin. Infect. Dis. 2004, 38, 1447–1453. [Google Scholar] [CrossRef]
- Worobey, M.; Levy, J.I.; Malpica Serrano, L.; Crits-Christoph, A.; Pekar, J.E.; Goldstein, S.A.; Rasmussen, A.L.; Kraemer, M.U.G.; Newman, C.; Koopmans, M.P.G.; et al. The Huanan Seafood Wholesale Market in Wuhan was the early epicenter of the COVID-19 pandemic. Science 2022, 377, 951–959. [Google Scholar] [CrossRef]
- Temmam, S.; Vongphayloth, K.; Baquero, E.; Munier, S.; Bonomi, M.; Regnault, B.; Douangboubpha, B.; Karami, Y.; Chretien, D.; Sanamxay, D.; et al. Bat coronaviruses related to SARS-CoV-2 and infectious for human cells. Nature 2022, 604, 330–336. [Google Scholar] [CrossRef]
- Cui, J.; Li, F.; Shi, Z.L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019, 17, 181–192. [Google Scholar] [CrossRef] [Green Version]
- Tyrrell, D.A.; Bynoe, M.L. Cultivation of a Novel Type of Common-Cold Virus in Organ Cultures. Br. Med. J. 1965, 1, 1467–1470. [Google Scholar] [CrossRef] [Green Version]
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Li, Y.; Zhang, A.L.; Wang, Y.; Molina, M.J. Identifying airborne transmission as the dominant route for the spread of COVID-19. Proc. Natl. Acad. Sci. USA 2020, 117, 14857–14863. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhu, J.; Wang, X.; Yang, J.; Liu, X.F.; Xu, X.K. Characterizing COVID-19 Transmission: Incubation Period, Reproduction Rate, and Multiple-Generation Spreading. Front. Phys. 2021, 8, 589963. [Google Scholar] [CrossRef]
- Wang, M.Y.; Zhao, R.; Gao, L.J.; Gao, X.F.; Wang, D.P.; Cao, J.M. SARS-CoV-2: Structure, Biology, and Structure-Based Therapeutics Development. Front. Cell. Infect. Microbiol. 2020, 10, 587269. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.Z.; Wang, S.Y.; Zheng, Z.Q.; Yi, H.; Li, W.W.; Xu, Z.S.; Wang, Y.Y. SARS-CoV-2 membrane glycoprotein M antagonizes the MAVS-mediated innate antiviral response. Cell. Mol. Immunol. 2021, 18, 613–620. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.R.; Yin, W.C.; Jiang, Y.; Xu, H.E. Structure genomics of SARS-CoV-2 and its Omicron variant: Drug design templates for COVID-19. Acta Pharmacol. Sin. 2022, 43, 3021–3033. [Google Scholar] [CrossRef] [PubMed]
- Rando, H.M.; MacLean, A.L.; Lee, A.J.; Lordan, R.; Ray, S.; Bansal, V.; Skelly, A.N.; Sell, E.; Dziak, J.J.; Shinholster, L.; et al. Pathogenesis, Symptomatology, and Transmission of SARS-CoV-2 through Analysis of Viral Genomics and Structure. mSystems 2021, 6, e0009521. [Google Scholar] [CrossRef]
- Decaro, N.; Martella, V.; Saif, L.J.; Buonavoglia, C. COVID-19 from veterinary medicine and one health perspectives: What animal coronaviruses have taught us. Res. Vet. Sci. 2020, 131, 21–23. [Google Scholar] [CrossRef]
- Decaro, N.; Lorusso, A. Novel human coronavirus (SARS-CoV-2): A lesson from animal coronaviruses. Vet. Microbiol. 2020, 244, 108693. [Google Scholar] [CrossRef]
- Zhang, Y.Z.; Holmes, E.C. A Genomic Perspective on the Origin and Emergence of SARS-CoV-2. Cell 2020, 181, 223–227. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Kruger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e8. [Google Scholar] [CrossRef] [PubMed]
- Cui, S.; Liu, Y.; Zhao, J.; Peng, X.; Lu, G.; Shi, W.; Pan, Y.; Zhang, D.; Yang, P.; Wang, Q. An Updated Review on SARS-CoV-2 Infection in Animals. Viruses 2022, 14, 1527. [Google Scholar] [CrossRef]
- Hancock, J.T.; Rouse, R.C.; Stone, E.; Greenhough, A. Interacting Proteins, Polymorphisms and the Susceptibility of Animals to SARS-CoV-2. Animals 2021, 11, 797. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Moore, M.J.; Vasilieva, N.; Sui, J.; Wong, S.K.; Berne, M.A.; Somasundaran, M.; Sullivan, J.L.; Luzuriaga, K.; Greenough, T.C.; et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 2003, 426, 450–454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Devaux, C.A.; Pinault, L.; Osman, I.O.; Raoult, D. Can ACE2 Receptor Polymorphism Predict Species Susceptibility to SARS-CoV-2? Front. Public Health 2020, 8, 608765. [Google Scholar] [CrossRef]
- Conceicao, C.; Thakur, N.; Human, S.; Kelly, J.T.; Logan, L.; Bialy, D.; Bhat, S.; Stevenson-Leggett, P.; Zagrajek, A.K.; Hollinghurst, P.; et al. The SARS-CoV-2 Spike protein has a broad tropism for mammalian ACE2 proteins. PLoS Biol. 2020, 18, e3001016. [Google Scholar] [CrossRef]
- Xu, X.G.; Wang, H.; Wang, S.Y.; Zhang, J.; Sun, C.; Wei, M.X.; Yan, J.; Mao, G.X. Predictive Analysis of Susceptibility of Different Species to SARS-CoV-2 based on ACE2 Receptors. Biomed. Environ. Sci. 2020, 33, 877–881. [Google Scholar]
- Hassan, S.S.; Ghosh, S.; Attrish, D.; Choudhury, P.P.; Aljabali, A.A.A.; Uhal, B.D.; Lundstrom, K.; Rezaei, N.; Uversky, V.N.; Seyran, M.; et al. Possible Transmission Flow of SARS-CoV-2 Based on ACE2 Features. Molecules 2020, 25, 5906. [Google Scholar] [CrossRef]
- Olivieri, E.R.; Heller, L.K.; Gillim-Ross, L.; Wentworth, D.E. Analysis of SARS-CoV receptor activity of ACE2 orthologs. Adv. Exp. Med. Biol. 2006, 581, 277–280. [Google Scholar]
- Damas, J.; Hughes, G.M.; Keough, K.C.; Painter, C.A.; Persky, N.S.; Corbo, M.; Hiller, M.; Koepfli, K.P.; Pfenning, A.R.; Zhao, H.; et al. Broad host range of SARS-CoV-2 predicted by comparative and structural analysis of ACE2 in vertebrates. Proc. Natl. Acad. Sci. USA 2020, 117, 22311–22322. [Google Scholar] [CrossRef]
- Lam, S.D.; Bordin, N.; Waman, V.P.; Scholes, H.M.; Ashford, P.; Sen, N.; van Dorp, L.; Rauer, C.; Dawson, N.L.; Pang, C.S.M.; et al. SARS-CoV-2 spike protein predicted to form complexes with host receptor protein orthologues from a broad range of mammals. Sci. Rep. 2020, 10, 16471. [Google Scholar] [CrossRef]
- Luan, J.; Jin, X.; Lu, Y.; Zhang, L. SARS-CoV-2 spike protein favors ACE2 from Bovidae and Cricetidae. J. Med. Virol. 2020, 92, 1649–1656. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Wang, H.; Tang, X.; Fang, S.; Ma, D.; Du, C.; Wang, Y.; Pan, H.; Yao, W.; Zhang, R.; et al. SARS-CoV-2 and Three Related Coronaviruses Utilize Multiple ACE2 Orthologs and Are Potently Blocked by an Improved ACE2-Ig. J. Virol. 2020, 94, e01283-20. [Google Scholar] [CrossRef]
- Zhao, X.; Chen, D.; Szabla, R.; Zheng, M.; Li, G.; Du, P.; Zheng, S.; Li, X.; Song, C.; Li, R.; et al. Broad and Differential Animal Angiotensin-Converting Enzyme 2 Receptor Usage by SARS-CoV-2. J. Virol. 2020, 94, e00940-20. [Google Scholar] [CrossRef]
- Wei, Y.; Aris, P.; Farookhi, H.; Xia, X. Predicting mammalian species at risk of being infected by SARS-CoV-2 from an ACE2 perspective. Sci. Rep. 2021, 11, 1702. [Google Scholar] [CrossRef]
- Elbe, S.; Buckland-Merrett, G. Data, disease and diplomacy: GISAID’s innovative contribution to global health. Glob. Chall. 2017, 1, 33–46. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.; Zhang, H.; Gao, J.; Huang, K.; Yang, Y.; Hui, X.; He, X.; Li, C.; Gong, W.; Zhang, Y.; et al. A serological survey of SARS-CoV-2 in cat in Wuhan. Emerg. Microbes Infect. 2020, 9, 2013–2019. [Google Scholar] [CrossRef]
- Sailleau, C.; Dumarest, M.; Vanhomwegen, J.; Delaplace, M.; Caro, V.; Kwasiborski, A.; Hourdel, V.; Chevaillier, P.; Barbarino, A.; Comtet, L.; et al. First detection and genome sequencing of SARS-CoV-2 in an infected cat in France. Transbound. Emerg. Dis. 2020, 67, 2324–2328. [Google Scholar] [CrossRef]
- Newman, A.; Smith, D.; Ghai, R.R.; Wallace, R.M.; Torchetti, M.K.; Loiacono, C.; Murrell, L.S.; Carpenter, A.; Moroff, S.; Rooney, J.A.; et al. First Reported Cases of SARS-CoV-2 Infection in Companion Animals—New York, March–April 2020. MMWR Morb. Mortal. Wkly. Rep. 2020, 69, 710–713. [Google Scholar] [CrossRef]
- Barrs, V.R.; Peiris, M.; Tam, K.W.S.; Law, P.Y.T.; Brackman, C.J.; To, E.M.W.; Yu, V.Y.T.; Chu, D.K.W.; Perera, R.; Sit, T.H.C. SARS-CoV-2 in Quarantined Domestic Cats from COVID-19 Households or Close Contacts, Hong Kong, China. Emerg. Infect. Dis. 2020, 26, 3071–3074. [Google Scholar] [CrossRef]
- Klaus, J.; Zini, E.; Hartmann, K.; Egberink, H.; Kipar, A.; Bergmann, M.; Palizzotto, C.; Zhao, S.; Rossi, F.; Franco, V.; et al. SARS-CoV-2 Infection in Dogs and Cats from Southern Germany and Northern Italy during the First Wave of the COVID-19 Pandemic. Viruses 2021, 13, 1453. [Google Scholar] [CrossRef]
- Patterson, E.I.; Elia, G.; Grassi, A.; Giordano, A.; Desario, C.; Medardo, M.; Smith, S.L.; Anderson, E.R.; Prince, T.; Patterson, G.T.; et al. Evidence of exposure to SARS-CoV-2 in cats and dogs from households in Italy. Nat. Commun. 2020, 11, 6231. [Google Scholar] [CrossRef]
- Klaus, J.; Palizzotto, C.; Zini, E.; Meli, M.L.; Leo, C.; Egberink, H.; Zhao, S.; Hofmann-Lehmann, R. SARS-CoV-2 Infection and Antibody Response in a Symptomatic Cat from Italy with Intestinal B-Cell Lymphoma. Viruses 2021, 13, 527. [Google Scholar] [CrossRef]
- Temmam, S.; Barbarino, A.; Maso, D.; Behillil, S.; Enouf, V.; Huon, C.; Jaraud, A.; Chevallier, L.; Backovic, M.; Perot, P.; et al. Absence of SARS-CoV-2 infection in cats and dogs in close contact with a cluster of COVID-19 patients in a veterinary campus. One Health 2020, 10, 100164. [Google Scholar] [CrossRef]
- Ruiz-Arrondo, I.; Portillo, A.; Palomar, A.M.; Santibanez, S.; Santibanez, P.; Cervera, C.; Oteo, J.A. Detection of SARS-CoV-2 in pets living with COVID-19 owners diagnosed during the COVID-19 lockdown in Spain: A case of an asymptomatic cat with SARS-CoV-2 in Europe. Transbound. Emerg. Dis. 2021, 68, 973–976. [Google Scholar] [CrossRef]
- Michelitsch, A.; Hoffmann, D.; Wernike, K.; Beer, M. Occurrence of Antibodies against SARS-CoV-2 in the Domestic Cat Population of Germany. Vaccines 2020, 8, 772. [Google Scholar] [CrossRef]
- Kleinerman, G.; Gross, S.; Topol, S.; Ariel, E.; Volokh, G.; Melloul, S.; Mergy, S.E.; Malamud, Y.; Gilboa, S.; Gal, Y.; et al. Low serological rate of SARS-CoV-2 in cats from military bases in Israel. Comp. Immunol. Microbiol. Infect. Dis. 2022, 90–91, 101905. [Google Scholar] [CrossRef]
- Genyk, S.N.; Baisa, A.V. Complications of hemosorption and the methods of their prevention. Sov. Med. 1986, 1, 48–50. [Google Scholar]
- Lenz, O.C.; Marques, A.D.; Kelly, B.J.; Rodino, K.G.; Cole, S.D.; Perera, R.; Weiss, S.R.; Bushman, F.D.; Lennon, E.M. SARS-CoV-2 Delta Variant (AY.3) in the Feces of a Domestic Cat. Viruses 2022, 14, 421. [Google Scholar] [CrossRef]
- Shi, J.; Wen, Z.; Zhong, G.; Yang, H.; Wang, C.; Huang, B.; Liu, R.; He, X.; Shuai, L.; Sun, Z.; et al. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS-coronavirus 2. Science 2020, 368, 1016–1020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bosco-Lauth, A.M.; Hartwig, A.E.; Porter, S.M.; Gordy, P.W.; Nehring, M.; Byas, A.D.; VandeWoude, S.; Ragan, I.K.; Maison, R.M.; Bowen, R.A. Experimental infection of domestic dogs and cats with SARS-CoV-2: Pathogenesis, transmission, and response to reexposure in cats. Proc. Natl. Acad. Sci. USA 2020, 117, 26382–26388. [Google Scholar] [CrossRef] [PubMed]
- Hobbs, E.C.; Reid, T.J. Animals and SARS-CoV-2: Species susceptibility and viral transmission in experimental and natural conditions, and the potential implications for community transmission. Transbound. Emerg. Dis. 2021, 68, 1850–1867. [Google Scholar] [CrossRef]
- Gaudreault, N.N.; Trujillo, J.D.; Carossino, M.; Meekins, D.A.; Morozov, I.; Madden, D.W.; Indran, S.V.; Bold, D.; Balaraman, V.; Kwon, T.; et al. SARS-CoV-2 infection, disease and transmission in domestic cats. Emerg. Microbes Infect. 2020, 9, 2322–2332. [Google Scholar] [CrossRef] [PubMed]
- van den Brand, J.M.; Haagmans, B.L.; Leijten, L.; van Riel, D.; Martina, B.E.; Osterhaus, A.D.; Kuiken, T. Pathology of experimental SARS coronavirus infection in cats and ferrets. Vet. Pathol. 2008, 45, 551–562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bao, L.; Song, Z.; Xue, J.; Gao, H.; Liu, J.; Wang, J.; Guo, Q.; Zhao, B.; Qu, Y.; Qi, F.; et al. Susceptibility and Attenuated Transmissibility of SARS-CoV-2 in Domestic Cats. J. Infect. Dis. 2021, 223, 1313–1321. [Google Scholar] [CrossRef] [PubMed]
- Halfmann, P.J.; Hatta, M.; Chiba, S.; Maemura, T.; Fan, S.; Takeda, M.; Kinoshita, N.; Hattori, S.I.; Sakai-Tagawa, Y.; Iwatsuki-Horimoto, K.; et al. Transmission of SARS-CoV-2 in Domestic Cats. N. Engl. J. Med. 2020, 383, 592–594. [Google Scholar] [CrossRef] [PubMed]
- Hosie, M.J.; Epifano, I.; Herder, V.; Orton, R.J.; Stevenson, A.; Johnson, N.; MacDonald, E.; Dunbar, D.; McDonald, M.; Howie, F.; et al. Detection of SARS-CoV-2 in respiratory samples from cats in the UK associated with human-to-cat transmission. Vet. Rec. 2021, 188, e247. [Google Scholar] [CrossRef]
- Chen, J.; Huang, C.; Zhang, Y.; Zhang, S.; Jin, M. Severe Acute Respiratory Syndrome Coronavirus 2-Specific Antibodies in Pets in Wuhan, China. J. Infect. 2020, 81, e68–e69. [Google Scholar] [CrossRef]
- Koley, T.; Madaan, S.; Chowdhury, S.R.; Kumar, M.; Kaur, P.; Singh, T.P.; Ethayathulla, A.S. Structural analysis of COVID-19 spike protein in recognizing the ACE2 receptor of different mammalian species and its susceptibility to viral infection. 3 Biotech 2021, 11, 109. [Google Scholar] [CrossRef]
- Stout, A.E.; Andre, N.M.; Jaimes, J.A.; Millet, J.K.; Whittaker, G.R. Coronaviruses in cats and other companion animals: Where does SARS-CoV-2/COVID-19 fit? Vet. Microbiol. 2020, 247, 108777. [Google Scholar] [CrossRef]
- Ekstrand, K.; Flanagan, A.J.; Lin, I.E.; Vejseli, B.; Cole, A.; Lally, A.P.; Morris, R.L.; Morgan, K.N. Animal Transmission of SARS-CoV-2 and the Welfare of Animals during the COVID-19 Pandemic. Animals 2021, 11, 2044. [Google Scholar] [CrossRef]
- Yilmaz, A.; Kayar, A.; Turan, N.; Iskefli, O.; Bayrakal, A.; Roman-Sosa, G.; Or, E.; Tali, H.E.; Kocazeybek, B.; Karaali, R.; et al. Presence of Antibodies to SARS-CoV-2 in Domestic Cats in Istanbul, Turkey, Before and After COVID-19 Pandemic. Front. Vet. Sci. 2021, 8, 707368. [Google Scholar] [CrossRef]
- Klaus, J.; Meli, M.L.; Willi, B.; Nadeau, S.; Beisel, C.; Stadler, T.; Eth Sars-Co, V.S.T.; Egberink, H.; Zhao, S.; Lutz, H.; et al. Detection and Genome Sequencing of SARS-CoV-2 in a Domestic Cat with Respiratory Signs in Switzerland. Viruses 2021, 13, 496. [Google Scholar] [CrossRef]
- Sit, T.H.C.; Brackman, C.J.; Ip, S.M.; Tam, K.W.S.; Law, P.Y.T.; To, E.M.W.; Yu, V.Y.T.; Sims, L.D.; Tsang, D.N.C.; Chu, D.K.W.; et al. Infection of dogs with SARS-CoV-2. Nature 2020, 586, 776–778. [Google Scholar] [CrossRef]
- Fritz, M.; Rosolen, B.; Krafft, E.; Becquart, P.; Elguero, E.; Vratskikh, O.; Denolly, S.; Boson, B.; Vanhomwegen, J.; Gouilh, M.A.; et al. High prevalence of SARS-CoV-2 antibodies in pets from COVID-19+ households. One Health 2021, 11, 100192. [Google Scholar] [CrossRef]
- Jairak, W.; Chamsai, E.; Udom, K.; Charoenkul, K.; Chaiyawong, S.; Techakriengkrai, N.; Tangwangvivat, R.; Suwannakarn, K.; Amonsin, A. SARS-CoV-2 delta variant infection in domestic dogs and cats, Thailand. Sci. Rep. 2022, 12, 8403. [Google Scholar] [CrossRef]
- Calvet, G.A.; Pereira, S.A.; Ogrzewalska, M.; Pauvolid-Correa, A.; Resende, P.C.; Tassinari, W.S.; Costa, A.P.; Keidel, L.O.; da Rocha, A.S.B.; da Silva, M.F.B.; et al. Investigation of SARS-CoV-2 infection in dogs and cats of humans diagnosed with COVID-19 in Rio de Janeiro, Brazil. PLoS ONE 2021, 16, e0250853. [Google Scholar] [CrossRef]
- Rivero, R.; Garay, E.; Botero, Y.; Serrano-Coll, H.; Gastelbondo, B.; Munoz, M.; Ballesteros, N.; Castaneda, S.; Patino, L.H.; Ramirez, J.D.; et al. Human-to-dog transmission of SARS-CoV-2, Colombia. Sci. Rep. 2022, 12, 7880. [Google Scholar] [CrossRef]
- Cui, H.; Zhang, L. Key Components of Inflammasome and Pyroptosis Pathways Are Deficient in Canines and Felines, Possibly Affecting Their Response to SARS-CoV-2 Infection. Front. Immunol. 2020, 11, 592622. [Google Scholar] [CrossRef]
- Boklund, A.; Hammer, A.S.; Quaade, M.L.; Rasmussen, T.B.; Lohse, L.; Strandbygaard, B.; Jorgensen, C.S.; Olesen, A.S.; Hjerpe, F.B.; Petersen, H.H.; et al. SARS-CoV-2 in Danish Mink Farms: Course of the Epidemic and a Descriptive Analysis of the Outbreaks in 2020. Animals 2021, 11, 164. [Google Scholar] [CrossRef]
- Sreenivasan, C.C.; Thomas, M.; Wang, D.; Li, F. Susceptibility of livestock and companion animals to COVID-19. J. Med. Virol. 2021, 93, 1351–1360. [Google Scholar] [CrossRef]
- Sharun, K.; Tiwari, R.; Natesan, S.; Dhama, K. SARS-CoV-2 infection in farmed minks, associated zoonotic concerns, and importance of the One Health approach during the ongoing COVID-19 pandemic. Vet. Q. 2021, 41, 50–60. [Google Scholar] [CrossRef]
- Oreshkova, N.; Molenaar, R.J.; Vreman, S.; Harders, F.; Oude Munnink, B.B.; Hakze-van der Honing, R.W.; Gerhards, N.; Tolsma, P.; Bouwstra, R.; Sikkema, R.S.; et al. SARS-CoV-2 infection in farmed minks, the Netherlands, April and May 2020. Eurosurveillance 2020, 25, 2001005. [Google Scholar] [CrossRef]
- Oude Munnink, B.B.; Sikkema, R.S.; Nieuwenhuijse, D.F.; Molenaar, R.J.; Munger, E.; Molenkamp, R.; van der Spek, A.; Tolsma, P.; Rietveld, A.; Brouwer, M.; et al. Transmission of SARS-CoV-2 on mink farms between humans and mink and back to humans. Science 2021, 371, 172–177. [Google Scholar] [CrossRef]
- Molenaar, R.J.; Vreman, S.; Hakze-van der Honing, R.W.; Zwart, R.; de Rond, J.; Weesendorp, E.; Smit, L.A.M.; Koopmans, M.; Bouwstra, R.; Stegeman, A.; et al. Clinical and Pathological Findings in SARS-CoV-2 Disease Outbreaks in Farmed Mink (Neovison vison). Vet. Pathol. 2020, 57, 653–657. [Google Scholar] [CrossRef] [PubMed]
- Bartlett, S.L.; Diel, D.G.; Wang, L.; Zec, S.; Laverack, M.; Martins, M.; Caserta, L.C.; Killian, M.L.; Terio, K.; Olmstead, C.; et al. Sars-Cov-2 Infection and Longitudinal Fecal Screening in Malayan Tigers (Panthera Tigris Jacksoni), Amur Tigers (Panthera Tigris Altaica), and African Lions (Panthera Leo Krugeri) at the Bronx Zoo, New York, USA. J. Zoo Wildl. Med. 2021, 51, 733–744. [Google Scholar] [CrossRef] [PubMed]
- Koeppel, K.N.; Mendes, A.; Strydom, A.; Rotherham, L.; Mulumba, M.; Venter, M. SARS-CoV-2 Reverse Zoonoses to Pumas and Lions, South Africa. Viruses 2022, 14, 120. [Google Scholar] [CrossRef] [PubMed]
- Melin, A.D.; Janiak, M.C.; Marrone, F., 3rd; Arora, P.S.; Higham, J.P. Comparative ACE2 variation and primate COVID-19 risk. Commun. Biol. 2020, 3, 641. [Google Scholar] [CrossRef]
- Lu, S.; Zhao, Y.; Yu, W.; Yang, Y.; Gao, J.; Wang, J.; Kuang, D.; Yang, M.; Yang, J.; Ma, C.; et al. Comparison of nonhuman primates identified the suitable model for COVID-19. Signal. Transduct. Target Ther. 2020, 5, 157. [Google Scholar] [CrossRef] [PubMed]
- Rockx, B.; Kuiken, T.; Herfst, S.; Bestebroer, T.; Lamers, M.M.; Oude Munnink, B.B.; de Meulder, D.; van Amerongen, G.; van den Brand, J.; Okba, N.M.A.; et al. Comparative pathogenesis of COVID-19, MERS, and SARS in a nonhuman primate model. Science 2020, 368, 1012–1015. [Google Scholar] [CrossRef] [Green Version]
- Delahay, R.J.; de la Fuente, J.; Smith, G.C.; Sharun, K.; Snary, E.L.; Flores Giron, L.; Nziza, J.; Fooks, A.R.; Brookes, S.M.; Lean, F.Z.X.; et al. Assessing the risks of SARS-CoV-2 in wildlife. One Health Outlook 2021, 3, 7. [Google Scholar] [CrossRef]
- Hale, V.L.; Dennis, P.M.; McBride, D.S.; Nolting, J.M.; Madden, C.; Huey, D.; Ehrlich, M.; Grieser, J.; Winston, J.; Lombardi, D.; et al. SARS-CoV-2 infection in free-ranging white-tailed deer. Nature 2022, 602, 481–486. [Google Scholar] [CrossRef] [PubMed]
- Palmer, M.V.; Martins, M.; Falkenberg, S.; Buckley, A.; Caserta, L.C.; Mitchell, P.K.; Cassmann, E.D.; Rollins, A.; Zylich, N.C.; Renshaw, R.W.; et al. Susceptibility of white-tailed deer (Odocoileus virginianus) to SARS-CoV-2. J. Virol. 2021, 95, e00083-21. [Google Scholar] [CrossRef] [PubMed]
- Ulrich, L.; Wernike, K.; Hoffmann, D.; Mettenleiter, T.C.; Beer, M. Experimental Infection of Cattle with SARS-CoV-2. Emerg. Infect. Dis. 2020, 26, 2979–2981. [Google Scholar] [CrossRef] [PubMed]
- Schlottau, K.; Rissmann, M.; Graaf, A.; Schon, J.; Sehl, J.; Wylezich, C.; Hoper, D.; Mettenleiter, T.C.; Balkema-Buschmann, A.; Harder, T.; et al. SARS-CoV-2 in fruit bats, ferrets, pigs, and chickens: An experimental transmission study. Lancet Microbe 2020, 1, e218–e225. [Google Scholar] [CrossRef] [PubMed]
- Suarez, D.L.; Pantin-Jackwood, M.J.; Swayne, D.E.; Lee, S.A.; DeBlois, S.M.; Spackman, E. Lack of Susceptibility to SARS-CoV-2 and MERS-CoV in Poultry. Emerg. Infect. Dis. 2020, 26, 3074–3076. [Google Scholar] [CrossRef]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sheleme, T.; Bekele, F.; Ayela, T. Clinical Presentation of Patients Infected with Coronavirus Disease 19: A Systematic Review. Infect. Dis. 2020, 13, 1178633720952076. [Google Scholar] [CrossRef]
- Elliott, J.; Whitaker, M.; Bodinier, B.; Eales, O.; Riley, S.; Ward, H.; Cooke, G.; Darzi, A.; Chadeau-Hyam, M.; Elliott, P. Predictive symptoms for COVID-19 in the community: REACT-1 study of over 1 million people. PLoS Med. 2021, 18, e1003777. [Google Scholar] [CrossRef]
- Li, J.; Huang, D.Q.; Zou, B.; Yang, H.; Hui, W.Z.; Rui, F.; Yee, N.T.S.; Liu, C.; Nerurkar, S.N.; Kai, J.C.Y.; et al. Epidemiology of COVID-19: A systematic review and meta-analysis of clinical characteristics, risk factors, and outcomes. J. Med. Virol. 2021, 93, 1449–1458. [Google Scholar] [CrossRef]
- Fernandez-de-Las-Penas, C.; Notarte, K.I.; Peligro, P.J.; Velasco, J.V.; Ocampo, M.J.; Henry, B.M.; Arendt-Nielsen, L.; Torres-Macho, J.; Plaza-Manzano, G. Long-COVID Symptoms in Individuals Infected with Different SARS-CoV-2 Variants of Concern: A Systematic Review of the Literature. Viruses 2022, 14, 2629. [Google Scholar] [CrossRef] [PubMed]
- Mahase, E. Covid-19: How many variants are there, and what do we know about them? BMJ 2021, 374, n1971. [Google Scholar] [CrossRef] [PubMed]
- Morioka, S.; Tsuzuki, S.; Suzuki, M.; Terada, M.; Akashi, M.; Osanai, Y.; Kuge, C.; Sanada, M.; Tanaka, K.; Maruki, T.; et al. Post COVID-19 condition of the Omicron variant of SARS-CoV-2. J. Infect. Chemother. 2022, 28, 1546–1551. [Google Scholar] [CrossRef] [PubMed]
- Lundberg, A.L.; Lorenzo-Redondo, R.; Ozer, E.A.; Hawkins, C.A.; Hultquist, J.F.; Welch, S.B.; Prasad, P.V.V.; Oehmke, J.F.; Achenbach, C.J.; Murphy, R.L.; et al. Has Omicron Changed the Evolution of the Pandemic? JMIR Public Health Surveill. 2022, 8, e35763. [Google Scholar] [CrossRef] [PubMed]
- Watson, O.J.; Barnsley, G.; Toor, J.; Hogan, A.B.; Winskill, P.; Ghani, A.C. Global impact of the first year of COVID-19 vaccination: A mathematical modelling study. Lancet Infect. Dis. 2022, 22, 1293–1302. [Google Scholar] [CrossRef] [PubMed]
- Safiabadi Tali, S.H.; LeBlanc, J.J.; Sadiq, Z.; Oyewunmi, O.D.; Camargo, C.; Nikpour, B.; Armanfard, N.; Sagan, S.M.; Jahanshahi-Anbuhi, S. Tools and Techniques for Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2)/COVID-19 Detection. Clin. Microbiol. Rev. 2021, 34, e00228-20. [Google Scholar] [CrossRef]
- Kevadiya, B.D.; Machhi, J.; Herskovitz, J.; Oleynikov, M.D.; Blomberg, W.R.; Bajwa, N.; Soni, D.; Das, S.; Hasan, M.; Patel, M.; et al. Diagnostics for SARS-CoV-2 infections. Nat. Mater. 2021, 20, 593–605. [Google Scholar] [CrossRef]
- Chen, Q.; He, Z.; Mao, F.; Peo, H.; Cao, H.; Liu, X. Diagnostic technologies for COVID-19: A review. RSC Adv. 2020, 10, 35257–35264. [Google Scholar] [CrossRef]
- Wu, S.Y.; Yau, H.S.; Yu, M.Y.; Tsang, H.F.; Chan, L.W.C.; Cho, W.C.S.; Shing Yu, A.C.; Yuen Yim, A.K.; Li, M.J.W.; Wong, Y.K.E.; et al. The diagnostic methods in the COVID-19 pandemic, today and in the future. Expert Rev. Mol. Diagn. 2020, 20, 985–993. [Google Scholar] [CrossRef]
- Tsujimoto, Y.; Terada, J.; Kimura, M.; Moriya, A.; Motohashi, A.; Izumi, S.; Kawajiri, K.; Hakkaku, K.; Morishita, M.; Saito, S.; et al. Diagnostic accuracy of nasopharyngeal swab, nasal swab and saliva swab samples for the detection of SARS-CoV-2 using RT-PCR. Infect. Dis. 2021, 53, 581–589. [Google Scholar] [CrossRef]
- Juscamayta-Lopez, E.; Valdivia, F.; Horna, H.; Tarazona, D.; Linares, L.; Rojas, N.; Huaringa, M. A Multiplex and Colorimetric Reverse Transcription Loop-Mediated Isothermal Amplification Assay for Sensitive and Rapid Detection of Novel SARS-CoV-2. Front. Cell. Infect. Microbiol. 2021, 11, 653616. [Google Scholar] [CrossRef] [PubMed]
- Dong, L.; Zhou, J.; Niu, C.; Wang, Q.; Pan, Y.; Sheng, S.; Wang, X.; Zhang, Y.; Yang, J.; Liu, M.; et al. Highly accurate and sensitive diagnostic detection of SARS-CoV-2 by digital PCR. Talanta 2021, 224, 121726. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Huang, S.; Zhou, L.; Wang, X.; Yang, H.; Li, W. Coronavirus Disease 2019 (COVID-19): Emerging detection technologies and auxiliary analysis. J. Clin. Lab. Anal. 2022, 36, e24152. [Google Scholar] [CrossRef] [PubMed]
- Piec, I.; English, E.; Thomas, M.A.; Dervisevic, S.; Fraser, W.D.; John, W.G. Performance of SARS-CoV-2 serology tests: Are they good enough? PLoS ONE 2021, 16, e0245914. [Google Scholar] [CrossRef]
- Dinnes, J.; Deeks, J.J.; Berhane, S.; Taylor, M.; Adriano, A.; Davenport, C.; Dittrich, S.; Emperador, D.; Takwoingi, Y.; Cunningham, J.; et al. Rapid, point-of-care antigen and molecular-based tests for diagnosis of SARS-CoV-2 infection. Cochrane Database Syst. Rev. 2021, 3, CD013705. [Google Scholar]
- Carobene, A.; Milella, F.; Famiglini, L.; Cabitza, F. How is test laboratory data used and characterised by machine learning models? A systematic review of diagnostic and prognostic models developed for COVID-19 patients using only laboratory data. Clin. Chem. Lab. Med. 2022, 60, 1887–1901. [Google Scholar] [CrossRef]
- McAloose, D.; Laverack, M.; Wang, L.; Killian, M.L.; Caserta, L.C.; Yuan, F.; Mitchell, P.K.; Queen, K.; Mauldin, M.R.; Cronk, B.D.; et al. From People to Panthera: Natural SARS-CoV-2 Infection in Tigers and Lions at the Bronx Zoo. mBio 2020, 11, e02220-20. [Google Scholar] [CrossRef]
- Fernandez-Bellon, H.; Rodon, J.; Fernandez-Bastit, L.; Almagro, V.; Padilla-Sole, P.; Lorca-Oro, C.; Valle, R.; Roca, N.; Grazioli, S.; Trogu, T.; et al. Monitoring Natural SARS-CoV-2 Infection in Lions (Panthera leo) at the Barcelona Zoo: Viral Dynamics and Host Responses. Viruses 2021, 13, 1683. [Google Scholar] [CrossRef]
- Garigliany, M.; Van Laere, A.S.; Clercx, C.; Giet, D.; Escriou, N.; Huon, C.; van der Werf, S.; Eloit, M.; Desmecht, D. SARS-CoV-2 Natural Transmission from Human to Cat, Belgium, March 2020. Emerg. Infect. Dis. 2020, 26, 3069–3071. [Google Scholar] [CrossRef]
- Grome, H.N.; Meyer, B.; Read, E.; Buchanan, M.; Cushing, A.; Sawatzki, K.; Levinson, K.J.; Thomas, L.S.; Perry, Z.; Uehara, A.; et al. SARS-CoV-2 Outbreak among Malayan Tigers and Humans, Tennessee, USA, 2020. Emerg. Infect. Dis. 2022, 28, 833–836. [Google Scholar] [CrossRef] [PubMed]
- Currie, D.W.; Shah, M.M.; Salvatore, P.P.; Ford, L.; Whaley, M.J.; Meece, J.; Ivacic, L.; Thornburg, N.J.; Tamin, A.; Harcourt, J.L.; et al. Relationship of SARS-CoV-2 Antigen and Reverse Transcription PCR Positivity for Viral Cultures. Emerg. Infect. Dis. 2022, 28, 717–720. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.W.; Chia, W.N.; Qin, X.; Liu, P.; Chen, M.I.; Tiu, C.; Hu, Z.; Chen, V.C.; Young, B.E.; Sia, W.R.; et al. A SARS-CoV-2 surrogate virus neutralization test based on antibody-mediated blockage of ACE2-spike protein-protein interaction. Nat. Biotechnol. 2020, 38, 1073–1078. [Google Scholar] [CrossRef] [PubMed]
- Goletic, S.; Goletic, T.; Softic, A.; Zahirovic, A.; Rukavina, D.; Kavazovic, A.; Omeragic, J.; Umihanic, S.; Hukic, M. The Evidence of SARS-CoV-2 Human-to-Pets Transmission in Household Settings in Bosnia and Herzegovina. Front. Genet. 2022, 13, 839205. [Google Scholar] [CrossRef]
- Zhao, S.; Schuurman, N.; Li, W.; Wang, C.; Smit, L.A.M.; Broens, E.M.; Wagenaar, J.A.; van Kuppeveld, F.J.M.; Bosch, B.J.; Egberink, H. Serologic Screening of Severe Acute Respiratory Syndrome Coronavirus 2 Infection in Cats and Dogs during First Coronavirus Disease Wave, the Netherlands. Emerg. Infect. Dis. 2021, 27, 1362–1370. [Google Scholar] [CrossRef]
- Thompson, A.; Kutz, S. Introduction to the Special Issue on ‘Emerging Zoonoses and Wildlife’. Int. J. Parasitol. Parasites Wildl. 2019, 9, 322. [Google Scholar] [CrossRef]
- Cutler, S.J.; Fooks, A.R.; van der Poel, W.H. Public health threat of new, reemerging, and neglected zoonoses in the industrialized world. Emerg. Infect. Dis. 2010, 16, 1. [Google Scholar] [CrossRef]
- Brown, C. Emerging zoonoses and pathogens of public health significance—An overview. Rev. Sci. Tech. 2004, 23, 435–442. [Google Scholar] [CrossRef] [Green Version]
- Cleaveland, S.; Laurenson, M.K.; Taylor, L.H. Diseases of humans and their domestic mammals: Pathogen characteristics, host range and the risk of emergence. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2001, 356, 991–999. [Google Scholar] [CrossRef]
- Westgarth, C.; Pinchbeck, G.L.; Bradshaw, J.W.; Dawson, S.; Gaskell, R.M.; Christley, R.M. Dog-human and dog-dog interactions of 260 dog-owning households in a community in Cheshire. Vet. Rec. 2008, 162, 436–442. [Google Scholar] [CrossRef]
- Fernandes, M.R.; Sellera, F.P.; Moura, Q.; Carvalho, M.P.N.; Rosato, P.N.; Cerdeira, L.; Lincopan, N. Zooanthroponotic Transmission of Drug-Resistant Pseudomonas aeruginosa, Brazil. Emerg. Infect. Dis. 2018, 24, 1160–1162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hubalek, Z. Emerging human infectious diseases: Anthroponoses, zoonoses, and sapronoses. Emerg. Infect. Dis. 2003, 9, 403–404. [Google Scholar] [CrossRef] [PubMed]
- Loeffler, A.; Boag, A.K.; Sung, J.; Lindsay, J.A.; Guardabassi, L.; Dalsgaard, A.; Smith, H.; Stevens, K.B.; Lloyd, D.H. Prevalence of methicillin-resistant Staphylococcus aureus among staff and pets in a small animal referral hospital in the UK. J. Antimicrob. Chemother. 2005, 56, 692–697. [Google Scholar] [CrossRef] [PubMed]
- Manian, F.A. Asymptomatic nasal carriage of mupirocin-resistant, methicillin-resistant Staphylococcus aureus (MRSA) in a pet dog associated with MRSA infection in household contacts. Clin. Infect. Dis. 2003, 36, e26–e28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Duijkeren, E.; Wolfhagen, M.J.; Box, A.T.; Heck, M.E.; Wannet, W.J.; Fluit, A.C. Human-to-dog transmission of methicillin-resistant Staphylococcus aureus. Emerg. Infect. Dis. 2004, 10, 2235–2237. [Google Scholar] [CrossRef] [PubMed]
- Overgaauw, P.A.; van Zutphen, L.; Hoek, D.; Yaya, F.O.; Roelfsema, J.; Pinelli, E.; van Knapen, F.; Kortbeek, L.M. Zoonotic parasites in fecal samples and fur from dogs and cats in The Netherlands. Vet. Parasitol. 2009, 163, 115–122. [Google Scholar] [CrossRef]
- Overgaauw, P.A.M.; Vinke, C.M.; Hagen, M.; Lipman, L.J.A. A One Health Perspective on the Human-Companion Animal Relationship with Emphasis on Zoonotic Aspects. Int. J. Environ. Res. Public Health 2020, 17, 3789. [Google Scholar] [CrossRef]
- Elad, D. Immunocompromised patients and their pets: Still best friends? Vet. J. 2013, 197, 662–669. [Google Scholar] [CrossRef]
- Santaniello, A.; Garzillo, S.; Amato, A.; Sansone, M.; Fioretti, A.; Menna, L.F. Occurrence of Pasteurella multocida in Dogs Being Trained for Animal-Assisted Therapy. Int. J. Environ. Res. Public Health 2020, 17, 6385. [Google Scholar] [CrossRef]
- Santaniello, A.; Sansone, M.; Fioretti, A.; Menna, L.F. Systematic Review and Meta-Analysis of the Occurrence of ESKAPE Bacteria Group in Dogs, and the Related Zoonotic Risk in Animal-Assisted Therapy, and in Animal-Assisted Activity in the Health Context. Int. J. Environ. Res. Public Health 2020, 17, 3178. [Google Scholar] [CrossRef]
- Santaniello, A.; Varriale, L.; Dipineto, L.; Borrelli, L.; Pace, A.; Fioretti, A.; Menna, L.F. Presence of Campylobacterjejuni and C. coli in Dogs under Training for Animal-Assisted Therapies. Int. J. Environ. Res. Public Health 2021, 18, 3717. [Google Scholar] [CrossRef] [PubMed]
- Decaro, N.; Vaccari, G.; Lorusso, A.; Lorusso, E.; De Sabato, L.; Patterson, E.I.; Di Bartolo, I.; Hughes, G.L.; Teodori, L.; Desario, C.; et al. Possible Human-to-Dog Transmission of SARS-CoV-2, Italy, 2020. Emerg. Infect. Dis. 2021, 27, 1981–1984. [Google Scholar] [CrossRef] [PubMed]
- Shaheen, M.N.F. The concept of one health applied to the problem of zoonotic diseases. Rev. Med. Virol. 2022, 32, e2326. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Santaniello, A.; Perruolo, G.; Cristiano, S.; Agognon, A.L.; Cabaro, S.; Amato, A.; Dipineto, L.; Borrelli, L.; Formisano, P.; Fioretti, A.; et al. SARS-CoV-2 Affects Both Humans and Animals: What Is the Potential Transmission Risk? A Literature Review. Microorganisms 2023, 11, 514. https://doi.org/10.3390/microorganisms11020514
Santaniello A, Perruolo G, Cristiano S, Agognon AL, Cabaro S, Amato A, Dipineto L, Borrelli L, Formisano P, Fioretti A, et al. SARS-CoV-2 Affects Both Humans and Animals: What Is the Potential Transmission Risk? A Literature Review. Microorganisms. 2023; 11(2):514. https://doi.org/10.3390/microorganisms11020514
Chicago/Turabian StyleSantaniello, Antonio, Giuseppe Perruolo, Serena Cristiano, Ayewa Lawoe Agognon, Serena Cabaro, Alessia Amato, Ludovico Dipineto, Luca Borrelli, Pietro Formisano, Alessandro Fioretti, and et al. 2023. "SARS-CoV-2 Affects Both Humans and Animals: What Is the Potential Transmission Risk? A Literature Review" Microorganisms 11, no. 2: 514. https://doi.org/10.3390/microorganisms11020514
APA StyleSantaniello, A., Perruolo, G., Cristiano, S., Agognon, A. L., Cabaro, S., Amato, A., Dipineto, L., Borrelli, L., Formisano, P., Fioretti, A., & Oriente, F. (2023). SARS-CoV-2 Affects Both Humans and Animals: What Is the Potential Transmission Risk? A Literature Review. Microorganisms, 11(2), 514. https://doi.org/10.3390/microorganisms11020514






